Abstract
Catechol estrogens are steroid metabolites that elicit physiological responses through binding to a variety of cellular targets. We show here that catechol estrogens directly inhibit soluble adenylyl cyclases and the abundant trans-membrane adenylyl cyclases. Catechol estrogen inhibition is non-competitive with respect to the substrate ATP, and we solved the crystal structure of a catechol estrogen bound to a soluble adenylyl cyclase from Spirulina platensis in complex with a substrate analog. The catechol estrogen is bound to a newly identified, conserved hydrophobic patch near the active center but distinct from the ATP-binding cleft. Inhibitor binding leads to a chelating interaction between the catechol estrogen hydroxyl groups and the catalytic magnesium ion, distorting the active site and trapping the enzyme substrate complex in a non-productive conformation. This novel inhibition mechanism likely applies to other adenylyl cyclase inhibitors, and the identified ligand-binding site has important implications for the development of specific adenylyl cyclase inhibitors.
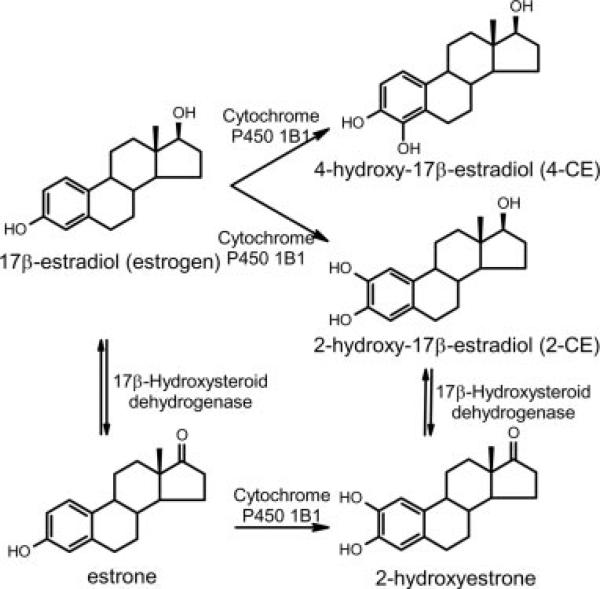
Oxidative hydroxylation of the steroid hormone estrogen, which results in the formation of catechol estrogens (CEs1; Fig. 1), is the first step in estrogen catabolism. CEs, however, are not only catabolites; they are involved in a multitude of physiological processes and may contribute to many human diseases such as cancer and cardiovascular conditions (1, 2). High concentrations of CEs are found in a number of tissues such as the brain, endocrine glands (3), and mammary tumors (4). Although CEs elicit some physiological responses via the classical estrogen receptor, other biological CE effects are not associated with activation of this receptor (5, 6). In the latter regard, it has long been known that CEs exert effects in the cAMP signaling system, as shown by their inhibition of cAMP accumulation in the hypothalamus (7).
Fig. 1. Chemical structures of estrogen and its metabolites.
The catechol derivatives 2-CE, 4-CE, and 2-hydroxyestrone are major catabolites of the steroid hormones estrogen and estrone. The hydroxyl group added to the sterol scaffold is needed for subsequent conjugation and decomposition reactions.
The conversion of ATP to cAMP in mammals is mediated by members of the class III adenylyl cyclase family (AC; E.C. 4.6.1.1), which in humans comprises nine trans-membrane AC enzymes (tmACs) and one soluble AC (sAC) (8, 9). tmACs are regulated by heterotrimeric G proteins in response to the stimulation of G protein-coupled receptors and therefore play a key role in the cellular response to extracellular signals (10). sAC, in contrast, is insensitive to G proteins and the additional tmAC regulator calmodulin as well as to the non-physiological diterpenic activator forskolin (11, 12). Instead, sAC is directly activated by calcium and the metabolite bicarbonate, rendering the enzyme an intracellular metabolic sensor (12, 13). Together, tmACs and sAC regulate a diverse set of essential biological processes such as differentiation and gene transcription.
All mammalian ACs comprise two catalytic domains, and the crystal structure of a tmAC enzyme revealed that these domains, C1 and C2, are structurally similar (14). The C1C2 heterodimer therefore resembles a homodimer, and the shared active site at the dimer interface has a pseudo-symmetric site that is catalytically inactive due to the lack of several catalytic residues. This pseudo-active site is partially exploited by the non-physiological tmAC activator forskolin. By solving crystal structures of the cyanobacterial sAC homolog CyaC, we recently showed that sAC enzymes, despite their unique regulation, have the same overall structure as tmACs and employ the same two-metal ion mechanism for catalysis (15, 16).
The biological importance of AC enzymes calls for AC inhibitors, but there are currently no potent specific inhibitors known. The best characterized AC inhibitors are the so-called P-site inhibitors that occupy the binding site for the substrate ATP, show low AC isoform specificity, and bear the potential to bind to a large number of cellular nucleotide-binding proteins (17). A second class of AC inhibitors, which contain an adenine linked to ion chelators, suffers from the same predicament (18). There are currently no efficient AC inhibitors that bind to a site other than the ATP-binding pocket and which would therefore promise a higher specificity for AC enzymes and among AC isoforms.
Given that CEs inhibited cAMP production in cell extracts rich in sAC (19), we attempted to elucidate the molecular mechanism of CE action. We found that CEs directly bind to and inhibit the catalytic domains of class III AC enzymes non-competitively with respect to ATP. The crystal structure of the trimeric complex between the catalytic domain of CyaC, an ATP analog, and a CE identifies a hydrophobic patch at the dimer center as the first AC inhibitor-binding site outside the active site. The adjacent hydroxyls of the CE inhibitor capture the catalytic magnesium ion via a pincer-like interaction and displace it from its “catalytic” position. This novel inhibitor-binding site shows variations between different AC isoforms, indicating its potential use for the development of highly specific AC inhibitors.
MATERIALS AND METHODS
Protein Purification and Activity Assay
[α-32P]ATP and [3H]cAMP were purchased from PerkinElmer Life Sciences, estrogen and derivatives were from Steraloids, and all other chemicals came from Sigma. A glutathione S-transferase fusion of human sAC (20) and the soluble C1 and C2 domains of human tmAC VII (21) were expressed and purified as described previously. Residues 998–1202 and 1005–1202, respectively, of CyaC from Spirulina platensis were expressed and purified with an N-terminal His tag as described (16). The protein was stored at –80 °C for later crystallization trials, and protein for activity assays was supplemented with 50% (v/v) glycerol and stored at –20 °C. Individual mammalian adenylyl cyclase isoforms 1 (bovine), 2 (rat), and 5 (dog) were recombinantly expressed in Sf9 cells, and the adenylyl cyclase-containing membranes were prepared as described (22).
Cyclase assays were performed in 100 μl of total volume in 50 mm Tris-HCl (pH 7.5) with [α-32P]ATP as substrate and 10 mm MgCl2. Assays for sAC activity were supplemented with 50 mm bicarbonate, and those for CyaC were also supplemented with 5 mm CaCl2; reactions with tmAC VII were supplemented with purified Gsα protein (kind gift of Shengyu Yang), 10 mm NaF, and 30 μm AlF3. Indicated amounts of catechol estrogen were added using 100 mm stock solutions in methanol. Assays were started by adding purified tmAC VII C1 plus C2, human GST-sAC, and CyaC-(998–1202), respectively. Reactions were incubated for 30 min at 30 °C and stopped by adding 200 μl of 2% SDS. [α-32P]cAMP was recovered using a two-column method described previously (23). Mixtures of equal amounts of sample and scintillation fluid were counted for 1 min in a Beckman scintillation counter. Initial velocity measurements were made under conditions where the conversion of [α-32P]ATP did not exceed 10%. Activity values are the averages of at least three independent determinations. Adenylyl cyclase activity of Sf9 membrane preparations was measured using the procedure described by Smigel (24). All assays were performed for 12 min at 30 °C in a final volume of 100 μl containing 20 μg of membrane protein, 100 μm forskolin, 10 mm MgCl2, and the indicated concentration of the inhibitor.
Crystallization and X-ray Data Collection
CyaC-(1005–1202) was crystallized in complex with the ATP analog α,β-methylene-adenosine-5′-triphosphate (α,β-Me-ATP) by mixing 1 μl of protein solution (6 mg/ml in 20 mm Tris/HCl, pH 7.8, 5 mm α,β-Me-ATP, 5 mm MgCl2, and 5 mm CaCl2) with 1 μl of reservoir solution and equilibrating against a 0.4-ml reservoir (100 mm cacodylate/NaOH, pH 6.7, 6% (v/v) isopropanol, and 11% (w/v) polyethylene glycol 4000). Crystal plates of ~150 × 100 × 10 μm3 were soaked with 5 mm 2-hydroxyestradiol (2-CE) for 10 min. Soaked crystals were frozen 60 s after the addition of two drop volumes of cryoprotectant solution (25% (v/v) isopropanol in reservoir solution supplemented with 5 mm 2-CE). A complete data set was collected at 100 K at the Brookhaven National Laboratory's beamline X4A (Table I). Diffraction data were processed with DENZO and SCALEPACK (25). Soaking with 2-CE changed the initial space group of the α,β-Me-ATP complex from P212121 with two monomers in the asymmetric unit to P21 with four monomers in the asymmetric unit.
Table I.
Data collection and refinement statistics
| CyaC + α,β-ATP + 2-CE | |
|---|---|
| Space group | P21 |
| Unit cell constants | a = 53.4 Å; b = 70.2 Å; c = 106.7 Å; β = 96.0° |
| Resolution (Å) | 38.2–2.3 |
| Unique reflections | 35,639 |
| 〈 I/σ 〉 | 8.7 |
| Completeness (outermost shell) | 98.5% (97.5%) |
| Rmergea (outermost shell) | 6.3% (35.2%) |
| Resolution range for refinement (Å) | 15.0–2.3 |
| Total reflections used | 28,605 |
| Number of atoms in asymmetric unit | |
| Protein | 6013 |
| Ligands | 174 |
| Water | 139 |
| Root mean square deviations | |
| Bond length (Å) | 0.01 |
| Bond angles (°) | 1.2 |
| Bonded B-factors (Å2) | 1.8 |
| Average B-factor (Å2) | 35.9 |
| Final R/Rfreeb,c (%) | 20.7/25.7 |
Rmerge = Σ(I – 〈I〉)/ΣI; I is the intensity of an individual measurement, and 〈I〉 is the corresponding mean value.
R-factor = Σ∥Fobs| – k|Fcalc∥/Σ|Fobs|; |Fobs| is the observed and |Fcalc| the calculated structure factor amplitude.
Rfree was calculated from 7% of the measured reflections omitted from refinement.
Structure Solution and Analysis
The structure was solved by using Patterson search techniques with the program MOLREP (26) using data between 20- and 3-Å resolution. The protein part of the CyaC·α,β-Me-ATP complex (16) was used as search model, yielding a prominent solution with four monomers in the asymmetric unit, a correlation coefficient of 45%, and an R-factor of 48%. The model was refined by simulated annealing and conjugate gradient minimization with CNS (27), and rebuilding was done in O (28). An overall anisotropic B-factor and bulk solvent correction were applied to the data. In a later stage of refinement, individual isotropic Debye-Waller factors were refined. The final refined model of the CyaC·α,β-Me-ATP/2-CE complex comprises residues 1002–1199 (monomer A), 1004–1108 and 1112–1199 (monomer B), 1004–1199 (monomer C), and 1005–1107 and 1110–1199 (monomer D) with 89.6% of the residues in the most favored Ramachandran plot regions and no residues in disallowed regions. The model further contains one α,β-Me-ATP molecule and two metal ions per monomer, a total of 139 solvent atoms, and one 2-CE molecule in monomer A and B, respectively. For analysis and comparison of the structure, O, CNS, and LSQMAN (29) were used. Structural figures were generated with MOL-SCRIPT (30) and RASTER3D (31) (Figs. 3B and 4A), SETOR (32) (Fig. 3C), GRASP (33) (Fig. 4B), and PyMol (DeLano Scientific; Figs. 3D and 4, C and D).
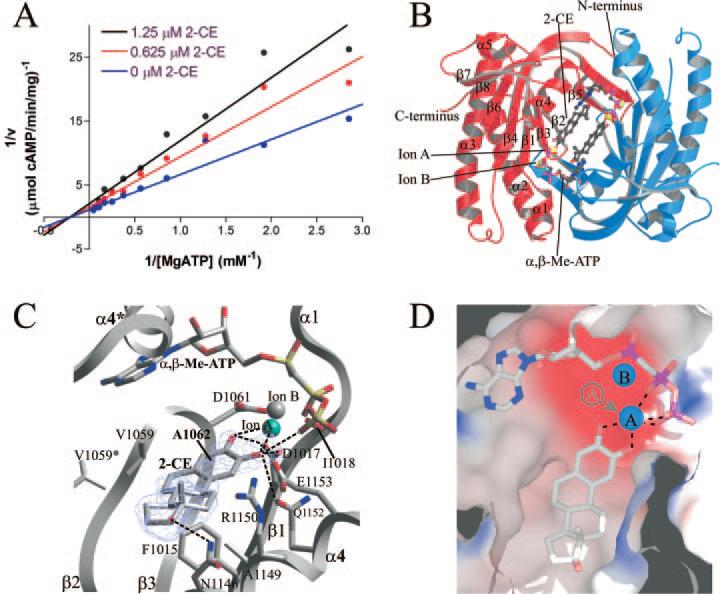
Fig. 3. Structure of CyaC in complex with CE.
A, kinetic mechanism for the inhibition of CyaC by 2-CE. AC activities determined at varying substrate and inhibitor concentrations are displayed in a double reciprocal plot. The linear extrapolations intersect on the x-axis, indicating that CE inhibition is non-competitive with the substrate ATP. B, overall structure of CyaC in complex with the substrate analog α,β-Me-ATP and the inhibitor 2-CE. The two monomers of the homodimer are shown in red and blue, respectively. Two inhibitor molecules occupy the center of the dimer, whereas the active sites are still occupied by the substrate analog and two metal ions. C, detailed view of the interactions between CyaC and the inhibitor 2-CE bound next to the active site, which harbors the substrate analog α,β-Me-ATP and two diva-lent metal ions (one magnesium ion colored cyan, and one calcium ion in gray). Fo – Fc omit electron density for the inhibitor is shown contoured at 2.5 σ. Single letter amino acid abbreviations are presented with position numbers. D, electrostatic surface of the 2-CE binding site showing its mainly hydrophobic nature (blue, positively charged; red, negatively charged). The hydroxyl groups of the inhibitor chelate the catalytic magnesium ion (ion A), removing it from its normal binding site (indicated in gray).
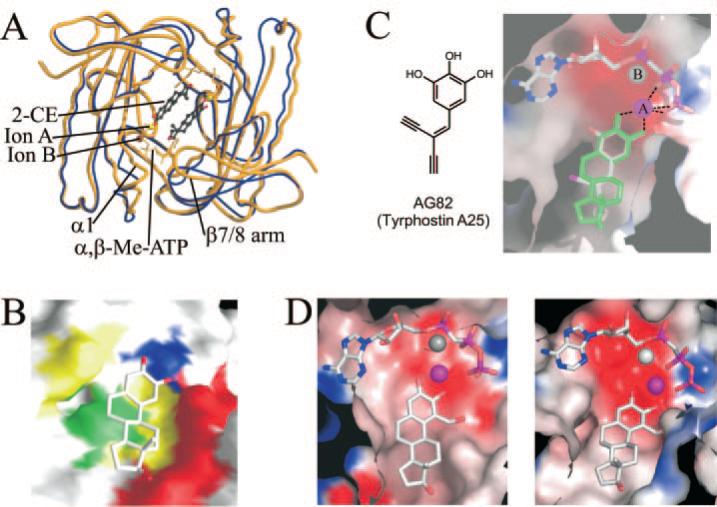
Fig. 4. Analysis of the CE binding site.
A, overlay of the structures of CyaC·α,β-Me-ATP without inhibitor (blue) and after the addition of 2-CE (yellow). Binding of the inhibitor induces large movements of β-strands 7 and 8 and of the small helix α1, resembling the bicarbonate-inducible active site closure observed during CyaC activation. B, molecular surface of CyaC in complex with 2-CE. CE-interacting residues conserved in all AC enzymes are colored blue, and those with low variance are in green. Residues that vary between sAC and tmACs are in yellow, and those that are not conserved between different AC enzymes are in red. C, model for binding of tyrphostin A25 (left) to an AC enzyme based on the CyaC·2-CE complex (right). Tyrphostin A25 (colored according to atom type) can be modeled on top of 2-CE (green) without generating steric conflicts with CyaC (electrostatic surface). D, model for an AC complex with 1-hydroxymethyl-2-hydroxy estrogen based on the CyaC·2-CE complex. The compound can bind in a 2-CE-like fashion to a tmAC V pocket (left), but its hydroxymethyl group at position one would generate steric hindrance when attempting to bind to CyaC (right) or several other tmACs. The catalytic ion A is shown as a magenta sphere, and ion B is colored gray.
RESULTS
CEs Directly Inhibit Class III Mammalian and Bacterial Adenylyl Cyclases
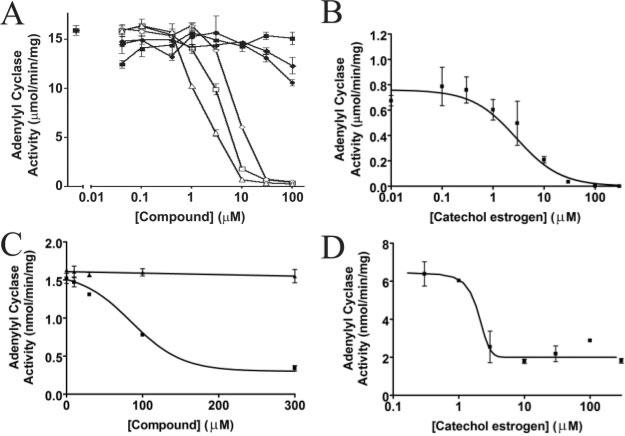
To determine whether CEs (Fig. 1) directly inhibit sACs, we used purified catalytic domains in adenylyl cyclase activity inhibition assays. CEs such as 2-CE, 4-hydroxyestradiol (4-CE), and 2-hydroxyestrone inhibited recombinant human sAC in a concentration dependent manner (Fig. 2A). In contrast the parent compounds estrone and estrogen did not influence the enzymatic activity. The inhibition was reversible (data not shown), and the IC50 values are in the low micromolar range. The cyanobacterial sAC enzyme CyaC was inhibited by 2-CE (the major form of CE) and 4-CE with similar efficacy (Fig. 2B and Supplemental Fig. 3, available in the on-line version of this article).
Fig. 2. Inhibition of CyaC and mammalian tmACs by catechol estrogens.
A, inhibition of purified recombinant human sAC enzyme by catechol estrogens. 2-CE (◇), 4-CE (△), and 2-hydroxy estrone (□) inhibit sAC with comparable affinity, whereas the parent compounds estrogen (◇) and estrone (■) as well as the metabolite 2-methoxy-2-CE (●) have no effect on sAC activity. B, inhibition of the cyanobacterial sAC homolog CyaC by 2-CE. The purified recombinant enzyme is inhibited with an IC50 of ~2 μm. C, dose-dependent inhibition of Sf9 cell membrane preparations containing human tmAC I by 2-CE. AC activity was assayed in duplicate in the presence of the indicated concentration of 2-CE (■) or estrogen (▲). D, inhibition of the purified soluble catalytic domains of human tmAC VII by CE. The tmAC VII activity is inhibited by 2-CE with an IC50 in the low micromolar range.
To reveal whether CEs can also act on tmACs, we first used membrane preparations of insect cells overexpressing human full-length tmACs I, II, and V, respectively. The adenylyl cyclase activity assays showed that CE inhibited the generation of cAMP by these membrane preparations in a concentration-dependent manner (Fig. 2C and Supplemental Fig. 1, available in the on-line version of this article). We then performed the enzymatic assay with purified soluble catalytic domains of human tmAC VII (21), which showed that 2-CE inhibited the enzyme with an IC50 value of ~2 μm (Fig. 2D). These results show that CEs are general inhibitors of class III AC activity and that they act directly on the AC enzymes. The increased IC50 values in our experiments with membrane preparations are likely artifacts due to insertion of the hydrophobic inhibitor into the membrane, which lowers the effective concentration of the compound.
2-CE Is Not Competitive with Respect to ATP and Binds to a Novel Hydrophobic Patch Next to the Active Site
To determine the mode of inhibition of CEs, we assayed adenylyl cyclase activity of CyaC as a function of ATP and 2-CE concentrations, which indicated that 2-CE inhibits the enzyme non-competitively with respect to the substrate ATP (Fig. 3A). This suggests that CEs do not bind to the hydrophobic adenosine recognition pocket of the active site. In addition, CEs do not appear to utilize the known forskolin-binding pocket, the degenerate site pseudo-symmetrically related to the active site in C1C2 heterodimeric ACs. This conclusion is drawn from the observation that CEs are equally potent at inhibiting homodimeric CyaC, which does not have the degenerate site. Therefore, we hypothesized that CEs may interact with an unknown binding site on AC enzymes.
To identify the CE-binding site and determine the molecular mechanism of CE inhibition, we solved the crystal structure of CyaC in complex with 2-CE and the ATP analog α,β-Me-ATP (Table I). Two inhibitor molecules, one per active site, interact symmetrically with a hydrophobic patch at the central depression of the dimer, adjacent to the active sites (Fig. 3B). The two molecules of the ATP analog α,β-Me-ATP still occupy the two active sites located at the dimer interface (16, 34), as they do in the absence of 2-CE. Although the depression at the dimer center is a prominent feature of the wreath-like structure of class III cyclases, it has not been shown previously to interact with any regulators. The identification of this site as the CE-binding site qualifies it as a novel inhibitor-binding site. There are two dimers in the crystallographic asymmetric unit, one with both of these binding sites occupied by 2-CE molecules and one without any bound inhibitor. No obvious reasons could be identified for this difference, such as lack of accessibility due to crystal lattice interactions.
The central helix α4 (Asn-1146, Ala-1149, Arg-1150, Gln1152, and Glu1153), the active site strands β2 and β3 (Val-1059, Ala-1062, and Val-1059*; where the asterisk (*) indicates the partner monomer within the dimer), and the neighboring strand β1 (Phe-1015 and Asp-1017) accommodate the non-polar ring system of 2-CE by interacting with its flat surface and edges (Fig. 3C). The other flat side of the inhibitor shows limited interaction with the protein; instead, in the CyaC complex this side faces the 2-CE molecule bound to the partner monomer and possibly forms weak hydrophobic interactions. However, we did not observe pronounced cooperativity in 2-CE inhibition of CyaC, suggesting that this interaction does not contribute significantly to 2-CE binding and inhibition. The potential cooperative behavior seen against soluble tmAC domains was not confirmed by our experiments with tmAC-containing membranes. This lack of cooperativity is also more consistent with the assumption that only one CE molecule can bind to a catalytic heterodimer (see below). In the CyaC·2-CE complex, the end of 2-CE distal to the catechol moiety also interacts with the adenine ring and the sugar from the symmetric active site; the interaction is fairly localized, centering at C16 of 2-CE.
2-CE Captures the Catalytic Ion to Inactivate the Enzyme
Comparison of the CyaC·α,β-Me-ATP complex structures with and without 2-CE reveals that this compound exploits a general feature of class III AC enzymes for inhibition. The active site of these enzymes comprises two divalent metal ions in the so-called ion A and ion B sites. The ion A site harbors a catalytic magnesium ion that acidifies the ribose 3′ hydroxyl and stabilizes the transition state, whereas the ion B serves as an anchoring point for ATP by coordinating and stabilizing its β and γ phosphates (15, 16). The two adjacent hydroxyl groups of 2-CE coordinate the magnesium ion at the A site (distances 2.2 and 2.5 Å, respectively) like a pair of pincers (Fig. 3D), stabilizing this ion at 3 Å away from its original position and preventing its participation in catalysis. In contrast, the position of ion B did not change significantly. The two adjacent inhibitor hydroxyls are also part of a hydrogen bond network involving Glu-1153, Asp-1017, and a γ-phosphate oxygen of the substrate analog (Fig. 3D). In addition, the 17-hydroxyl group of 2-CE is in H-bond distance to Asn-1146.
In the 2-CE-inhibited structure, the α-phosphate of the ATP analog exhibits a non-productive conformation not suitable for in-line attack, likely as a result of the coordination of its β- and γ-phosphate oxygen atoms to the shifted ion A. This change in phosphate conformation is also accompanied by active site closure (Fig. 4A), reflected in the movement of helix α1 by ~4 Å and the movement of the small sheet β7/β8 by 2 Å toward the dimer center (16, 34). Although active site closure is also a prominent feature of productive catalysis, the conformation of the phosphates and the ion positions renders the enzyme inactive rather than active.
The importance of the observed inhibitor-ion interaction suggests that only one CE molecule binds to heterodimeric mammalian ACs (all known tmACs and sAC). Although the pocket analog to the CE binding site exists on both C1 and C2, the inhibitor likely occupies only the pocket on the surface of C1 that connects to the active site rather than the degenerate site, which lacks the ion-binding aspartates. Without the metal ion interactions, the affinity of the inhibitor to the C2 site will be predictably much lower. In addition, placing 2-CE to the C2 pocket of tmACs would create steric hindrance to the bound forskolin, which interacts with tmACs at nanomolar affinity (35). The addition of 100 μm forskolin, however, has no effect on the inhibition by 2-CE (Supplemental Fig. 2, available in the on-line version of this article). The importance of the 2-CE-ion interaction also suggests that 4-CE binds 180° rotated along its long axis, which would keep its hydrophobic part in the non-polar binding cleft and position its hydroxyl groups similarly to those of 2-CE for ion binding.
The CE Binding Site Contains Both Conserved and Variable Elements
The CE-binding site is largely formed by a surface depression at the center of the dimeric class III AC enzymes that is present in all known crystal structures of this enzyme family (14, 16, 36, 37). Although the existence of the patch is conserved, a sequence comparison between CyaC and the mammalian heterodimeric ACs (Supplemental Fig. 4, available in the on-line version of this paper) shows that among residues directly in contact with 2-CE, only one is strictly conserved in the analogous C1 patch (residue Asp-1017), five are mostly conserved (Phe-1015, Val-1059, Ala-1062, Gln-1152, and Ala-1149), and the remaining three (Asn-1146, Arg-1150, and Glu-1153) are highly variable (Fig. 4B).
Almost all contacts between the protein and the mainly hydrophobic inhibitor are apolar, with the exception of polar interactions with the 2-, 3-, and 17-hydroxyls in 2-CE. The dominance of hydrophobic contacts in CE binding rationalizes the observed promiscuity of CEs for the different AC isoforms. However, the inhibitor-binding pocket is formed and flanked by several variable polar residues (Fig. 4B), suggesting that the CE-binding site bears the potential for a more specific inhibition that has not been fully exploited by CEs. In addition, the observed CE/enzyme interaction shows a non-ideal shape complementarity, indicating another factor that could be exploited to yield more effective and specific AC inhibitors.
The CE Inhibition Mechanism Predicts the Binding Mode for Other AC Inhibitors
The mode of AC inhibition by 2-CE predicts that, as in a catechol moiety, two adjacent hydroxyl groups coupled to a hydrophobic scaffold are required for efficient AC binding and inhibition. This insight on the structural requirement of this class of AC inhibitors explains previous observations on a series of catechol estrogens and related physiological compounds on cAMP production from sAC-containing extracts (19). Those that contain a catechol moiety (2-CE, 4-CE, 2-hydroxyestrone, apomorphine, catechol, dopamine, l-dopa, norepinephrine, and isoproterenol) inhibited sAC activity, although with varying efficacies depending on the hydrophobicity of the compound parts linked to the catechol ring system (Supplemental Fig. 5, available in the on-line version of this paper). In contrast, similar compounds that do not have two adjacent hydroxyl groups (tyrosine, phenylephrine, estradiol, and 6α-hydroxyestradiol) are much less effective (~100 ×).
More recently two other AC inhibitors, tyrphostins (38) and calmidazolium (39), were identified as being non-competitive with ATP. Tyrphostins are a family of aromatic tyrosine kinase inhibitors. The tyrphostins effective in AC inhibition also possess a catechol moiety and likely use similar molecular mechanisms for AC inhibition as those observed here for CEs. A model of the most effective tyrphostin A25 bound to CyaC may be generated without creating steric clash by superposition of the tyrphostin on the CE of the CyaC·2-CE complex (Fig. 4C). Calmidazolium is a largely hydrophobic calcium chelator, suggesting it might also exploit the same binding site and inhibition mechanism, although the details of this interaction are more difficult to predict from the CE-bound CyaC structure. Therefore, it appears that the binding site identified from the CyaC·2-CE complex structure may be a common site utilized by an emerging family of non-competitive AC inhibitors.
DISCUSSION
The 10 mammalian AC subclasses control diverse sets of essential physiological processes, rendering them interesting targets for therapeutic intervention. Successful drug development based on this target has to date been limited, however, as most known AC inhibitors bind to the enzyme's ATP binding site (10, 17). These nucleotide mimics tend to bind to a wide range of nucleotide-binding proteins, potentially causing many unwanted side effects (e.g. the potent P-site inhibitor 3′-dATP also inhibits DNA polymerase) (40). This binding mode thus promises little opportunity for specific inhibition of ACs or AC subclasses. In contrast, inhibitors binding outside the active site (allosteric inhibitors) tend to exploit more specific features of a target protein and have been described, e.g. for protein tyrosine phosphatase 1B (41) and G protein-coupled receptors (42). Examples of the successful application of this principle to drug development are the non-nucleoside inhibitors (nevirapine, delavirdine, and efavirenz) of reverse transcriptase (43) and the inhibition of chemokine receptors by Repertaxin (44).
The CE-binding pocket described here constitutes a novel ligand-binding site in AC enzymes specific to this protein family. Sequence variations in this binding pocket indicate that it constitutes a promising starting point for the development of inhibitors specific to ACs and even to AC subclasses. Adding a hydroxymethyl group at 2-CE position 1, for example, would yield a compound compatible with the tmAC V-binding site but incompatible with CyaC and sAC (Fig. 4D). In addition, the non-ideal complementarity between 2-CE and its binding site suggests that modifications may be made to the estrogen scaffold to increase both specificity and affinity. Consistently, compound classes completely different from the steroids seem to be able to exploit the CE-binding site and inhibition mechanism. Tyrphostins (38) as well as epinephrine and related compounds inhibiting ACs (19, 45) still carry a catechol moiety, and the parts attached to the catechol ring strongly influence the affinity of the respective compound. Calmidazolium (39) and the benzimidazole derivative KH7,2 finally, are AC inhibitors that lack any structural similarity to CEs. This structural diversity indicates that a large set of compounds and modifications should be considered in order to identify inhibitors that further exploit the subclass-specific features of this binding site and can potentially serve as highly specific AC-targeted drugs.
Are CEs physiological regulators of ACs? The affinity of the compounds we tested indicates that AC inhibition can contribute to their physiological effects in tissues with elevated CE levels. However, we cannot exclude the possibility that other unknown physiological compounds with a hydrophobic scaffold might exploit the CE-binding site and constitute physiological AC regulators of higher efficacy and specificity. This situation resembles the findings for the tmAC-activating diterpene forskolin isolated from plant extracts (46). Its hydrophobic binding cleft is conserved in eight of the nine human tmAC subclasses, but a physiological ligand, probably displaying higher specificity, has not yet been identified.
In summary, we described here CEs as novel, non-competitive inhibitors of AC enzymes. The crystal structure of a sAC CE complex reveals that the inhibitor binds to a pocket near to the active site and “hijacks” the catalytic magnesium ion, a mechanism that likely also applies to the CE inhibition of tyrosine hydroxylase (47) and 5-lipoxygenase (48). Variations in the inhibitor-binding pockets of the various AC isoforms suggest that exploiting this pocket might enable the development of highly specific AC-targeted drugs.
Supplementary Material
Acknowledgments
We thank R. Abramowitz and X. Yang for technical support at National Synchrotron Light Source beamline X4A.
Footnotes
This work was supported in part by funds from the National Institutes of Health (to L. R. L. and J. B.), the Hirschl Weill-Caulier Trust (to L. R. L.), the Ellison Medical Foundation (to J. B.), and United States Public Health Service Grant GM53645 (to R. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains Supplemental Figs. 1–5, which present further data on the effects of catechol estrogen.
The abbreviations used are: CE, catechol estrogen; AC, adenylyl cyclase; sAC, soluble AC; tmAC, trans-membrane AC; 2-CE, 2-hydroxyestradiol; 4-CE, 4-hydroxyestradiol; CyaC, cyanobacterial sAC; α,β-Me-ATP, α,β-methylene-adenosine-5′-triphosphate.
The atomic coordinates and structure factors (code 2BW7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
J. H. Zippin, K. Hess, M. Kamenetsky, T. E. McGraw, J. Buck, and L. R. Levin, submitted for publication.
REFERENCES
- 1.Zhu BT, Conney AH. Carcinogenesis. 1998;19:1–27. doi: 10.1093/carcin/19.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Dubey RK, Tofovic SP, Jackson EK. J. Pharmacol. Exp. Ther. 2004;308:403–409. doi: 10.1124/jpet.103.058057. [DOI] [PubMed] [Google Scholar]
- 3.Paul SM, Axelrod J. Science. 1977;197:657–659. doi: 10.1126/science.877577. [DOI] [PubMed] [Google Scholar]
- 4.Liehr JG, Ricci MJ. Proc. Natl. Acad. Sci. U. S. A. 1996;93:3294–3296. doi: 10.1073/pnas.93.8.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das SK, Taylor JA, Korach KS, Paria BC, Dey SK, Lubahn DB. Proc. Natl. Acad. Sci. U. S. A. 1997;94:12786–12791. doi: 10.1073/pnas.94.24.12786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philips BJ, Ansell PJ, Newton LG, Harada N, Honda S, Ganjam VK, Rottinghaus GE, Welshons WV, Lubahn DB. Biochemistry. 2004;43:6698–6708. doi: 10.1021/bi036154j. [DOI] [PubMed] [Google Scholar]
- 7.Paul SM, Skolnick P. Nature. 1977;266:559–561. doi: 10.1038/266559a0. [DOI] [PubMed] [Google Scholar]
- 8.Linder JU, Schultz JE. Cell. Signal. 2003;15:1081–1089. doi: 10.1016/s0898-6568(03)00130-x. [DOI] [PubMed] [Google Scholar]
- 9.Buck J, Sinclair ML, Schapal L, Cann MJ, Levin LR. Proc. Natl. Acad. Sci. U. S. A. 1999;96:79–84. doi: 10.1073/pnas.96.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanoune J, Defer N. Annu. Rev. Pharmacol. Toxicol. 2001;41:145–174. doi: 10.1146/annurev.pharmtox.41.1.145. [DOI] [PubMed] [Google Scholar]
- 11.Buck J, Sinclair ML, Levin LR. Methods Enzymol. 2002;345:95–105. doi: 10.1016/s0076-6879(02)45009-4. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Cann MJ, Litvin TN, Iourgenko V, Sinclair ML, Levin LR, Buck J. Science. 2000;289:625–628. doi: 10.1126/science.289.5479.625. [DOI] [PubMed] [Google Scholar]
- 13.Zippin JH, Levin LR, Buck J. Trends Endocrinol. Metab. 2001;12:366–370. doi: 10.1016/s1043-2760(01)00454-4. [DOI] [PubMed] [Google Scholar]
- 14.Tesmer JJ, Sunahara RK, Gilman AG, Sprang SR. Science. 1997;278:1907–1916. doi: 10.1126/science.278.5345.1907. [DOI] [PubMed] [Google Scholar]
- 15.Tesmer JJ, Sunahara RK, Johnson RA, Gosselin G, Gilman AG, Sprang SR. Science. 1999;285:756–760. doi: 10.1126/science.285.5428.756. [DOI] [PubMed] [Google Scholar]
- 16.Steegborn C, Litvin TN, Levin LR, Buck J, Wu H. Nat. Struct. Mol. Biol. 2005;12:32–37. doi: 10.1038/nsmb880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dessauer CW, Tesmer JJ, Sprang SR, Gilman AG. Trends Pharmacol. Sci. 1999;20:205–210. doi: 10.1016/s0165-6147(99)01310-3. [DOI] [PubMed] [Google Scholar]
- 18.Levy DE, Bao M, Cherbavaz DB, Tomlinson JE, Sedlock DM, Homcy CJ, Scarborough RM. J. Med. Chem. 2003;46:2177–2186. doi: 10.1021/jm0205604. [DOI] [PubMed] [Google Scholar]
- 19.Braun T. Proc. Soc. Exp. Biol. Med. 1990;194:58–63. doi: 10.3181/00379727-194-43055. [DOI] [PubMed] [Google Scholar]
- 20.Litvin TN, Kamenetsky M, Zarifyan A, Buck J, Levin LR. J. Biol. Chem. 2003;278:15922–15926. doi: 10.1074/jbc.M212475200. [DOI] [PubMed] [Google Scholar]
- 21.Yan SZ, Tang WJ. Methods Enzymol. 2002;345:231–241. doi: 10.1016/s0076-6879(02)45019-7. [DOI] [PubMed] [Google Scholar]
- 22.Taussig R, Tang WJ, Gilman AG. Methods Enzymol. 1994;238:95–108. doi: 10.1016/0076-6879(94)38009-0. [DOI] [PubMed] [Google Scholar]
- 23.Salomon Y. Adv. Cyclic Nucleotide Res. 1979;10:35–55. [PubMed] [Google Scholar]
- 24.Smigel MD. J. Biol. Chem. 1986;261:1976–1982. [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. DENZO: A Film Processing Program for Macromolecular Crystallography. Yale University; New Haven, CT: 1993. [Google Scholar]
- 26.Vagin A, Teplyakov A. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 27.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Jones TA, Kjelgaard M. O: The Manual. University of Uppsala; Uppsala, Sweden: 1991. [Google Scholar]
- 29.Kleywegt GJ, Jones TA. ESF/CCP4 Newsletter. 1994;31:9–14. [Google Scholar]
- 30.Kraulis PJ. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 31.Merrit EA, Murphy MEP. Acta Crystallogr. Sect. D Biol. Crystallogr. D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 32.Evans SV. J. Mol. Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 33.Nicholls A, Bharadwaj R, Honig B. Biophys. J. 1993;64:A166. (abstr.) [Google Scholar]
- 34.Tesmer JJ, Sprang SR. Curr. Opin. Struct. Biol. 1998;8:713–719. doi: 10.1016/s0959-440x(98)80090-0. [DOI] [PubMed] [Google Scholar]
- 35.Dessauer CW, Scully TT, Gilman AG. J. Biol. Chem. 1997;272:22272–22277. doi: 10.1074/jbc.272.35.22272. [DOI] [PubMed] [Google Scholar]
- 36.Zhang G, Liu Y, Ruoho AE, Hurley JH. Nature. 1997;386:247–253. doi: 10.1038/386247a0. [DOI] [PubMed] [Google Scholar]
- 37.Bieger B, Essen LO. EMBO J. 2001;20:433–445. doi: 10.1093/emboj/20.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaleel M, Shenoy AR, Visweswariah SS. Biochemistry. 2004;43:8247–8255. doi: 10.1021/bi036234n. [DOI] [PubMed] [Google Scholar]
- 39.Haunso A, Simpson J, Antoni FA. Mol. Pharmacol. 2003;63:624–631. doi: 10.1124/mol.63.3.624. [DOI] [PubMed] [Google Scholar]
- 40.Englund PT, Huberman JA, Jovin TM, Kornberg A. J. Biol. Chem. 1969;244:3038–3044. [PubMed] [Google Scholar]
- 41.Wiesmann C, Barr KJ, Kung J, Zhu J, Erlanson DA, Shen W, Fahr BJ, Zhong M, Taylor L, Randal M, McDowell RS, Hansen SK. Nat. Struct. Mol. Biol. 2004;11:730–737. doi: 10.1038/nsmb803. [DOI] [PubMed] [Google Scholar]
- 42.Soudijn W, Van Wijngaarden I, Ijzerman AP. Drug Discov. Today. 2004;9:752–758. doi: 10.1016/S1359-6446(04)03220-9. [DOI] [PubMed] [Google Scholar]
- 43.Esnouf R, Ren J, Ross C, Jones Y, Stammers D, Stuart D. Nat. Struct. Biol. 1995;2:303–308. doi: 10.1038/nsb0495-303. [DOI] [PubMed] [Google Scholar]
- 44.Bertini R, Allegretti M, Bizzarri C, Moriconi A, Locati M, Zampella G, Cervellera MN, Di Cioccio V, Cesta MC, Galliera E, Martinez FO, Di Bitondo R, Troiani G, Sabbatini V, D'Anniballe G, Anacardio R, Cutrin JC, Cavalieri B, Mainiero F, Strippoli R, Villa P, Di Girolamo M, Martin F, Gentile M, Santoni A, Corda D, Poli G, Mantovani A, Ghezzi P, Colotta F. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11791–11796. doi: 10.1073/pnas.0402090101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ebina T, Toya Y, Oka N, Schwencke C, Kawabe J, Ishikawa Y. J. Mol. Cell. Cardiol. 1997;29:1247–1254. doi: 10.1006/jmcc.1996.0362. [DOI] [PubMed] [Google Scholar]
- 46.Tang WJ, Hurley JH. Mol. Pharmacol. 1998;54:231–240. doi: 10.1124/mol.54.2.231. [DOI] [PubMed] [Google Scholar]
- 47.Lloyd T, Weisz J. J. Biol. Chem. 1978;253:4841–4843. [PubMed] [Google Scholar]
- 48.Alanko J, Sievi E, Lahteenmaki T, Mucha I, Vapaatalo H, Parantainen J. Biochem. Pharmacol. 1998;55:101–104. doi: 10.1016/s0006-2952(97)00398-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.