Abstract
miRNA-22 was previously reported to be a tumor suppressor. The aim of this study was to explore the expression and function of miRNA-22 in esophageal squamous cell carcinoma (ESCC). Expression of miRNA-22 in 100 ESCC tissues was examined by q-PCR. The correlation between miRNA-22 level and clinicopathological features was analyzed using SPSS16.0 statistical software. Moreover, the effect of miRNA-22 expression on radiosensitivity of ESCC cells was examined. miRNA-22 expression decreased in ESCC tissues, and statistical analyses showed that the expression of miRNA-22 was associated with the stage of clinical classification. No correlation was found between miRNA-22 expression and the overall survival of ESCC patients. However, significant positive correlation was found between miRNA-22 expression and the survival of patients who received radiotherapy (P < 0.05). Increased expression of miRNA-22 sensitized ESCC cells to γ-ray radiation and promoted the apoptosis of ESCC cells induced by γ-ray radiation. Increased expression level of miRNA-22 had effects on Rad51 expression after irradiation. These results demonstrate for the first time that decreased miRNA-22 expression correlates with increased radiotherapy resistance of ESCC, and that this effect is mediated, at least in part, by the Rad51 pathway.
Keywords: miRNA-22, ESCC, γ-ray, radiosensitivity, Rad51
INTRODUCTION
Esophageal carcinoma (EC) is a highly malignant disease with an overall 5-year survival rate of less than 10%. The worldwide incidence of EC is increasing, particularly for adenocarcinoma located in the lower esophagus and gastroesophageal junction. In contrast to that of western countries, esophageal squamous cell carcinoma (ESCC) is the most prevalent type of EC in China. Radiotherapy has the widest application to ESCC patients, and plays a central role in contemporary treatment concepts [1, 2]. Unfortunately, local failure remains a major concern, with persistent or recurrent disease being reported in around 40–60% of patients [3, 4]. So, the radiosensitization is a promising approach to improving the therapeutic ratio for irradiation.
miRNAs are conserved 19–22 nucleotide non-coding RNAs. They repress protein expression at the post-transcriptional level, mainly by annealing with the 3′ UTR of the target mRNA and interfering with its translation and/or stability [5]. The precise number of miRNA genes in the human genome is still unknown, but estimates of the total number of miRNA precursors are as high as 25 000. These molecules are thought to regulate approximately 30% of the genes in the human genome [6, 7]. Studies have shown that some miRNAs regulate cellular differentiation, proliferation and apoptotic processes that are important in cancer aggravation. miRNA profiling across multiple human cancers has suggested that miRNAs are potential indicators of the developmental origin of poorly differentiated cancers. In the present study, we found that miRNA-22 expression decreased in ESCC tissues, and that its expression was positively associated with the survival of patients who received radiotherapy. In addition, increased miRNA-22 expression promoted radiosensitivity of ESCC cells.
MATERIALS AND METHODS
Tissue specimens
One hundred fresh tissue samples, which contained ESCC and the adjacent histologically normal tissue, were procured from surgical resection specimens collected by the Department of Tumor Medicine, Shandong Provincial Chest Hospital from 2001 to 2007. Primary tumor regions and the corresponding histologically normal tissues from the same patients were separated by experienced pathologists and immediately stored at –70°C until use. None of the patients received treatment before surgery, and all signed informed consent forms for sample collection. Of these 100 patients, 58 received concurrent chemotherapy with radiation therapy after surgery. Radiation therapy was administered in daily divided doses with linear accelerators, and all patients received thoracic irradiation with daily fraction doses of 2.0 Gy for a total dose 50 Gy. The chemotherapy program was Cisplatin (CDDP) + 5-fluorouracil (5-FU). The other 42 patients were treated with chemotherapy only. Use of patient samples of tumors and adjacent histologically normal tissues was approved by the institutional Ethics Committee.
RNA extraction
Total RNA was extracted from ESCC tissue and the corresponding normal tissue using the Absolutely RNA™ Miniprep kit (Stratagene, Santa Clara, CA) according to the manufacturer's instructions. RNA quantification was performed with a DUVR 800 UV/Vis Spectrophotometer (Beckman Coulter, Fullerton, CA).
Quantitative reverse transcription-polymerase chain reaction
TaqMan miRNA assays (ABI PRISM, Carisbad, CA) were used to detect the expression levels of mature miRNA-22. For the reverse transcription (RT) reactions, 10 ng of total RNA was used in each reaction and mixed with the RT primer. RT reactions were performed at 16°C for 30 min, 42°C for 30 min, 85°C for 5 min and then maintained at 4°C. Following the RT reactions, 1.5 µl of cDNA was used for a polymerase chain reaction (PCR) using 2 µl of the TaqMan primers. The PCR was conducted at 95°C for 10 min followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec in an ABI 7500 real-time PCR system. The real-time PCR results were analyzed and expressed as the relative miRNA level using the U6 snRNA for normalization purposes. The RT and PCR primers for miRNA-22 were purchased from ABI PRISM (ABI PRISM, Carisbad, CA). The fold change in the miRNA expression in each tumor sample relative to the average expression in the non-cancerous control was calculated based on the threshold cycle (CT) value using the following formula:
Relative gene expression = 2−ΔΔCt, where –ΔΔCt = (Ct gene of interest – Ct internal control gene) Treated – (Ct gene of interest – Ct internal control gene) Untreated [8]. A 2-fold change in either direction was considered to be significant.
Cell lines
The ESCC cell lines EC9706 and KYSE150 were purchased from the tumor cell bank of the Chinese Academy of Medical Science. These cells were cultured in RPMI 1640 (Invitrogen) supplemented with 10% FBS.
Ionizing radiation
KYSE150 or EC9706 cells were exposed to different doses of irradiation in a JL Shepherd Model 143 and 137Cesium γ-irradiated at a rate of 2.4 Gy/min.
miRNA northern blots
For miRNA northern blots, 15 µg of total RNA was separated on a 15% denaturing polyacrylamide gel, electro-transferred onto a GeneScreen Plus hybridization transfer membrane (PerkinElmer, Waltham, Mass) and hybridized using ULTRAhyb-Oligo buffer (Ambion, Austin, TX). Oligonucleotides complementary to the mature miRNA-22 were end-labeled with T4 polynucleotide kinase (Invitrogen, Carlsbad, CA) and used as probes. Hybridization was performed at 42°C overnight, and the membranes were washed twice in 0.1 × SSPE and 0.1% SDS at 42°C for 15 min each. The membranes were then exposed to a storage phosphor screen (GE Healthcare Bio-Sciences, Piscataway, NJ) for 8 h and imaged using a Typhoon 9410 Variable Mode Imager (GE Healthcare Bio-Sciences, Piscataway, NJ)). Saved images were cropped using Photoshop 6.0 (Adobe Systems Inc., San Jose, CA).
Western blot analysis
Whole cell extracts were prepared from ESCC tissues and cultured cells by homogenizing cells in a lysis buffer (10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP40) containing a cocktail of protease inhibitors. After centrifugation at 15 000 rpm for 30 min at 4°C, supernatants were recovered and used for immunoblot analysis. The proteins were separated by SDS-PAGE and then transferred to polyvinylidene difluoride membranes (Millipore). Blots were blocked and then probed with Rad51 (1:500, Santa Cruz), Ku70 (1:500, Santa Cruz) and β-actin (1:5000, Abcam). After washing, the blots were incubated with horseradish peroxidase-conjugated secondary antibodies and visualized by super ECL detection reagent (Applygen, Beijing, China).
Plasmid construction and cell transfection
The miRNA-22 expression vector was constructed by cloning of annealed oligonucleotides that contained the pre-miRNA-22 sequence into the pSuppressorNeo expression vector. A scrambled sequence without significant homology to any rat, mouse or human gene was used as a negative control (vector group) (ABI PRISM, Carisbad, CA). Anti-miRTM miRNA-22 inhibitor (ABI PRISM, Carisbad, CA) was used to knockdown miRNA-22 expression. The expression constructs or inhibitor were transfected into the KYSE150 or EC9706 cell line, respectively, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. A northern blot was used to confirm the overexpression of miRNA-22.
Colony formation assay
A total of 400 cells were aliquoted into each well of a 6-well plate in triplicate and exposed to γ-rays with different irradiation dose. After 10 days of incubation, the colonies were stained with Giemsa stain, and a minimum of 50 viable cells were counted. Quantity One software (version 4.6.2) was used to analyze the results.
γ-H2AX foci formation assay
Briefly, cells were grown in cover slips kept in 35 mm petri plates and irradiated with different dosages (0, 2, 4 and 6 Gy) of γ-rays. Cell layers were washed with PBS and fixed for 20 min in 4% paraformaldehyde at room temperature; afterwards the cells were washed twice in PBS. For immunofluorescence staining, cells were permeabilized for 3 min in 0.25% Triton X-100 in PBS, washed two times in PBS and blocked for 1 h with 5% BSA in PBS. Antibody was diluted (1:200) in 1% BSA in PBS. Cells were incubated with primary antibody for 1 h at room temperature, washed three times in PBS and incubated with secondary antibody for 1 h at room temperature. Finally, cells were rinsed and mounted with ProLong Gold antifede with DAPI mounting media (Molecular Probe, USA). Images were captured using a Carl Zeiss confocal microscope. Acquisition settings were optimized to obtain maximal signal in immunostained cells with minimal background. A total of 200 nuclei were counted. All experiments were carried out in triplicates, independently from each other. The primary antibodies used were rabbit anti-γH2AX (Cell Signaling, USA). Secondary antibodies used were Alexa Fluor 488 goat anti-rabbit IgG (Molecular Probe, USA).
Flow cytometry
Cells were harvested 8 h post-irradiation with a dose of 0, 2, 4 and 6 Gy for apoptosis detection using the annexin V-FITC apoptosis detection kit (Sigma), and subsequently analyzed by flow cytometry.
Statistical analysis
All statistical analyses were performed using SPSS16.0 software. We statistically evaluated experimental results using a x2 test and an ANOVA test. Survival curves were constructed using the Kaplan–Meier method. P < 0.05 was considered statistically significant.
RESULTS
The expression of miRNA-22 in ESCC tissue and its correlation with clinicopathological features
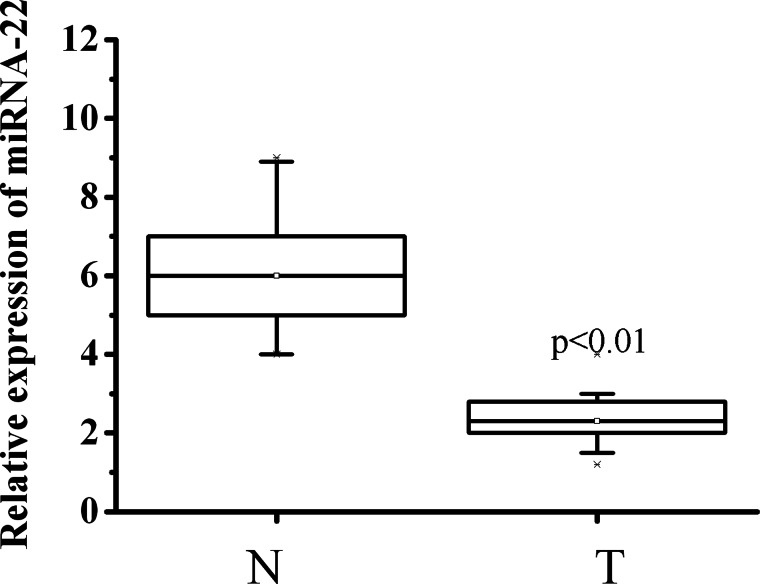
Expression of miRNA-22 was detected in 100 ESCC samples and the adjacent histologically normal tissues using RT-qPCR. miRNA-22 expression was normalized to the control U6B small nuclear RNA gene (RNU6B). The results showed that miRNA-22 expression levels were significantly lower in ESCC tissues compared with the corresponding non-cancerous tissues (Fig. 1).
Fig. 1.
Expression of miRNA-22 is down-regulated in ESCC tissues. The miRNA-22 expression was analyzed by qRT-PCR in normal epithelium (N, n = 100), and tumor tissues (T, n = 100). RNU6B served as an internal control.
Statistical analyses showed that the expression of miRNA-22 was only associated with the stage of clinical classification (Table 1). To determine if miRNA-22 expression was association with prognosis, a Kaplan–Meier survival analysis and a log-rank test were performed. No correlation was found between miRNA-22 expression and overall survival (P = 0.237, Fig. 2A). In the patients who did not receive radiotherapy (42 out of 100), no difference was found between miRNA-22 high-expression and low-expression patients (P = 0.425, Fig. 2B). In contrast, in patients who received concurrent chemotherapy with radiation therapy (58 out of 100), the survival rate of miRNA-22 high-expression patients was higher than that of miRNA-22 low-expression patients (P = 0.042, Fig. 2C). These results showed that the miRNA-22 expression status may have effects on the radiotherapy outcome of ESCC patients.
Table 1.
Relationship between miRNA-22 expression and tumor clinicopathological features
| Clinicopathological features | Number of cases | miRNA-22 expression |
||
|---|---|---|---|---|
| Low | High | P | ||
| Age, years | ||||
| ≥60 | 35 | 24 | 11 | 0.226 |
| <60 | 65 | 52 | 13 | |
| TNM classification pT | ||||
| pT1 | 12 | 9 | 3 | 0.843 |
| pT2 | 33 | 24 | 9 | |
| pT3 | 55 | 43 | 12 | |
| Lymph node metastasis | ||||
| N0 | 45 | 35 | 10 | 0.815 |
| N1 | 55 | 41 | 14 | |
| Stage | ||||
| I | 10 | 10 | 0 | 0.005 |
| II | 12 | 5 | 7 | |
| III | 36 | 31 | 5 | |
| IV | 42 | 30 | 12 | |
| Grade | ||||
| G1 | 25 | 20 | 5 | 0.689 |
| G2 | 30 | 21 | 9 | |
| G3 | 45 | 35 | 10 | |
Fig. 2.
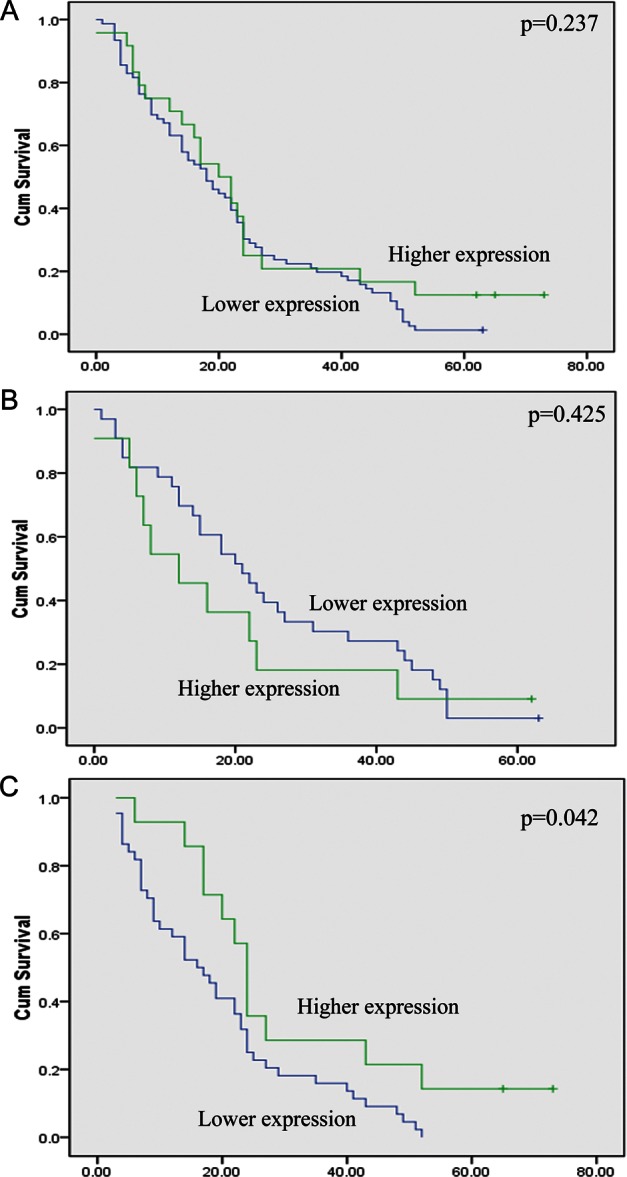
Kaplan-Meier survival curve of esophageal squamous cell carcinoma (ESCC) patients sub-grouped as miRNA-22 negative or positive. (A) No difference was found between miRNA-22 expression and the overall survival of all 100 ESCC patients. (B) No difference was found between miRNA-22 expression and the overall survival of patients who received only chemotherapy. (C) Significant difference was found between miRNA-22 expression and the overall survival of patients who received concurrent chemotherapy with radiotherapy. The prognosis of miRNA-22 positive cases was significantly longer than that of miRNA-22 negative cases (P = 0.042).
Effects of miRNA-22 expression on the radiosensitivity of ESCC cells
Expression of miRNA-22 in four ESCC cell lines was measured using RT-qPCR. As shown in Fig. 3, the sequence of miRNA-22 level was EC9706 > KYSE510 > KYSE450 >KYSE150. In the next experiments, EC9706 and KYSE150 were selected as cell models to study.
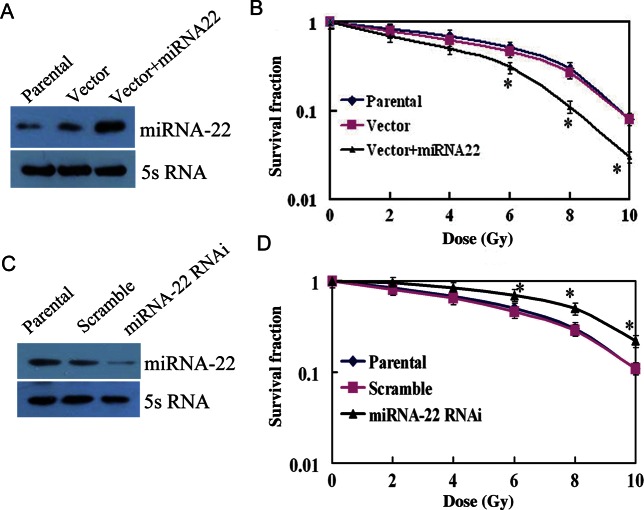
A colony formation assay was performed to examine the effects of miRNA-22 expression on the radiosensitivity of ESCC cells. The survival fraction of KYSE150 cells with forced miRNA-22 expression was lower than that of control groups (Fig. 4A and B). Conversely, the survival fraction of miRNA-22 knockdown EC9706 cells was significantly higher than that of control groups (Fig. 4C and D). These results indicate a positive correlation between miRNA-22 expression and radiosensitivity of ESCC cells to gamma radiation.
Fig. 3.
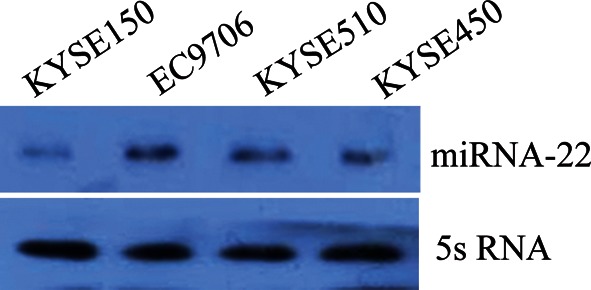
Expression of miRNA-22 in four kinds of ESCC cells. The sequence of miRNA-22 level was EC9706 > KYSE510 > KYSE450 > KYSE150.
Fig. 4.
Effects of miRNA-22 expression on the radiosensitivity of ESCC cells. (A) Expression of miRNA-22 increased after positive transfection of KYSE150 cells. (B) The colony formation assay was performed after parental, vector and vector + miRNA-22 cells were radiated using γ-rays with different dose irradiation. Data points shown represent the mean ± SD of three independent experiments. Asterisks indicate P < 0.05. (C) Expression of miRNA-22 decreased in miRNA-22 RNAi EC9706 cells. (D) Data points shown represent the mean ± SD of three independent experiments. Asterisks indicate P < 0.05.
We also examined the effects of miRNA-22 expression on the proliferation of ESCC cells using MTT experiments. The results obtained indicated that the proliferative ability of cells increased in low miRNA-22 level cells and decreased in miRNA-22 high-expression cells (Fig. 5A and B).
Fig. 5.
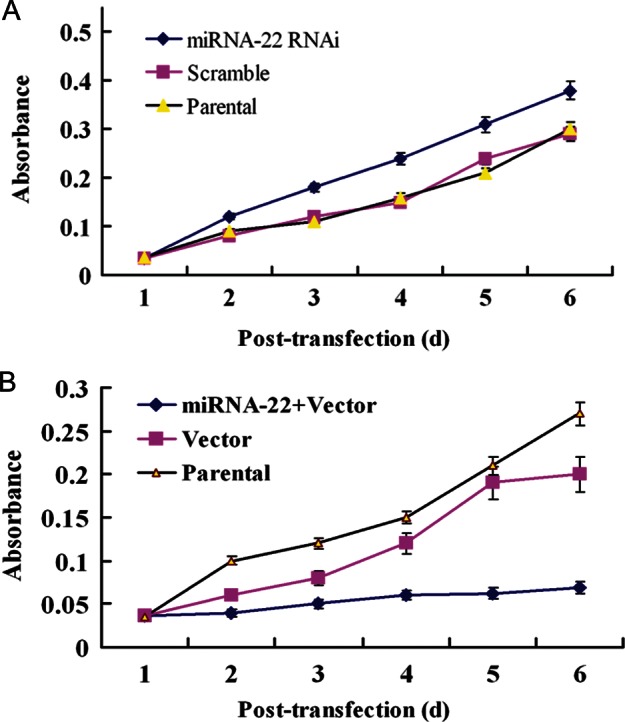
Effects of miRNA-22 expression on the proliferation of ESCC cells. MTT assay was performed after three groups of cells were radiated using γ- ray with a dose of 6 Gy. A, Proliferation ability of EC9706 cells with decreased miRNA-22 level was significant higher than that of control cells (P < 0.05). B, Proliferation ability of KYSE150 cells with increased miRNA-22 level was significant lower than that of control cells (P < 0.05).
Expression of miRNA-22 had an effect on the repair of DNA double-strand breaks induced by irradiation
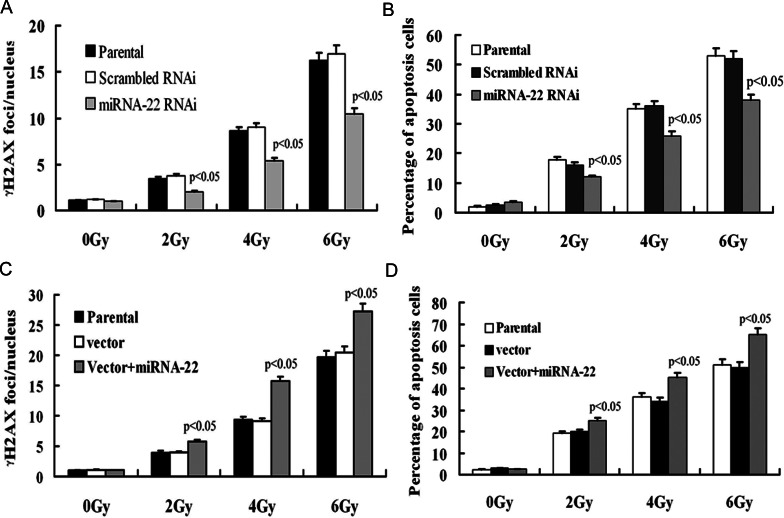
To investigate whether miRNA-22 expression has an effect on the repair ability of DNA damage induced by γ-rays, we performed γ-H2AX foci formation assays 1 h post-irradiation at different dosages. The number of γ-H2X foci in miRNA-22 knockdown cells was significantly lower than in control cells after irradiation (P < 0.01) (Fig. 6A). In accord with this result, the percentage of apoptotic cells with a low miRNA-22 level was significantly lower than that of control cells (P < 0.05) (Fig. 6B). Conversely, the number of γ-H2X foci in forced miRNA-22 expression cells was significantly increased (Fig. 6C). The percentage of apoptotic cells with a high miRNA-22 level was significantly higher than that of control cells (P < 0.05) (Fig. 6D). These data show that miRNA-22 expression can affect the repair ability of DSBs induced by irradiation.
Fig. 6.
(A) The number of γH2AX foci per nucleus in miRNA-22 knockdown EC9706 cells was significantly lower than that of control cells after 1 h post-IR (P < 0.01). Data bars shown represent the mean ± SD of γH2AX foci in unirradiated and irradiated cells. Two hundred nuclei were analyzed per condition. (B) The percentage of apoptotic EC9706 cells in the miRNA-22 knockdown group was significantly lower than in control groups after radiation (P < 0.05). (C) The number of γH2AX foci per nucleus in KYSE150 cells with miRNA-22 higher expression was significantly higher than in control cells (P < 0.05). (D) The percentage of apoptotic cells in the forced miRNA-22 expression group was significantly higher than in control groups (P < 0.05).
We next examined the expression of two key proteins Rad51 and Ku70, which were correlated with DNA double-strand breaks (DSBs) repair. As shown in Fig. 7A, Rad51 expression increased after miRNA-22 knockdown after irradiation, but the Ku70 level did not change. Forced expression of miRNA-22 decreased Rad51 expression after irradiation and the Ku70 expression was not affected (Fig. 7B).
Fig. 7.
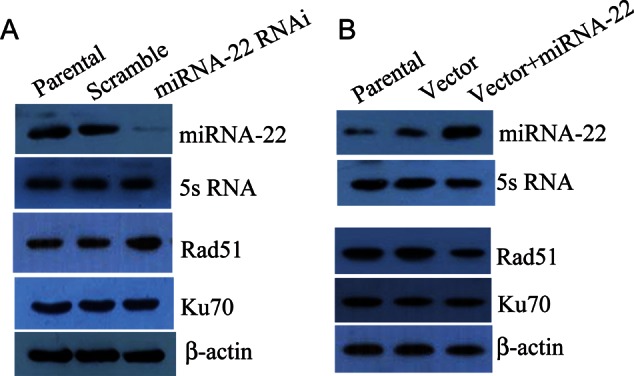
Effects of miRNA-22 on Rad51 and Ku70 expression in ESCC cells after radiation. (A) Downregulation of miRNA-22 in EC9706 cells led to increased expression of Rad51 after radiation with a dose of 6 Gy. No changes in Ku70 expression were observed. (B) Upregulation of miRNA-22 in KYSE150 cells led to decreased expression of Rad51 after radiation with a dose of 6 Gy. No changes in Ku70 expression were observed.
DISCUSSION
Studies have shown that miRNA expression fingerprints correlate with the clinical and biological characteristics of tumors, including tissue type, differentiation, aggression, response to therapy and prognosis [9]. Thus, a large amount of diagnostic information is encoded in a relatively small amount of miRNA. Liu et al. found that the miRNA-143 and miRNA-145 clusters were statistically different between esophageal cancer tissues and matched controls. The combined expression of miRNA-143 and miRNA-145 was significantly associated with a risk for EC [10]. miRNA-25 was found to be upregulated in 60 ESCC tissues compared with matched adjacent non-cancerous tissues. The upregulation of miRNA-25 was significantly correlated with status of lymph node metastasis and TNM stage, and markedly promoted migration and invasion of ESCC cells [11]. Kurashiqe et al. reported that the serum concentration of miRNA-21 in ESCC patients was significantly higher than that in healthy controls. A significant reduction in serum miRNA-21 levels was observed in post-operative samples vs pre-operative samples [12]. Other miRNA, such as miRNA-223 [13], miR-142-3p [14] and let-7 [15] were all found to have effects on ESCC progress. MiRNA-22 was previously reported to be a tumor suppressor. The relative expression level of miRNA-22 in lung cancer tissues was lower than that in normal tissues. Overexpression of miRNA-22 was shown to significantly inhibit the proliferation and invasion of lung cancer cell lines [16]. miRNA-22 expression was downregulated in hepatic cell cancer (HCC), and low miRNA-22 expression was predictive of poor survival in HCC patients. Overexpression of miRNA-22 significantly inhibited HCC cell proliferation and tumourigenicity [17]. However, the expression status of miRNA-22 in ESCC has not been reported until recently. Here, we detected the expression levels of miRNA-22 and investigated whether miRNA-22 expression also correlated with the clinicopathological features and prognosis of ESCC patients. Our results showed that miRNA-22 levels were decreased in ESCC patients, and a statistical analysis showed that the expression of miRNA-22 was negatively associated with the stage of clinical classification. Because some patients in our study received radiotherapy treatment and others did not, moreover, the chemotherapy project for all 100 patients was the same, we want to know if the miRNA-22 expression has an effect on the radiotherapy outcome for ESCC patients. Our results showed that the survival time of patients with high miRNA-22 expression was longer than that of patients with low miRNA-22 expression following radiotherapy, but miRNA-22 had no effect on survival time in patients who did not receive radiotherapy. This is the first report of a correlation between miRNA-22 expression and radiotherapy outcome.
To confirm the clinical data, we explored the correlation between miRNA-22 expression and radiosensitivity in ESCC cells. The colony formation and MTT assay proved that an increased miRNA-22 level prompted radiation sensitization of EC9706 cells. The radiosensitivity of cells is influenced by many factors. One of the most important factors is the repair ability of DNA DSBs. If the damage induced by radiation can be efficiently repaired, the cell will survive, otherwise the cell will undergo apoptosis. Given that our data proved overexpression of miRNA-22 increased the radiation sensitivity of ESCC cells, we hypothesize that this is due to miRNA-22 affecting the repair ability of DSBs induced by radiation in ESCC cells. The results of our γH2X foci experiment proved our hypothesis. There are two pathways for repairing DSBs, homologous recombination (HR) and non-homologous end-joining (NHEJ). Rad51 is a key protein in the HR DNA repair pathway. It is the major strand-transferase required for mitotic recombination [18]. The NHEJ pathway repairs the DNA by a homology-independent mechanism, rejoining broken ends irrespective of the sequence. Upon exposure to different genotoxic insults, the Ku70/80 heterodimer (Ku) binds to the broken DNA ends and recruits the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs). The formation of this complex results in the subsequent recruitment and phosphorylation of other proteins, such as XRCC4, DNA Ligase IV, Cernunnos/XLF and Artemis, so as to ligate the broken DNA ends [19]. To further explore the molecular mechanism of miRNA-22 affecting DSBs repair, we then measured the expression levels of Rad51 and Ku70 in miRNA-22 knockdown cells by forced expression after irradiation. The results indicated that miRNA-22 only affected the expression of Rad51. Overexpression of the Rad51 protein has been reported to stimulate homologous recombination and increase resistance of mammalian cells to ionizing radiation [20]. This result, combined with our data, suggests that downregulation of miRNA-22 can promote the repair ability of DSBs induced by γ-ray irradiation in ESCC cells, and that this effect is mediated, at least in part, by the Rad51 pathway. Tan et al. reported that miRNA-22 was upregulated in cells exposed to UV radiation, protecting cells from apoptosis by repressing PTEN expression. This UV stress response may inhibit the acute activation of the caspase signaling cascade and provide a time window for cells to repair the DNA damage induced by UV [21]. These various results show that miRNA-22 can play different roles in the context of different malignancies and different types of irradiation. Further studies should be implemented to explore the potential mechanism.
CONCLUSION
In conclusion, we have found, for the first time, that miRNA-22 expression is correlated with the prognosis of ESCC patients who receive radiotherapy. An increased miRNA-22 level inhibited the DSBs repair ability of ESCC cells, at least in part through the HR pathway, thereby improving the radiation sensitivity of ESCC cells.
SUPPLEMENTARY DATA
Supplementary data is available at the Journal of Radiation Research online.
FUNDING
Grant support was received from the National Natural Science Foundation of China (30901723, 81272511), the Natural Science Foundation of Tianjin (11JCYBJC13700) and the Institute Fund of Radiation Medicine (SF1103, SF1106).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Tian Jing for the collection of tumor samples.
REFERENCES
- 1.Berger B, Belka C. Evidence-based radiation oncology: oesophagus. Radiother Oncol. 2009;92:276–90. doi: 10.1016/j.radonc.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Borghesi S, Hawkins MA, Tait D. Oesophagectomy after definitive chemoradiation in patients with locally advanced oesophageal cancer. Clin Oncol (R Coll Radiol) 2008;20:221–6. doi: 10.1016/j.clon.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Minsky BD, Pajak TF, Ginsberg RJ, et al. INT 0123 (Radiation Therapy Oncology Group 94-05) phase III trial of combined-modality therapy for esophageal cancer: high-dose versus standard-dose radiation therapy. J Clin Oncol. 2002;20:1167–74. doi: 10.1200/JCO.2002.20.5.1167. [DOI] [PubMed] [Google Scholar]
- 5.Valencia-Sanchez MA, Liu J, Hannon GJ, et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 6.Berezikov E, Guryev V, van de Belt J, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–4. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 7.Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 8.Schmittgen TD, Lee EJ, Jiang J. High-throughput real-time PCR. Methods Mol Biol. 2008;429:89–98. doi: 10.1007/978-1-60327-040-3_7. [DOI] [PubMed] [Google Scholar]
- 9.Garzon R, Volinia S, Liu CG, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–9. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu R, Liao J, Yang M, et al. The cluster of miR-143 and miR-145 affects the risk for esophageal squamous cell carcinoma through co-regulating fascin homolog 1. PLoS One. 2012;7:e33987. doi: 10.1371/journal.pone.0033987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu X, Chen Z, Zhao X, et al. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2012;421:640–5. doi: 10.1016/j.bbrc.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 12.Kurashige J, Kamohara H, Watanabe M, et al. Serum microRNA-21 is a novel biomarker in patients with esophageal squamous cell carcinoma. J Surg Oncol. 2012;106:188–92. doi: 10.1002/jso.23064. [DOI] [PubMed] [Google Scholar]
- 13.Kurashige J, Watanabe M, Iwatsuki M, et al. Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer. 2012;106:182–8. doi: 10.1038/bjc.2011.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin RJ, Xiao DW, Liao LD, et al. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J Surg Oncol. 2012;105:175–82. doi: 10.1002/jso.22066. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q, Lv GD, Qin X, et al. Role of microRNA let-7 and effect to HMGA2 in esophageal squamous cell carcinoma. Mol Biol Rep. 2012;39:1239–46. doi: 10.1007/s11033-011-0854-7. [DOI] [PubMed] [Google Scholar]
- 16.Ling B, Wang GX, Long G, et al. Tumor suppressor miR-22 suppresses lung cancer cell progression through post-transcriptional regulation of ErbB3. J Cancer Res Clin Oncol. 2012;138:1355–61. doi: 10.1007/s00432-012-1194-2. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Yang Y, Yang T, et al. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer. 2010;103:1215–20. doi: 10.1038/sj.bjc.6605895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Tondravi M, Liu J, et al. Cortactin potentiates bone metastasis of breast cancer cells. Cancer Res. 2001;61:6906–11. [PubMed] [Google Scholar]
- 19.Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Vispe S, Cazaux C, Lesca C, et al. Overexpression of Rad51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res. 1998;26:2859–64. doi: 10.1093/nar/26.12.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan G, Shi Y, Wu ZH. MicroRNA-22 promotes cell survival upon UV radiation by repressing PTEN. Biochem Biophys Res Commun. 2012;417:546–51. doi: 10.1016/j.bbrc.2011.11.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.