Abstract
Benzo[a]pyrene (B[a]P), a major human carcinogen in combustion products such as cigarette smoke and diesel exhaust, is metabolically activated into DNA-reactive metabolites via three different enzymatic pathways. The pathways are the anti-(+)-benzo[a]pyrene 7,8-diol 9, 10-epoxide pathway (P450/ epoxide hydrolase catalyzed) (B[a]PDE), the benzo[a]pyrene o-quinone pathway (aldo ketose reductase (AKR) catalyzed) and the B[a]P radical cation pathway (P450 peroxidase catalyzed). We used a yeast p53 mutagenesis system to assess mutagenesis by B[a]P radical cations. Because radical cations are short-lived, they were generated in situ by reacting B[a]P with cumene hydroperoxide (CuOOH) and horse radish peroxidase (HRP) and then monitoring the generation of the more stable downstream products, B[a]P-1,6-dione and B[a]P-3,6-dione. Based on the B[a]P-1,6 and 3,6-dione formation, approximately 4µM of radical cation was generated. In the mutagenesis assays, the radical cations produced in situ showed a dose-dependent increase in mutagenicity from 0.25 µM to 10 µM B[a]P with no significant increase seen with further escalation to 50 µM B[a]P. However, mutagenesis was 200-fold less than with the AKR pathway derived B[a]P, 7–8 dione. Mutant p53 plasmids, which yield red colonies, were recovered from the yeast to study the pattern and spectrum of mutations. The mutation pattern observed was G to T (31%) > G to C (29%) > G to A (14%). The frequency of codons mutated by the B[a]P radical cations was essentially random and not enriched at known cancer hotspots. The quinone products of radical cations, B[a]P-1,6-dione and B[a]P-3,6-dione were more mutagenic than the radical cation reactions, but still less mutagenic than AKR derived B[a]P-7,8-dione. We conclude that B[a]P radical cations and their quinone products are weakly mutagenic in this yeast-based system compared to redox cycling PAH o-quinones.
Keywords: Radical cation, carcinogenesis, quinone, polycyclic aromatic hydrocarbon, PAH
Introduction
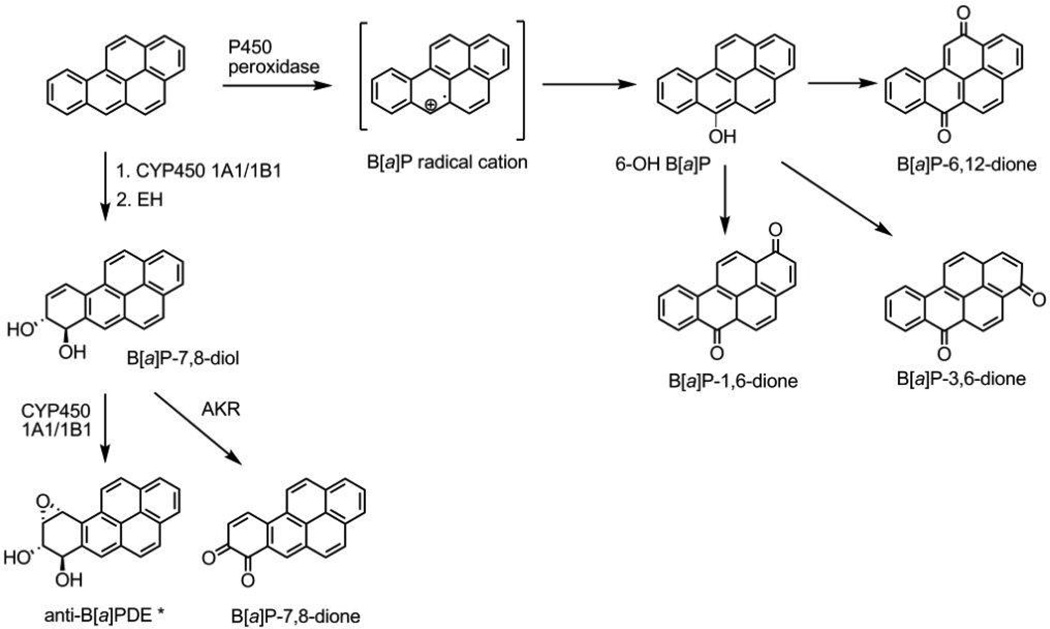
The leading cause of cancer death, annually resulting in more than one million deaths worldwide, is lung cancer. About 90% of lung cancer is caused by exposure to tobacco smoke.1 Tobacco smoke consists of more than 4000 chemicals with about 60 known carcinogens. One of the major classes of carcinogens found in tobacco smoke are the polycyclic aromatic hydrocarbons (PAH) such as benzo[a]pyrene (B[a]P).2, 3 PAH are ubiquitous combustion products so even though tobacco smoke is the leading cause of lung cancer, exposure to other environmental PAHs contribute to lung carcinomas. B[a]P is unreactive with DNA and must be metabolically activated to ultimate carcinogens that form DNA-adducts. B[a]P has three major routes of metabolic activation, the diol-epoxide pathway, the o-quinone pathway and the radical cation pathway (Scheme 1).4–7
Scheme 1. B[a]P metabolism.
The three pathways of activation, (i) the diol epoxide pathway, (ii) the PAH o-quinone pathway and (iii) the radical cation pathway. Only one of the four enantiomers of B[a]PDE is shown.
The product of the cytochrome P450 (P450)/epoxide hydrolase (EH) pathway (±)-anti-7β,8α-dihydroxy-9α10α-oxo-tetrahydrobenzo[a]pyrene ((±)-anti-B[a]PDE or B[a]PDE, a diol-epoxide has been the most extensively studied. B[a]PDE is an ultimate carcinogen that forms stable DNA adducts, primarily at guanine bases. B[a]PDE adducts are preferentially formed at codons in the p53 tumor suppressor which coincide with hotspots mutated in human lung cancer.8
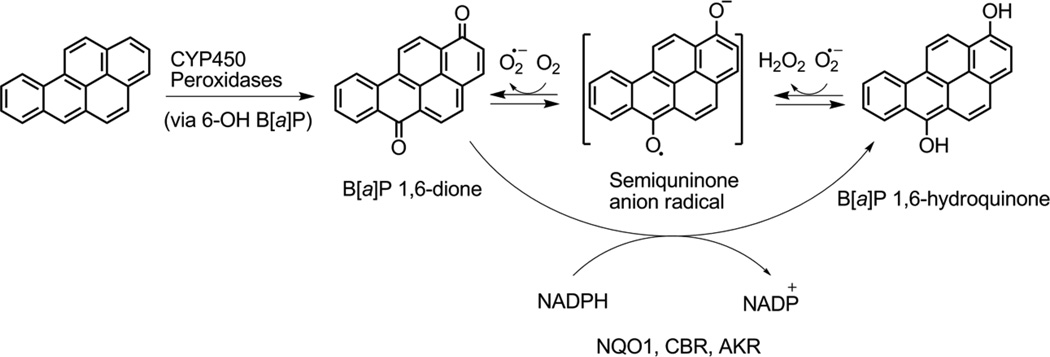
The aldo keto reductase (AKR) pathway forms o-quinones such as B[a]P-7,8-dione (BPQ). B[a]P-7,8-dione can act as a Michael acceptor and form stable covalent DNA adducts in vitro.9, 10 However, in the presence of NADPH, B[a]P-7,8-dione enters futile redox cycles to generate reactive oxygen species (ROS).11 This process is exacerbated by enzymatic two electron redox-cycling catalyzed by NQO1 and AKRs themselves.12 ROS damages DNA by the formation of 8-oxo-2’-deoxyguanosine (8-oxo-dGuo), which can lead to G to T transversions (Scheme 2). While PAH o-quinones can form stable and depurinating adducts in vitro, they are predominantly mutagenic by generating ROS to form 8-oxo-dGuo. Comparative mutagenesis studies suggest that the primary route to DNA damage by B[a]P-7,8-dione and other o-quinones is through ROS.13, 14
Scheme 2. Redox cycling of quinones.
Example of BP-1,6-dione redox cycling is shown.
The radical cation pathway of B[a]P activation is catalyzed by P450 through a ‘peroxidase-like’ activity. P450 can catalyze one-electron oxidation of B[a]P in addition to catalyzing two-electron oxidations. One-electron oxidation of B[a]P by P450 generates a radical cation at the C6 position of B[a]P, which is DNA reactive. The radical cations also rapidly undergo hydroxylation and air-oxidation to form three quinones, B[a]P-1,6-dione, B[a]P-3,6 dione and B[a]P-6,12 dione.15 Prior to their conversion to diones, radical cations are reactive electrophilic species which bind to DNA and form predominantly depurinating DNA adducts.16 The depurinating adducts include 7-(benzo[a]pyrene-6-yl)-guanine (B[a]P-6N7-Gua), 7-(benzo[a]pyrene-6-yl)-adenine (B[a]P-6-N7-Ade) and 8-(benzo[a]pyrene-6-yl)-guanine (B[a]P-6-C8-Gua).17–19 In mouse skin about 80% of the radical cation DNA adducts are depurintating adducts, while 20% are stable adducts such as 10-(gaunin-2-yl)-7,8,9-trihydroxy-,7,8,9,10-tetrahydrobenzo[a]pyrene (anti-B[a]PDE-N2-dGuo).20
Reactive metabolites of PAHs cause mutations in tumor suppressors such as p53, which is the most commonly mutated gene in human lung cancer.21 More than 20,000 mutations in p53 have been identified in tumors and compiled into a database that can be used to identify patterns (the frequency of specific base changes) and spectra (the frequency of mutations by codon). There are three distinct characteristics of the pattern and spectrum of p53 mutations in lung cancer.22 The first is the predominance of G to T transversions in the pattern of mutations. Other types of cancers show different mutational patterns, generally dominated by G to A (C to T) transitions. The predominance of G to T transversions is the most unambiguous hallmark of lung cancer mutations. The second feature, called a strand bias, is that guanine residues are preferentially mutated in the non-transcribed strand when compared to the transcribed strand.23 The third is seen in the spectrum of mutations, where preferential mutations of about 23 codons account for 50% all the reported mutations. The most common of these hotspots are codons 248, 273, 249, 245, 158 and 157 but because most of these codons are mutated in other cancers, this characteristic is not as unique to lung cancer.21, 23 The pathways through which tobacco smoke carcinogens mutate p53 must account for the specific mutational pattern and spectrum in lung cancer.
Testing p53 mutagenesis is difficult in mammalian cells because cells containing mutant p53 are not easily identified. Moreover, all three pathways of metabolic activation were found to occur to a comparable extent in human bronchoalveolar (H358) cells making it difficult to assess the contribution of each pathway to mutagenesis.24 We have employed a p53 mutagenicity assay that scores for functional p53 using a yeast reporter system.13, 25, 26 The primary advantage of this assay over bacterial systems is that the assay system utilizes the transcriptional activity of a p53 expression plasmid to isolate biologically relevant mutations. In the yeast mutagenesis assay, a plasmid expressing p53 is exposed to a mutagen in vitro. The treated plasmid is then transformed in to the host yeast strain YIG397, an ade2- strain, which has multiple p53 binding sites engineered into a promoter driving expression of ADE2. When wild-type p53 is expressed, it binds DNA and drives the expression of ADE2, which will turn the host yeast strain from red to white. Mutant p53 colonies remain red. The ratio of red to white colonies is used to quantify mutagenic frequency and the p53 plasmid can be isolated and sequenced to determine patterns and spectra.
In the yeast system, B[a]P-7,8-dione is 25 to 80 times more mutagenic than (±)-(anti)-B[a]PDE, provided redox-cycling conditions were present.13, 14 The mutational pattern observed with B[a]P-7,8-dione showed a predominance of G to T transversions. The primary lesion responsible for these mutations is likely 8-oxo-dGuo since there was a linear correlation between the mutagenic frequency of the dione and the amount of 8-oxo-dGuo formed.27,28 The spectra of p53 mutations are random, without an enrichment of known cancer hotspots. However, if an additional level selectivity is added to detect dominant mutations, then the spectrum closely matches that seen in lung cancer.27,29
Herein we report the results from p53 mutagenesis conducted with the yeast reporter system using B[a]P radical cations generated in situ as the mutagen. Radical cations were poor mutagens when compared to o-quinones, even when the radical cation-derived quinone products B[a]P-1,6-dione and B[a]P-3,6 dione were tested under redox cycling conditions. The majority of the mutations were G to T (31%) transversions followed by G to C transversions (29%). Mutations were randomly found in the DNA binding domain without enrichment for hotspots.
Materials and Methods
Caution
All PAHs are potentially hazardous and should be handled in accordance with NIH Guidelines for the Laboratory Use of Chemical Carcinogens.
Chemicals and Regents
Adenine, L-leucine, L-tryptophan, Horseradish Peroxidase Type VI-A, Cumene hydroperoxide (80%) and benzo[a]pyrene (BP) were purchased from Sigma (St. Louis, MO). YEASTMAKER Yeast Transformation kit and all yeast culture media were obtained from CLONTECH (Palo Alto, CA). Zymoprep Yeast Plasmid Miniprep II from Zymo Research (Irvine, CA) was used to isolate the yeast plasmid and Bacterial Plasmid Isolation kit was purchased from Qiagen. B[a]P-7,8 dione, B[a]P-1,6 dione, and B[a]P-3,6 dione was purchased from MRIGlobal Chemical Carcinogen Repository (Kansas City, MO). [13C4]-B[a]P-16,-dione and [13C4]-B[a]P-3,6-dione were synthesized as described.30 The purity of all chemicals was assessed by HPLC and UV spectroscopy. Other reagents were of the highest grade available.
Yeast Strains, Media and Plasmids
The ade reporter yeast strain, yIG397 was kindly provided by Dr. Richard Iggo (Swiss Institute for Experimental Cancer Research, 1066 Epalinges, Basel, Switzerland).31 Basic methods for yeast manipulations were carried out as described.32
p53 reporter gene assay
About 1 µg of p53 was treated for 2h at 37 °C with the indicated amounts of B[a]P, 2 mM Cumene hydroperoxide and 1 mg/mL HRP in 67mM Sorenson’s buffer with the final reaction volume of 100 µL. Final reactions contained 10% DMSO to enhance B[a]P solubility. After the 2h incubation, the DNA was precipitated using 20 µL of 5M NaCl and 2.5 volumes of 95% ethanol at −80 °C for 2h. The DNA was isolated by microcentrifugation and washed twice with 70% ethanol. The pellets were then dried using a speed vacuum and suspended in deionized water to be further used in host strain yIG397 (cultured until an A600 of 0.9–1.1 was obtained) for transformations. Where indicated, incubations containing B[a]P-7,8-diones, B[a]P-1,6-diones and B[a]P-3,6-diones were supplemented with 100 µM CuCl2 plus 1 mM NADPH. Incubations with the B[a]P-diones were performed for 2 h at 37 °C. The treated p53 and 15 µL (150 µg) of the carrier (herring testis) DNA were co-transformed into the yeast host strain yIG397 strain (grown to an OD 600 of 0.9–1.1) using the lithium acetate procedure according to the YEASTMAKER Yeast Transformation System Kit (CLONTECH). The cells were collected, suspended in 300 µL 0.9% NaCl, plated on synthetic minimal medium minus leucine plus minimal adenine (5 µg/mL), and incubated for 3 days at 30 °C. Strain yIG397 has an ADE2 reporter gene under the control of a p53 dependent promoter stably integrated into its genome. Wild type p53 stimulates reporter gene expression, whereas change-in-function mutations of p53 do not. Yeast colonies expressing wild-type p53 are white and yeast colonies expressing mutant p53 are red. Red colonies were clearly identifiable after 3 days at 30 °C, but the color is more intense after an additional 2 days at 4 °C. The mutation frequency was expressed as (number of red colonies – number of spontaneous red colonies/ total number of colonies) × 100.
Recovery of p53 plasmids from yeast and DNA sequencing
p53 expression plasmids were rescued from the mutant yeast, transformed into E. coli, isolated and then sequenced as described.13 In some cases p53 was amplified by PCR from yeast DNA and then sequenced as described.33 The DNA binding domain of p53 (codons 102 to 292) was sequenced on both strands with S6 (5’-dCTGGGACAGCCAAGTCTGT-3’) and R6 (5’-dCCTCATTCAGCTCTCGGAA-3’) primers. These primers allow double stranded sequencing from amino acids 126 to 339 encompassing almost the entire DNA binding domain. Sequencing was performed on an Applied Biosystems 373A automated sequencer in the DNA sequencing facility at the Cell Center at the Perelman School of Medicine, University of Pennsylvania.
DNA strand scission assay
p53 plasmid DNA was treated with mutagens for 2 h at 37 °C and about 20 µL reaction mixture was directly run on a 1% agarose gel with 10 µL of ethidium bromide. The gel was run for 35 min in TAE buffer and directly analyzed after the run on a BioRad UV detection system (Milan, Italy) with Chemidoc analysis software.
HPLC Analysis
The analysis of B[a]P radical cation derived metabolites and standards was conducted using a Waters Alliance 2695 chromatographic HPLC system coupled to a Waters 996 photodiode array (PDA) detector. Individual standards as well as a mixture, B[a]P 1,6-dione, B[a]P 3,6-dione and 6-OH-B[a]P were analyzed by an Agilent Zorbax ODS-C18 column, 5um, 4.5 mm × 150 mm (Agilent technology, Santa Clara, CA) by RP-HPLC. The mobile phase A was water and the mobile phase B was methanol. Initial elution conditions were 55% B which increased to 70% B in a linear gradient over 20 min at a flow rate of 0.5mL/ min. The mobile phase was increased to 80% B over the next 10 min and kept at 80% B for the next 20 min. Then the mobile phase was increased to 98% B over 10 min and was held at 98% B for 30 min, the gradient was then adjusted to 55% B over 5 min. All the product peaks were identified by their UV spectrum and compared with purified standards.
Liquid Chromatography-Tandem Mass Spectrometric Analysis
LC-MS analysis was performed in the Biomarker core facility of the Center of Excellence in Environmental Toxicology according to previously published methods. Briefly, a Waters Alliance 2690 HPLC system (Waters Corporation, Milford, MA) interfaced with a Finnigan TSQ Quantum Ultra spectrometer (Thermo Fisher, San Jose, CA) was used for the analysis. A HPLC gradient system with solvents A (5 mM ammonium acetate in water containing 0.02% (v/v) formic acid) and B (5 mM ammonium acetate in methanol containing 0.02% (v/v) formic acid) at a flow rate of 0.5 mL/min was adopted. The linear gradient increased from 55% B to 85% B over 15 min, the mobile phase was then increased to 99% B over 20 min and was held at 99% B for 20 min, the gradient was then adjusted to 55% B over 5 min. The eluent from the HPLC column was directly introduced into a standard APCI source equipped with a corona discharge pin. SRM transitions of the protonated molecules for B[a]P-1,6-dione and B[a]P-3,6-dione were: m/z 283 to m/z 226; and the corresponding MS/MS transition for the internal standards, [13C4]-B[a]P-1,6-dione and [13C4]-B[a]P-3,6-dione, were m/z 287 to m/z 229, demonstrating the loss of two carbonyl groups.
Statistical Analysis
There are 12 different possible base substitution mutations. Each of the four bases can mutate to any of the other three. Since our mutagenicity assay does not distinguish between whether the initial hit was on the coding or non-coding strand, the number of possible base substitutions is reduced to six: G→T (transversion), G→A (transition), G→C (transversion), A→G (transition), A→C (transversion) and A→T (transversion). The test was based on the probability that any one of the mutations is preferred over a random distribution. To test whether the radical cations caused G→T transversions more than any other base substitution, we used a χ2 goodness of fit test. Under the null hypothesis that all the mutations occur randomly, all mutations should occur with equal frequency. For our primary test, we tested the general fit of the data to the randomness assumption. This gave us a χ2 value with 5 degrees of freedom. For the second part of our analysis we tested whether the samples contained more G→T transversions vs. all others combined. This gives us a χ2 value with one degree of freedom. We repeated the analysis for G→T transversions, and for G→C transversions, we also asked if in some sample sets, were they significantly underrepresented vs. all others combined using the same strategy we used to test whether G→T mutations were significantly more.
Results
In situ B[a]P Radical Cation Generation
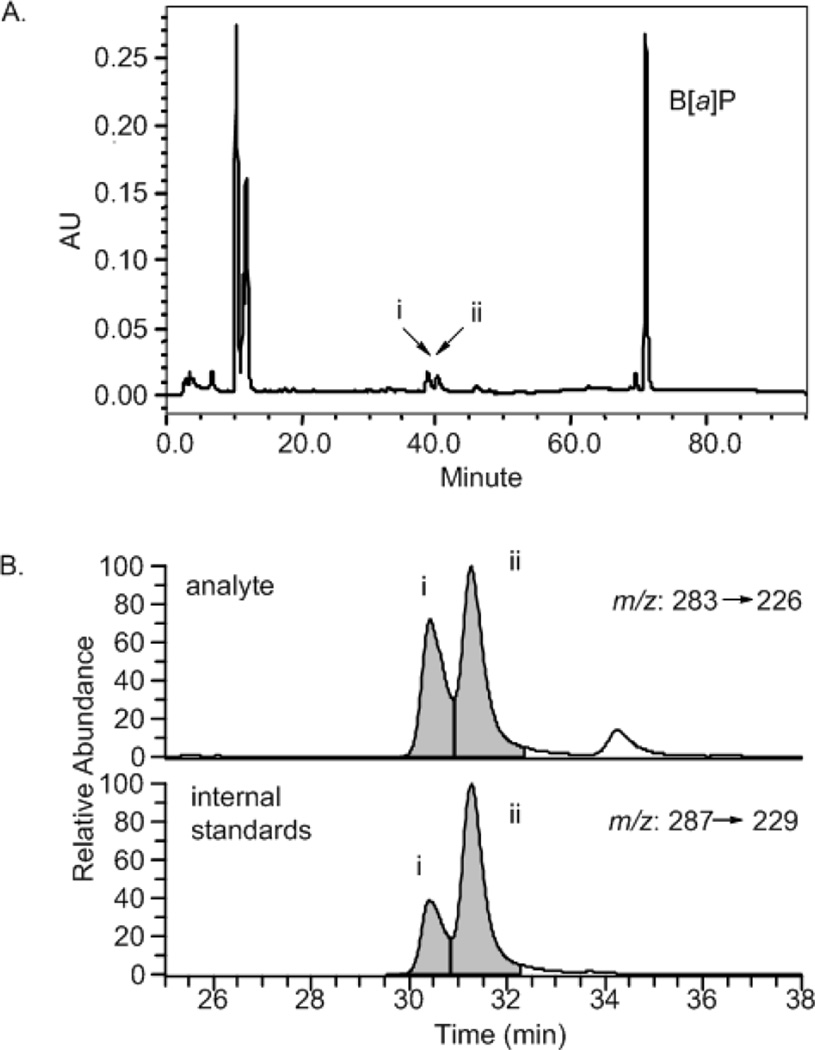
Because B[a]P radical cations have very short half-lives we generated them in situ by modifying a previously published procedure.34,35 To establish optimized conditions for the most efficient formation of radical cations, we compared purified horse radish peroxidase (HRP) with rat liver microsomes and cumene hydroperoxide (CuOOH) with hydrogen peroxide (H2O2). Formation of the radical cations was monitored by the generation of its downstream products, B[a]P-1,6-dione and B[a]P-3,6-dione using HPLC within line UV detection (Figure 1A). We injected B[a]P 1,6 dione, B[a]P 3,6-dione and 6-OH-B[a]P standards individually as well as a cocktail of mixed standards into a Zorbax ODS-C18 column to optimize chromatography and identify product peaks based on their retention times. The product peaks were also identified by either using their corresponding UV spectra (data not shown) or by comparing their retention times, molecular ions, and mass transitions with those of the authentic standards by liquid chromatography/APCI/MS/MS/ (Figure 1B). The chromatographic peaks were quantified either by HPLC-UV method using purified standards to develop a calibration curve and stable isotope dilution liquid chromatography mass spectrometry using the [13C4]-internal standards.24
Figure 1. Characterization and quantification of HRP radical cation reaction products.
(A) HPLC-UV chromatogram of HRP oxidation of B[a]P. i, B[a]P-1,6-dione; ii, B[a]P-3,6-dione. (B) LC-MS chromatogram of B[a]P-1,6-dione and B[a]P-3,6-dione from the HRP reaction. Lower panel shows shows the analysis of 13C-labeled internal standards.
We found that horseradish peroxidase (HRP) catalyzed the one electron oxidation of B[a]P most efficiently and cumene hydropeoxide (CuOOH) was a superior substrate to H2O2. By contrast, rat liver microsomes were not used since they contain P450 enzymes which react with B[a]P to give products such as B[a]PDE thus limiting their ability to study only the radical cation pathway. The microsomes also rapidly degrade plasmids, becuase of high levels of contaminating nucleases. Therefore, we chose CuOOH for its higher oxidizing property and HRP for its specificity. Incubation of B[a]P with 2 mM CuOOH in the presence of 1mg/mL HRP at 37 °C for 2 hours was found to be the most optimal reaction conditions (Figure 1). Incubation of B[a]P with only HRP or only CuOOH under the same reaction conditions did not yield the diones, indicating that the complete system was required for radical cation production (data not shown).
Mutagenesis of p53 with B[a]P Radical Rations
We measured mutagenesis with a yeast reporter system that scores for a functional protein based on p53 transcriptional activation of the yeast ADE2 gene.13, 26, 31 Wild-type p53 plasmid is first exposed to a mutagen in vitro, and then the treated plasmid is transformed in yeast. The assay generates three types of data: frequency, pattern, and spectrum of mutations. From the frequency of red colonies formed, we can analyze the potency of the carcinogen. Analysis of the specific base pair change yields the pattern of mutations, whereas plots of mutations against codon number yield the spectrum of mutations. B[a]P-7,8-dione was previously analyzed in our lab in this assay system; therefore, it was used as a positive control for the experiments.13
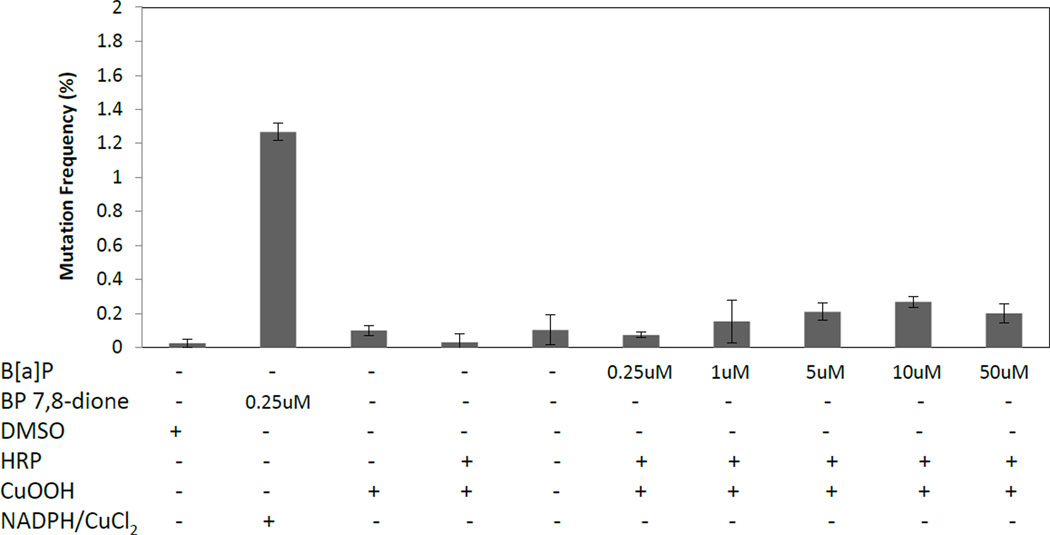
At concentrations as high as 50 µM, very low mutation frequencies (0.2–0.5% red colonies) were observed when the B[a]P radical cations were generated in situ (Figure 2). For comparison 250 nM B[a]P-7,8-dione yielded mutation frequencies of 1.5–2%, under redox cycling conditions. Quantification by HPLC, using standard curves, indicated that typically about 2 µM of B[a]P-1,6-dione was generated in a reaction. Assuming that about 40% of the radical cation is converted into B[a]P 1,6-dione (the rest is converted to other products such as B[a]P-3,6-dione and B[a]P-6,12-dione),36 we calculated the radical cation concentration in a typical reaction to be 3–5 µM (~8% conversion of B[a]P). Similar levels were calculated using stable isotope dilution LC-MS to quantify the B[a]P-1,6-dione and B[a]P-3,6-dione produced in reactions. Using the internal standard ratio method we find that 50 µM B[a]P gave rise to 1.95 µM and .90 µM B[a]P-1,6-dione and B[a]P-3,6-dione respectively. These data suggest that even though a considerable amount of radical cations are formed, they are poorly mutagenic.
Figure 2.
Mutation frequency of p53 due to in situ generated B[a]P radical cations. p53 treated with 250 nM B[a]P-7,8-dione is used as a positive control and treatment with DMSO is used as a negative control. Data is presented as the percentage of red mutant colonies. Incubation conditions are described in Materials and Methods.
Mutational Patterns and spectrums observed with B[a]P Radical Cations
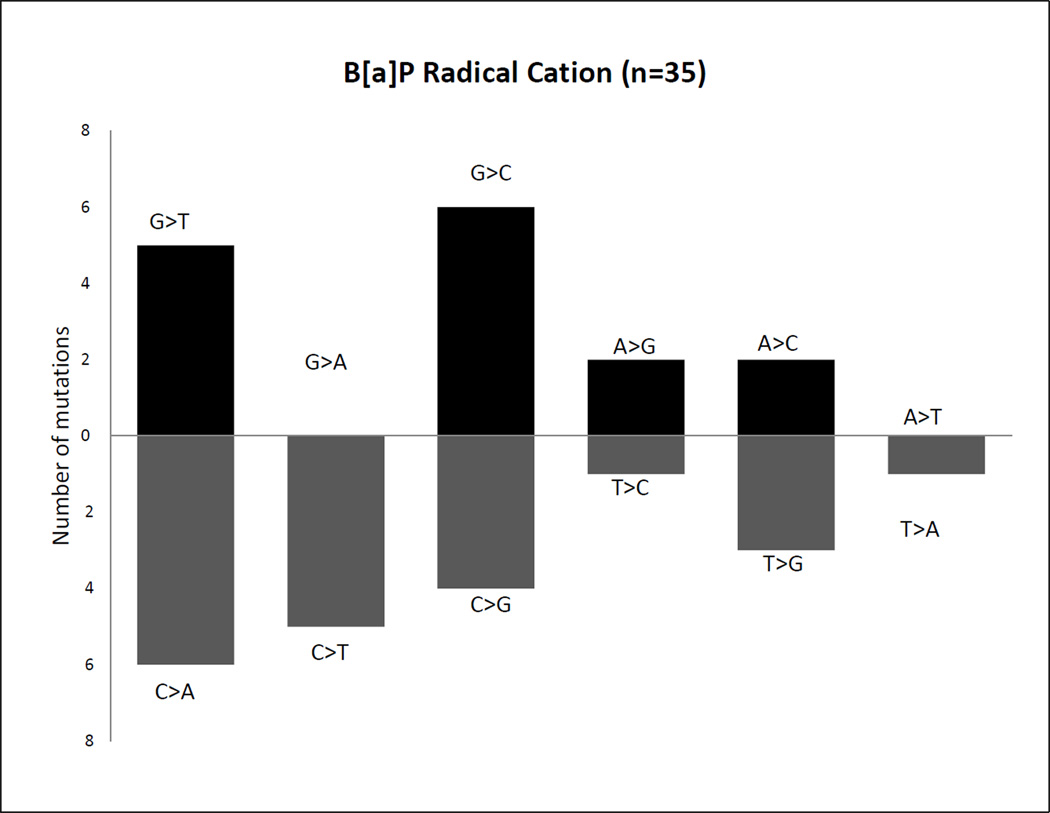
To analyze the pattern and spectrum of the mutation, we rescued the plasmid DNA from red colonies and then sequenced the DNA binding domain of p53 (Figure 3 and table S1). For comparison and detection of background mutations, DNA was also isolated from yeast treated with CuOOH and HRP in the absence of B[a]P (data not shown). In most instances (~90%), only single point mutations were detected in the DNA binding domain. The changes in the coding sequence are plotted with complementary base changes below the base line. For the B[a]P radical cation, the pattern was dominated by G>T transversions (or its complement C>A) (31%; p = 0.019), G>C transversions (or its complement C>G) (29%; p =.06) and 14% G >A transversions (Figure 3). Although the predominant mutations were G>T transversions as seen in lung cancer, there was a trend towards an increase in G>C transversions. There was no significant strand bias. The mutation pattern obtained from B[a]P radical cations were thus different from B[a]P-7,8-dione. It was closer to what we observed with (+)-anti-B[a]PDE,14 which also had a substantial number of G>C transversions. It is possible that the DNA- lesion that causes mutations by the radical cation pathway may be a bulky adduct and not an apurinic site.
Figure 3.
p53 mutational pattern induced by the radical cation of B[a]P. The mutations observed in red colonies following treatment of wild-type p53 with in situ generated B[a]P radical cations. The 12 possible changes are reduced to 6 when complimentary changes are plotted together because the original base lesion cannot be determined from the mutant sequence.
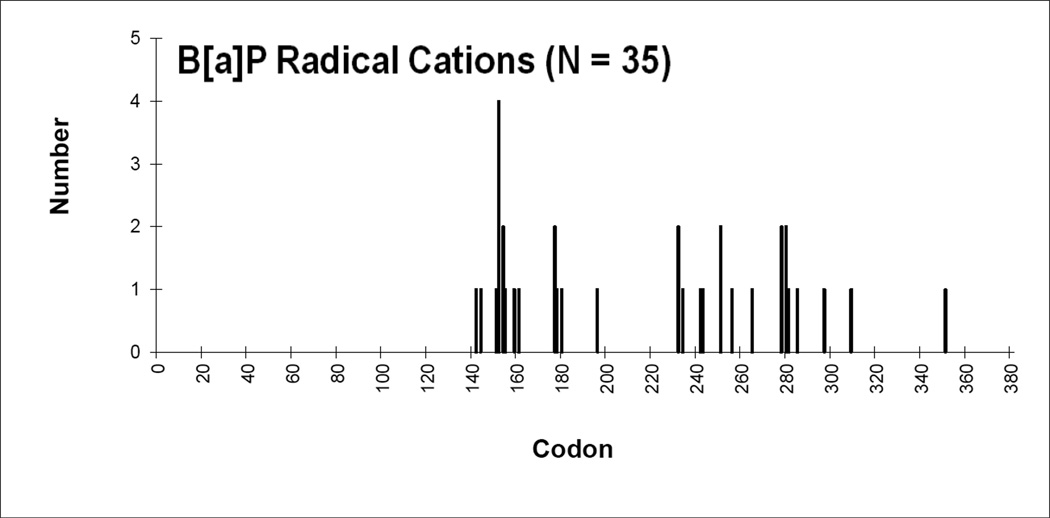
To obtain the mutational spectrum for radical cations, single point mutations were plotted against codon number (Figure 4). The region sequenced was from codons 126 to 339 which encompass most of the DNA binding domain (amino acids 102 to 292). An analysis of IARC database identified 23 codons that account for ~50% of the reported mutations in lung cancers. If the 23 hotspots were assumed to be randomly mutated, then they would be predicted to hit 23/213 or 10.8% of the time. We did not find any mutations on the hotspot codons observed in lung cancer with B[a]P radical cations. The mutations were randomly distributed in the DNA binding domain. About 91% of the mutations were missense mutations. Only one nonsense mutation (3%) and 2 (6%) silent mutations were found. In conclusion, we could not find significant similarities between the pattern and spectrum of B[a]P radical cations to the pattern and spectrum found in lung cancer.
Figure 4.
Mutation spectra induced in p53 by B[a]P radical cations. The occurrence of point mutations are plotted against codon number for the B[a]P radical cation treated p53 plasmid.
Mutagenesis of p53 with B[a]P-1,6 and B[a]P-3,6 Diones
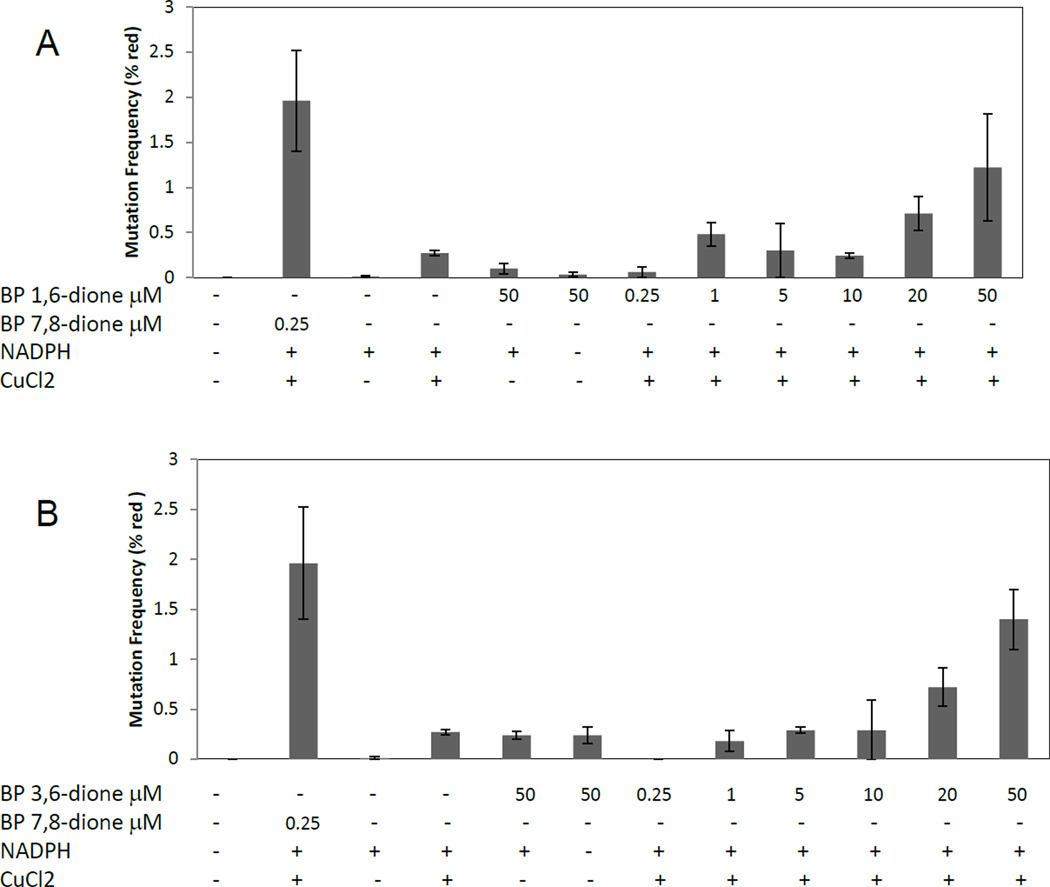
Since B[a]P radical cations can be rapidly converted to B[a]P-1,6 and B[a]P-3,6 dione, this represents an alternative mechanism by which radical cations may be mutagenic. To address the mutagenicity of these quinone metabolites, we compared them to the AKR derived quinone, B[a]P-7,8-dione.37 B[a]P-1,6 and B[a]P-3,6 dione were poorly mutagenic even at concentrations up to 50 µM (Figure 5A and 5B). Next, we tested them under redox-cycling conditions by including NADPH and CuCl2. A concentration dependent mutation of p53 was seen both for both B[a]P-1,6-dione and B[a]P 3,6-dione under redox-cycling conditions using NADPH and CuCl2. Very few mutant colonies (0.2–0.5%) were detected when the plasmid was incubated with quinones alone or the quinones with NADPH. For both radical cation derived quinones, concentrations up to 50 µM only yielded 1.2–1.4% mutation frequency, whereas only 250 nM B[a]P-7,8-dione yielded ~2% mutational frequency.
Figure 5.
Mutation frequency of p53 treated with radical cation derived quinones. p53 treated with 250nM B[a]P-7,8-dione is used as a positive control treatment with DMSO is used as a negative control. Data are presented as the percentage of mutation frequency vs the treatment indicated. (A) Dose response with B[a]P-1,6-dione. (B) Dose response using B[a]P-3,6-dione.
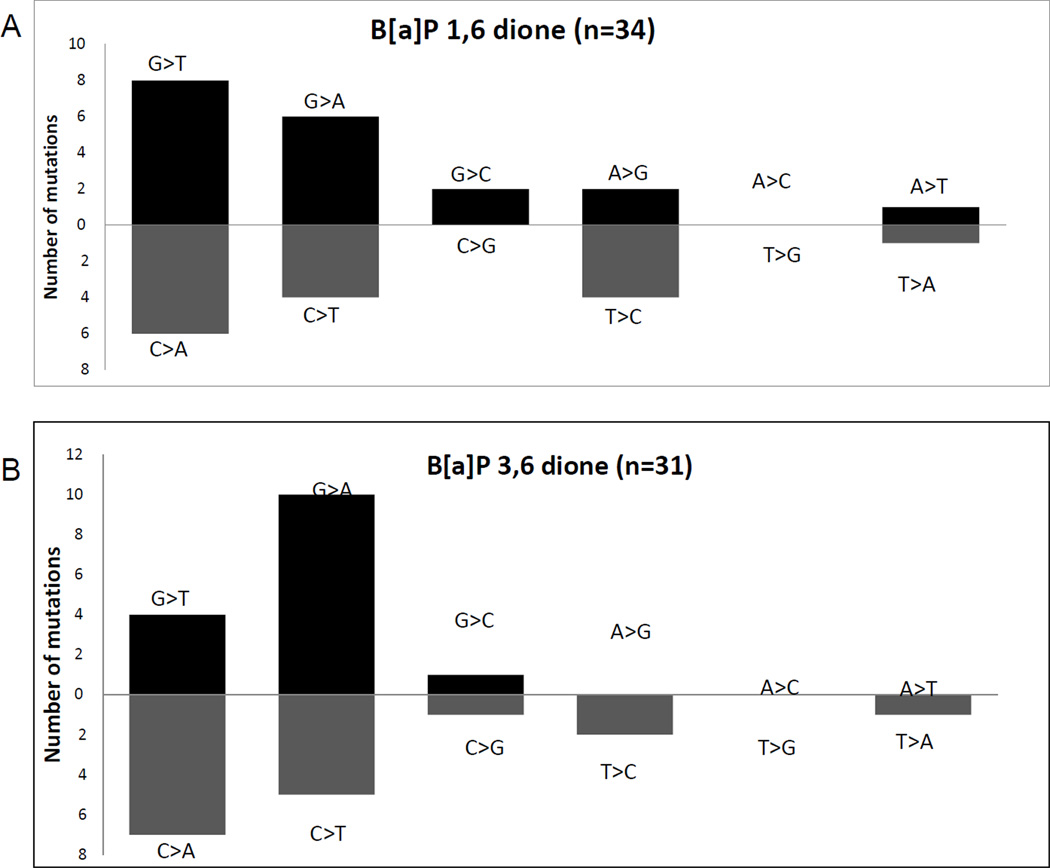
The mutation pattern of the B[a]P-1,6 and B[a]P-3,6 diones were both dominated by G>T transversions and G>A transitions (Figure 6A and 6B and tables S1 and S2), similar to what is seen with B[a]P-7,8-dione under oxidizing conditions. Although we usually observe more G>T transversions, G>A transitions are common and in some experiments they predominate. Of note, unlike radical cation mutagenesis, both B[a]P 1,6 and 3,6 diones did not yield significant numbers of G>C transversions consistent with different mechanism of mutagenesis. The spectrum of mutations was essentially random. This indicated that like B[a]P-7,8-dione, B[a]P-1,6 and B[a]P-3,6 dione were damaging DNA through a redox mechanism, but were significantly less potent than B[a]P-7,8-dione.
Figure 6.
Mutation patterns with (A) B[a]P-1,6-dione and (B) B[a]P-3,6-dione.
DNA damage and NADPH oxidation
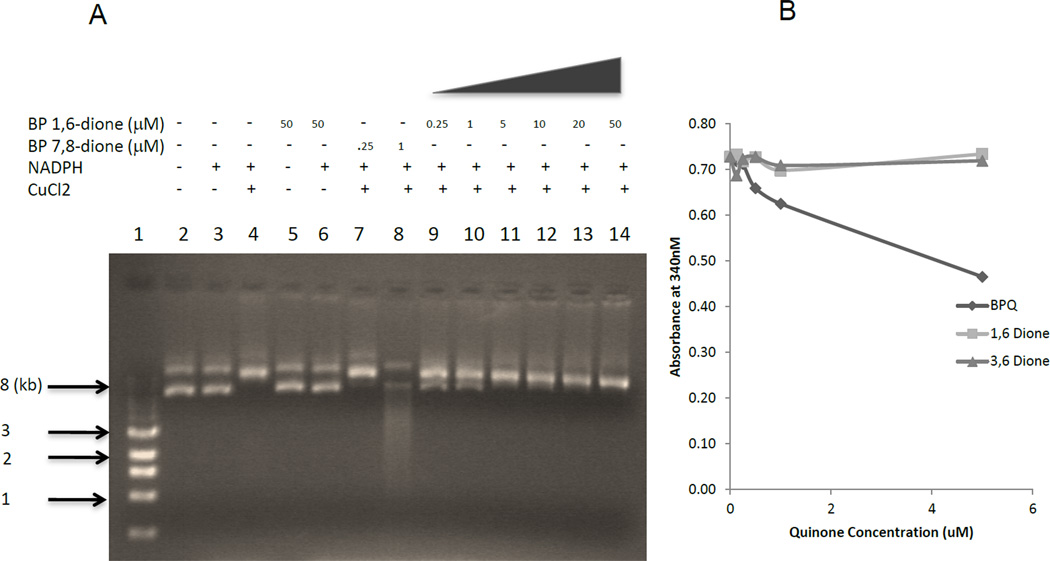
Another way to measure DNA damage is to measure DNA strand scission. B[a]P-7,8 dione damages and eventually cleaves DNA in the presence of NADPH and CuCl2 through redox-cycling and ROS generation.13 We incubated p53 plasmid with the diones alone or in the presence of either NADPH, or NADPH and CuCl2 and analyzed the products using agarose gel electrophoresis (Figure 7A and supplementary Figure 1). As controls, p53 plasmid DNA was also treated with solvent (DMSO), NADPH, and NADPH plus CuCl2 in absence of quinones. B[a]P-7,8-dione was used as a control at concentrations of 250 nM and 1 µM. As previously reported, we saw substantial degradation of the DNA when treated with 1 µM B[a]P-7,8-dione. In comparison, both B[a]P-1,6 and 3,6 diones showed less DNA degradation even up to the concentrations of 50 µM. The DNA strand scission assay correlates with the poor mutagenesis data generated with higher concentrations of these diones. The weak mutagenesis and strand scission by the radical cation derived quinones suggested that they may redox cycle inefficiently when compared to PAH o-quinones. To test this we measured NADPH oxidation Spectrophotometrically by measuring the decrease in absorbance at 340 nm. We found that redox cycling was inefficient compared to B[a]P-7,8-dione (Figure 7B).
Figure 7.
DNA strand scission and NADPH consumption by PAH quinones (A) p53 cDNA plasmid was incubated with vehicle (DMSO) alone, (lane 1); 0.25 µM and 1 µM B[a]P-7,8-dione in the presence of 10 mM NADPH and 100 µM CuCl2 for 2h at 37 °C (lanes 7 and 8); 0.25 µM, 1 µM, 5 µM, 10 µM, 20 µM and 50 µM B[a]P-1,6-dione in the presence of 10 mM NADPH and 100 µM CuCl2 for 2h at 37 °C (lanes 9–14). The treated p53 was directly analyzed by electrophoresis on a 1% agarose gel. (B) NADPH oxidation was analyzed by absorbance at 340 nm.
Discussion
Radical cations can damage DNA and cause mutations through two mechanisms (1) depurination and (2) redox cycling of their quinone products. We addressed both mechanisms in this study, and compared them to the AKR-derived o-quinone B[a]P-7,8-dione. The B[a]P radical cations were much less mutagenic than B[a]P-7,8-dione. We calculated that 50 µM of B[a]P was converted to 2–5 µM of radical cation, and was a weak mutagen, while 250 nM of B[a]P-7,8-dione was highly mutagenic. In our assay system, the radical cation derived quinones were also less mutagenic than B[a]P-7,8-dione, although their mutagenicity was comparable to levels reported for them in Ames tests.37 It is likely that radical cation derived quinones were less efficient at redox cycling than B[a]P-7,8-dione because they were considerably less active than B[a]P-7,8-dione in causing DNA strand scission assays and NADPH oxidation.
Cavalieri et al provided evidence that PAH radical cations bind to N7-, or C8- positions of guanine and adenine to form DNA adducts both in vivo and in vitro.38–40 These DNA adducts are unstable because of the presence of a labile N-glycosidic bond, resulting in spontaneous depurination to yield apurinic sites. The formation of DNA adducts from the radical cation pathway were observed in vivo in mouse skin and rat mammary gland. Several adducts were induced after topical application of B[a]P for 4h on mouse skin.18 Depurinating DNA adducts B[a]P-6-C8Gua (34%), B[a]P-6-N7-Ade (22%), B[a]P-C6-N7-Gua (10%), B[a]PDE-C10-N7-Ade (3%) and B[a]PDE-C10-N7-Gua (2%) accounted for 71% of the total adducts produced, 66% of which originated from the radical cation pathway. The DNA adduct profile was also determined after injection of B[a]P into rat mammary gland.41 Two unstable depurinating adducts identified were, B[a]P-6-C8-Gua and B[a]P-6-N7Ade, whereas the stable adduct was essentially B[a]PDE-10-N2−dGuo (64%). From these observations they proposed that the formation and inefficient repair of these apurinic sites are primarily responsible for the carcinogenic and mutagenic effects of PAHs such as B[a]P, DMBA or DB[a,l]P both in vitro and in mouse skin. However, another laboratory was not able to find apurinic sites in HL-60 cells treated with B[a]P, despite high levels of P450 peroxidases in the cells.42, 43
The propensity to remove an electron from an already electron deficient carbon in a PAH is related to its ionization potential. In vitro studies with peroxidases led to the suggestions that PAHs with ionization potentials lower than 7.34 ev may be activated in biological systems by one-electron oxidation. Thus for B[a]P the electron is removed from C6. One-electron oxidation of PAHs was investigated chemically by treating PAH with iodine, and manganese acetate; electrochemically, and by the use of either horseradish peroxidase plus H2O2 or by rat liver microsomes plus CuOOH. Recently a more efficient chemical method44 reported a conversion of 50% B[a]P into its radical cation. However, these studies did not find any correlation between the ionization potentials of different PAH and their mutagenic potency, leading the authors to conclude that radical cation formation does not correlate with mutagenesis.
The second mechanism by which radical cations can damage DNA is through the redox cycling of their quinone products B[a]P-1,6-dione and B[a]P-3,6-dione. Indeed, these have been shown to be mutagenic in Ames assays and generate ROS in H358 cells.12, 37 While we did find 1,6-dione and 3,6-dione mutagenic at levels comparable to those reported by Chesis et. al., they were considerable less mutagenic than B[a]P-7,8-dione. We speculate that the reduced level of mutagenicity is because the carbonyl groups are on different aromatic rings. Highly redox active quinones, such as B[a]P-7,8-dione, naphthalene 1,2-dione, and 9,10 anthraquinone are characterized by the presence of adjacent carbonyl groups in an ortho-arrangement (Scheme 1). We note that the assays in our study were done with CuCl2 catalyzed redox cycling in vitro, while in cells, the inefficient redox cycling by 1, 6-dione and 3,6-dione is likely to be enhanced by enzymes such as NAD(P)H:quinone oxidoreductase (NQO1) and carbonyl reductases (CBR1 and CBR3). Therefore, we have not ruled out the possibility that the B[a]P-1,6-dione and B[a]P-3,6-dione might be more redox active in cells than in our in vitro assays.
Depurinating DNA adducts that arise from radical cations are unstable and rapidly decompose to yield apurinic sites.. Abasic sites, when unrepaired, stall DNA and RNA polymerases, resulting in a disruption of DNA replication and gene transcription.45 An unrepaired apurinic site can be read through by translesion DNA synthesis, but this is error prone because DNA polymerases will often insert a base which is not complementary to the one lost by depurination. Thus, abasic sites produced by an adduct depurination can cause mutations by substitutions of a single base. In mammalian cells, the different DNA bases are inserted in the following order: A (33%) > C (28%) > T (26%) > G (12%).46 In yeast, the base substitutions at apurinic sites varies, with some studies suggesting that guanines are inserted opposite the lesion this would cause A>C and G>C transversions (G-rule), while other studies suggest that adenines are inserted yielding A>T and G>T transversions (A-rule).45 Most of the mutations we observed were at guanine residues suggesting that adducts were forming at guanine bases. The mutational patterns that we observed were predominantly G>T and G>C consistent with both the A-rule and G-rule. We did not observe many G>C transversions with oxidative damage in this study or in our other studies. The spectrum of mutations were random throughout the DNA binding domain, as we have previously observed in the system; if the additional filter of selecting for dominant mutations is used, then spectra more closely match those seen in lung cancer.27, 29, 47
Conclusion
In an in vitro yeast reporter assay, the Aldo ketose reductase derived quinone B[a]P 7,8 dione is at least 200 times more mutagenic than B[a]P radical cations produced in situ. Additionally, the radical cation-derived quinones 1,6-dione and 3,6-dione are less efficient at redox cycling than B[a]P-7,8-dione, although redox cycling may be more efficient in cells where NQO1 and AKRs will assist reduction.12
Supplementary Material
Acknowledgements
We would like to extend our gratitude to Dr. Mary Kushman and Dr. Li Zhang for their kind help with the HPLC experiments and Junseok Moon for help with mutagenesis.
Funding Support
This work was supported by grant R01 GM48241 and R01 NIEHS to J.F and P30 ES013508. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH.
Abbreviations
- PAH
Polycyclic Aromatic Hydrocarbons
- (B[a]P)
Benzo[a]pyrene
- (±)-anti-BPDE
(±)anti-benzo[a]pyrene 7,8-diol 9,10-epoxide
- BP-7,8-diol
B[a]P-7,8-trans-dihydrodiol
- BPQ
benzo[a]pyrene-7,8-dione
- CuOOH
Cumene hydroperoxide
Footnotes
Supporting information
Supplementary tables 1–3 are available free of charge at http://pubs.acs.org
References
- 1.General S. How tobacco smoke causes disease. 2010 [Google Scholar]
- 2.Roe FJ. Role of 3,4-benzopyrene in carcinogenesis by tobacco smoke condensate. Nature. 1962;194:1089–1090. doi: 10.1038/1941089a0. [DOI] [PubMed] [Google Scholar]
- 3.Hecht SS. Tobacco smoke carcinogens and lung cancer. J. Natl. Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 4.Gelboin HV. Benzo[a]pyrene metabolism, activation and carcinogenesis: Role and regulation of mixed function oxidases and related enzymes. Physiol. Rev. 1980;60:1107–1166. doi: 10.1152/physrev.1980.60.4.1107. [DOI] [PubMed] [Google Scholar]
- 5.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons. G.H.A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 6.Cavalieri EL, Rogan EG. Central role of radical cations in metabolic activation of polycyclic aromatic hydrocarbons. Xenobiotica. 1995;25:677–688. doi: 10.3109/00498259509061885. [DOI] [PubMed] [Google Scholar]
- 7.Penning TM, Burczynski ME, Hung CF, McCoull KD, Palackal NT, Tsuruda LS. Dihydrodiol dehydrogenases and polycyclic aromatic hydrocarbon activation: generation of reactive and redox-active o-quinones. Chem. Res Toxicol. 1999;12:1–18. doi: 10.1021/tx980143n. [DOI] [PubMed] [Google Scholar]
- 8.Hussain SP, Amstad P, Raja K, Sawyer M, Hofseth L, Shields P, Hewer A, Phillips DH, Ryberg D, Huagen A, Harris CC. Mutability of p53 hotspot codons to benzo[a]pyrene diol epoxide (BPDE) and the frequency of p53 mutations in nontumorous human lung. Cancer Res. 2001;61:6350–6355. [PubMed] [Google Scholar]
- 9.Shou M, Harvey RG, Penning TM. Reactivity of benzo[a]pyrene-7,8-dione with DNA. Evidence for the formation of deoxyguanosine adducts. Carcinogenesis. 1993;14:475–482. doi: 10.1093/carcin/14.3.475. [DOI] [PubMed] [Google Scholar]
- 10.Balu N, Padgett WT, Lambert GR, Swank AE, Richard AM, Nesnow S. Identification and characterization of novel stable deoxyguanosine and deoxyadenosine adducts of benzo[a]pyrene-7,8-quinone from reactions at physiological pH. Chem. Res. Toxicol. 2004;17(6):827–838. doi: 10.1021/tx034207s. [DOI] [PubMed] [Google Scholar]
- 11.Penning TM, Ohnishi ST, Ohnishi T, Harvey RG. Generation of reactive oxygen species during the enzymatic oxidation of polycyclic aromatic hydrocarbon trans-dihydrodiols catalyzed by dihydrodiol dehydrogenase. Chem. Res. Toxicol. 1996;9:84–92. doi: 10.1021/tx950055s. [DOI] [PubMed] [Google Scholar]
- 12.Shultz CA, Quinn AM, Park JH, Harvey RG, Bolton JL, Maser E, Penning TM. Specificity of Human Aldo-Keto Reductases, NAD(P)H:Quinone Oxidoreductase, and Carbonyl Reductases to Redox-Cycle Polycyclic Aromatic Hydrocarbon Diones and 4-Hydroxyequilenin-o-quinone. Chem. Res. Toxicol. 2011 doi: 10.1021/tx200294c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu D, Berlin JA, Penning TM, Field J. Reactive oxygen species generated by PAH o-quinones cause change-in-function mutations in p53. Chem Res. Toxicol. 2002;15(6):832–842. doi: 10.1021/tx010177m. [DOI] [PubMed] [Google Scholar]
- 14.Shen YM, Troxel AB, Vedantam S, Penning TM, Field J. Comparison of p53 Mutations Induced by PAH o-Quinones with Those Caused by anti-Benzo[a]pyrene Diol Epoxide in Vitro: Role of Reactive Oxygen and Biological Selection. Chem. Res. Toxicol. 2006;19(11):1441–1450. doi: 10.1021/tx0601206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavalieri EL, Rogan EG, Cremonesi P, Devanesan PD. Radical cations as precursors in the metabolic formation of quinones from benzo[a]pyrene and 6-fluorobenzo[a]pyrene. Fluoro substitution as a probe for one-electron oxidation in aromatic substrates. Biochem. Pharmacol. 1988;37(11):2173–2182. doi: 10.1016/0006-2952(88)90578-3. [DOI] [PubMed] [Google Scholar]
- 16.Rogan EG, Cavalieri EL, Tibbels SR, Cremonesi P, Warner CD, Nagel DL, Tomer KB, Cerny RL, Gross ML. Synthesis and identification of benzo[a]pyrene-guanine nucleoside adducts formed by electrochemical oxidation and by horseradish peroxidase catalyzed reaction of benzo[a]pyrene with DNA. J. Am. Chem. Soc. 1988;110(12):4023–4029. [Google Scholar]
- 17.Devanesan PD, RamaKrishna NVS, Todorovic R, Rogan EG, Cavalieri EL, Jeong H, Jankowiak R, Small GJ. Idenitification and quantitation of Benzo[a]pyrene-DNA adducts formed by rat liver microsomes in vitro. Chem. Res. Toxicol. 1992;5:302–309. doi: 10.1021/tx00026a024. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Devanesan PD, Higginbotham S, Ariese F, Jankowiak R, Small GJ, Rogan EG, Cavalieri EL. Expanded analysis of benzo[a]pyrene-DNA adducts formed in vitro and in mouse skin: their significance in tumor initiation. Chem. Res. Toxicol. 1996:897–903. doi: 10.1021/tx960004a. [DOI] [PubMed] [Google Scholar]
- 19.Devanesan PD, Higginbotham S, Ariese F, Jankowiak R, Suh M, Small GJ, Cavalieri EL, Rogan EG. Depurinating and stable benzo[a]pyrene-DNA adducts formed in isolated rat liver nuclei. Chem. Res. Toxicol. 1996;9(7):1113–1116. doi: 10.1021/tx9600513. [DOI] [PubMed] [Google Scholar]
- 20.RamaKrishna NV, Gao F, Padmavathi NS, Cavalieri EL, Rogan EG, Cerny RL, Gross ML. Model adducts of benzo[a]pyrene and nucleosides formed from its radical cation and diol epoxide. Chem. Res. Toxicol. 1992;5(2):293–302. doi: 10.1021/tx00026a023. [DOI] [PubMed] [Google Scholar]
- 21.Beroud C, Soussi T. p53 gene mutation: software and database. Nucleic Acids Res. 1998;26:200–204. doi: 10.1093/nar/26.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 23.Toyooka S, Tsuda T, Gazdar AF. The TP53 gene, tobacco exposure, and lung cancer. Hum. Mutat. 2003;21(3):229–239. doi: 10.1002/humu.10177. [DOI] [PubMed] [Google Scholar]
- 24.Lu D, Harvey RG, Blair IA, Penning TM. Quantitation of Benzo[a]pyrene Metabolic Profiles in Human Bronchoalveolar (H358) Cells by Stable Isotope Dilution Liquid Chromatography-Atmospheric Pressure Chemical Ionization Mass Spectrometry. Chem. Res. Toxicol. 2011;24(11):1905–1914. doi: 10.1021/tx2002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flaman JM, Frebourg T, Moreau V, Charbonnier F, Martin C, Chappuis P, Sappino AP, Limacher JM, Bron L, Benhattar J, Tada M, Van Meir EG, Estreicher A, Iggo RD. A simple p53 functional asay for screening cell lines, blood, and tumors. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3963–3967. doi: 10.1073/pnas.92.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu D, Penning TM, Field JM. Benzo[a]pyrene-7,8-dione is more mutagenic than anti-BPDE on p53 and is dependent upon the generation of ROS. Polycyclic Aromatic Compounds. 2002;22:881–891. [Google Scholar]
- 27.Park JH, Gelhaus S, Vedantam S, Oliva AL, Batra A, Blair IA, Troxel AB, Field J, Penning TM. The pattern of p53 mutations caused by PAH o-quinones is driven by 8-oxo-dGuo formation while the spectrum of mutations is determined by biological selection for dominance. Chem. Res. Toxicol. 2008;21(5):1039–1049. doi: 10.1021/tx700404a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park JH, Troxel AB, Harvey RG, Penning TM. Polycyclic Aromatic Hydrocarbon (PAH) o-Quinones Produced by the Aldo-Keto-Reductases (AKRs) Generate Abasic Sites, Oxidized Pyrimidines, and 8-Oxo-dGuo via Reactive Oxygen Species. Chem Res. Toxicol. 2006;19(5):719–728. doi: 10.1021/tx0600245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brachmann RK, Vidal M, Boeke JD. Dominant-negative p53 mutations selected in yeast hit cancer hot spots. Proc. Natl. Acad. Sci. U.S.A. 1996;93(9):4091–4095. doi: 10.1073/pnas.93.9.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu A, Xu D, Lu D, Penning TM, Blair IA, Harvey RG. Synthesis of 13C4labelled oxidized metabolites of the carcinogenic polycyclic aromatic hydrocarbon Benzoapyrene. Tetrahedron. 2012 doi: 10.1016/j.tet.2012.05.130. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishioka C, Frebourg T, Yan Y-X, Vidal M, Friend SH, Schmidt S, Iggo R. Screening patients for heterozygous p53 mutations using a functional assay in yeast. Nat. Genet. 1993;5:124–129. doi: 10.1038/ng1093-124. [DOI] [PubMed] [Google Scholar]
- 32.Guthrie C, Fink GR. Methods in Enzymology. Vol. 194. New York: Academic Press Inc.; 1991. Guide to yeast genetics and molecular biology. [PubMed] [Google Scholar]
- 33.Yoon JH, Lee CS, Pfeifer GP. Simulated sunlight and benzo[a]pyrene diol epoxide induced mutagenesis in the human p53 gene evaluated by the yeast functional assay: lack of correspondence to tumor mutation spectra. Carcinogenesis. 2003;24(1):113–119. doi: 10.1093/carcin/24.1.113. [DOI] [PubMed] [Google Scholar]
- 34.Cavalieri EL, Devanesan PD, Rogan EG. Radical cations in the horseradish peroxidase and prostaglandin H synthase mediated metabolism and binding of benzo[a]pyrene to deoxyribonucleic acid. Biochem. Pharmacol. 1988;37(11):2183–2187. doi: 10.1016/0006-2952(88)90579-5. [DOI] [PubMed] [Google Scholar]
- 35.Rogan EG, Katomski PA, Roth RW, Cavalieri EL. Horseradish peroxidase/hydrogen peroxide-catalyzed binding of aromatic hydrocarbons to DNA. J. Biol. Chem. 1979;254(15):7055–7059. [PubMed] [Google Scholar]
- 36.Lesko S, Caspary W, Lorentzen R, Ts'o POP. Enzymic Formation of 6-Oxobenzo[a]pyrene Radical in Rat Liver Homogenates from Carcinogenic Benzo[a]pyrene. Biochemistry. 1975;14(18):7. [Google Scholar]
- 37.Chesis PL, Levin DE, Smith MT, Ernster L, Ames BN. Mutagenicity of quinones: pathways of metabolic activation and detoxification. Pro. Natl. Acad. Sci. U.S.A. 1984;81(6):1696–1700. doi: 10.1073/pnas.81.6.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cavalieri EL, Rogan EG. Radical cations in aromatic hydrocarbon carcinogenesis. Free Radical Res. 1990;11(1–3):77–87. doi: 10.3109/10715769009109670. [DOI] [PubMed] [Google Scholar]
- 39.Cavalieri EL, Rogan EG, Devanesan PD, Cremonesi P, Cerny RL, Michael L, Gross ML, Bodell WJ. Binding of benzo[a]pyrene to DNA by cytochrome P 450 catalyzed one-electron oxidation in rat liver microsomes and nuclei. Biochemistry. 1990;29(20):8. doi: 10.1021/bi00472a011. [DOI] [PubMed] [Google Scholar]
- 40.Chakravarti D, Pelling JC, Cavalieri EL, Rogan EG. Relating aromatic hydrocarbon-induced DNA adducts and c-H-ras mutations in mouse skin papillomas: the role of apurinic sites. Proc. Natl. Acad. Sci. U.S.A. 1995:10422–10426. doi: 10.1073/pnas.92.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Todorovic R, Ariese F, Devanesan P, Jankowiak R, Small GJ, Rogan E, Cavalieri E. Determination of benzo[a]pyrene- and 7,12-dimethylbenz[a]anthracene-DNA adducts formed in rat mammary glands. Chem. Res. Toxicol. 1997;10(9):941–947. doi: 10.1021/tx970003y. [DOI] [PubMed] [Google Scholar]
- 42.Melendez-Colon VJ, Luch A, Seidel A, Baird WM. Comparison of cytochrome P450- and peroxidase-dependent metabolic activation of the potent carcinogen dibenzo[a,l]pyrene in human cell lines: formation of stable DNA adducts and absence of a detectable increase in apurinic sites. Cancer Res. 1999;59(7):1412–1416. [PubMed] [Google Scholar]
- 43.Melendez-Colon VJ, Luch A, Seidel A, Baird WM. Formation of stable DNA adducts and apurinic sites upon metabolic activation of bay and fjord region polycyclic aromatic hydrocarbons in human cell cultures. Chem. Res. Toxicol. 2000;13(1):10–17. doi: 10.1021/tx9802724. [DOI] [PubMed] [Google Scholar]
- 44.Lehner AF, Horn J, Flesher JW. Formation of radical cations in a model for the metabolism of aromatic hydrocarbons. Biochem. Biophys. Res. Commun. 2004;322(3):1018–1023. doi: 10.1016/j.bbrc.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 45.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair. 2004;3(1):1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Gentil A, Cabral-Neto JB, Mariage-Samson R, Margot A, Imbach JL, Rayner B, Sarasin A. Mutagenicity of a unique apurinic/apyrimidinic site in mammalian cells. J. Mol. Biol. 1992;227(4):981–984. doi: 10.1016/0022-2836(92)90513-j. [DOI] [PubMed] [Google Scholar]
- 47.Rodin SN, Rodin AS. Human lung cancer and p53: the interplay between mutagenesis and selection. Proc. Natl. Acad. Sci. U.S.A. 2000;97(22):12244–12249. doi: 10.1073/pnas.180320897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.