Abstract
The single amino acid mutation G26R in human apolipoprotein A-I (apoA-IIOWA) leads to the formation of β-secondary structure rich amyloid fibrils in vivo. Here we show that full-length apoA-IIOWA has a decreased lipid binding capability, an increased amino terminal sensitivity to protease, and a propensity to form annular protofibrils visible by electron microscopy. The molecular basis for the conversion of apolipoprotein A-I to a pro-amyloidogenic form was examined by electron paramagnetic resonance spectroscopy. Our recent findings [Lagerstedt, J. O., Budamagunta, M. S., Oda, M. N., and Voss, J. C. (2007) J Biol Chem, 282, 9143–9149] indicate that Gly26 in native apo-protein separates a preceding β-strand structure (residues 20–25) from a downstream largely α-helical region. The current study demonstrates that the G26R variant promotes a structural transition of positions 27–56 to a mixture of coil and β-strand secondary structure. Microscopy and staining by amyloidophilic dyes suggest that this alteration extends throughout the protein within one week of incubation in vitro, leading to insoluble aggregates of distinct morphology. The severe consequences of the Iowa mutation likely arise from the combination of losing the contribution of the native Gly residue in terminating β-strand propagation and the promotion of β structure when an Arg is introduced adjacent to succeeding residue of identical charge and size, Arg27.
Apolipoprotein A-I (apoA-I)1 is the primary protein component of high density lipoprotein (HDL), where it plays a key role in reverse cholesterol transport, a primary mediator of cholesterol efflux and phospholipid metabolism. In reverse cholesterol transport, apoA-I interacts with several members of this pathway, including ATP-binding cassette transporters (ABCA1, ABCG1, and ABCG4), lecithin:cholesterol acyltransferase (LCAT), and scavenger receptor BI (SR-B1) [reviewed in (1) ]. The conformational adaptability of apoA-I (2),(3, 4) facilitates the processing of HDL by these distinct receptors and enzymes. For example with lipid binding, apoA-I undergoes a substantial change in structure, wherein the apoA-I helix bundle unfurls into an extended alpha helical “looped belt” conformation that resides on the periphery of the lipid particle [reviewed in (3, 5)]. This structural plasticity has made apoA-I difficult to examine and only recently has detailed structural information become available. Notably, the crystal structure of lipid-free apoA-I provides important new insight into the organization of the lipid-free protein, illuminating a N-terminal four-helix bundle followed by a more flexible C-terminal domain (6). However, features of the structure, particularly in the first 60 amino acids, differ from earlier solution measurements of apoA-I dynamics and secondary structure content (5, 7, 8). Thus important features of the structure in solution related to lipid binding and protein stability can be overlooked in the crystal structure.
Specific variants of human apoA-I aggregate in vivo and form amyloid fibril protein deposits containing high contents of β-structure. These fibrils accumulate in tissues and organs, causing severe pathophysiological consequences such as renal or liver failure (9). The first reported apoA-I variant associated with amyloidosis was found in an Iowa kindred (apoA-IIOWA) (10), and later shown to have a single heterozygous G26R substitution mutation (11, 12). As with most other amyloidogenic apoA-I variants (13), the N-terminal fragment (amino acids 1–83) of apoA-IIOWA is the predominant form of the protein found in amyloid fibril deposits (11). While several point mutations (along with their pathological consequences) have been identified in apoA-I, the structural consequences of these mutations have not been examined.
We have applied electron paramagnetic resonance (EPR) spectroscopy of site directed spin labels to examine the structural consequences of the G26R mutation. The results of these studies support the formation of β-strand secondary structure in the amino-terminus as well as pore-like annular protofibrils. The accumulation of β-strand content leading to insolubility is a common feature among fibril-forming proteins implicated in neurodegenerative diseases such as Parkinson’s and Alzheimer’s. We show that molecular mapping of the mis-folding process by EPR spectroscopy can elucidate regions within proteins responsible for the generation of protein deposits.
METHODS
Materials
Thio-specific nitroxide spin label ((1–Oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate) was received as a kind gift from Dr K Hideg (University of Pecs, Hungary). Standard molecular biology procedures were used to create the different plasmid constructs. All mutagenic primers were purchased from Sigma Genosys (The Woodlands, TX) and MWG Biotech (High Point, NC) and Davis Sequencing (Davis, CA) performed DNA sequence determination.
Expression, purification and labeling of recombinant apoA-I
Mutant human apoA-I proteins were expressed from pNFXex plasmid in bacterial Top10F’ cells (Invitrogen, Carlsbad, CA), extracted and purified by immobilized metal affinity chromatography, and labeled with the methanethiosulfonate spin label, which specifically reacts with the sulfhydryl of the targeted Cys residue (14), as previously described (4). The purified apoA-I proteins were concentrated by use of 30 kDa MWCO Amicon Ultra centrifugal filter devices (Millipore Corp., Billerica, MA) and maintained and analyzed in phosphate-buffered saline (20 mM sodium phosphate, pH 7.4, 500 mM NaCl).
Lipid clearance assay
Binding of wild-type apoA-I (apoA-IWT) and apoA-IIOWA to 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) vesicles was monitored by 90° light scattering. DMPC (Avanti Polar Lipids Inc., Alabastr, AL, 10 mg) was dissolved in chloroform:methanol (3:1 v/v), dried under N2 stream and the residual solvent removed by overnight lyophilization. The dried lipid was dispersed to a final concentration of 5 mg/ml using buffer (20 mM sodium phosphate, pH 7.4, 500 mM NaCl) pre-warmed to 37 °C and vortexing. The suspension was extruded using 200-nm filters (15) to yield unilamellar vesicles of ~200 nm in diameter. The binding of apoA-I to the lipids and the consequent clearance of the phospholipid vesicles solution as a result of the formation of smaller protein-lipid disc complexes was monitored by a Perking-Elmer spectrofluorometer (model LS 50B). Excitation and emission wavelengths were set at 600 nm with a slit width of 3 nm. All the solutions were preincubated at 23.6 °C and the cuvette holder maintained at the same temperature. The DMPC vesicles (100 µg) solution was equilibrated in the cuvette holder for 10 min; apoA-IWT or apoA-IIOWA in phosphate-buffered saline buffer was added to a final volume of 400 µl (0.25 mg/ml DMPC vesicles), rapidly mixed and the light scattering monitored as a function of time. The lipid binding efficiency of apoA-IWT and apoA-IIOWA was compared at phospholipid-protein ratios (w/w) of 0.5:1 (0.5 mg/ml protein), 1:1 (0.25 mg/ml protein), and 2:1 (0.125 mg/ml protein).
Limited proteolysis
Limited proteolysis was performed using chymotrypsin or V8 protease as described previously (16). A 2000:1 (w/w) ratio of unlabeled apoA-I (0.25 mg/ml) to protease was used and the reactions were quenched by the addition of protease inhibitor cocktail (Novagen). Proteolytic fragment sizes were estimated from SDS-PAGE migration. Fragment sequences were determined from SDS-PAGE bands blotted onto PVDF by Edman degradation at the Biomolecular Resources Facility, University of Texas Medical Branch (Galveston, TX).
Fourier transform infrared (FTIR) spectroscopy
FTIR spectra of apoA-I proteins (20 mg/ml in phosphate-buffered saline) were recorded on a Perkin-Elmer FTIR model 2000 spectrometer (Norwalk, CT), interfaced to a computer running Perkin-Elmer's Spectrum 3.1 software. Secondary structure distribution was determined by integration of peaks from the deconvoluted spectra in the amide I region (1620–1700 cm−1). After deconvolution of the spectra, the amide I components were assigned to the secondary structure elements as described elsewhere (17, 18).
EPR spectroscopy
EPR measurements were carried out in a JEOL X-band spectrometer fitted with a loop-gap resonator as previously described (4). Briefly, an aliquot (5 µl) of purified, spin-labeled protein (1 mg/ml) in phosphate-buffered saline was loaded into a sealed quartz capillary and placed in the resonator. Spectra were acquired at room temperature (20–22°C) from a single 60-s scan over a field of 100 G at a microwave power of 2 mW and a modulation amplitude optimized to the natural line width of the individual spectrum (0.5 – 1.5 G). The spectra as displayed are all normalized to the same number of spins using the sample unfolded in SDS, which reduces line broadening and thereby facilitates integrated intensity calculations. Based on the spin count for the protein concentrations measured, all sites labeled with an efficiency of >90%, except position 28, where labeling efficiency was lower than 70%. Molecular accessibility of spin-labeled side chains to CrOx was determined using successive power saturation scans as described (19). Π1/2 value was calculated using software provided by C. Altenbach, where the error for any sample did not exceed 3% of the calculated P1/2 value.
FSB staining
Five µl of samples (3.3 µM apoA-IWT and 3.3 µM apoA-IIOWA, incubated for 10 days at RT, and 10 µM Aβ1–40, incubated for 5 days at RT) were spotted on glass slides. After air drying, slides were rinsed with water 3 times and then stained with 10 µM FSB [(E,E)-1-Fluoro-2,5-bis(3–hydroxycarbonyl-4-hydroxy)styrylbenzene, Calbiochem] at RT for 20 min. To remove excess dye, samples were washed 3 times with 70% EtOH, and then photographed with a Zeiss Axioscop microscope equipped with AxioCam MR digital imaging system (Thornwood, NY).
Thioflavin T (ThT) binding assay
Samples of apoA-IWT and apoA-IWT in PBS (3.3 µM) were incubated at RT for the time periods indicated. ThT binding was then measured on 10 µl of sample mixed with 100 µl of ThT solution (5 µM in 50 mM glycine-NaOH at pH 8.5) and incubated for 5 min. The fluorescence intensity at 485 nm was collected on a spectrofluorimeter (Spectra MAXgeminiXPS, Molecular Devices, Sunnyvale, CA). Measurements were recorded at room temperature with excitation of 437 nm and an emission cut-off filter of 455 nm.
Electron microscopy studies
Samples of protein (10 µl; ~2 mg/ml) were placed on 300 mesh, carbon-formvar coated grids for 1 minute. Most of the sample was wicked away using filter paper, and replaced briefly with 10 µl of distilled water. The water was removed and replaced with 10 µl of 1% aqueous uranyl acetate for 30 seconds. The uranyl acetate was removed and the grid was air dried. Specimens were examined by routine transmission electron microscopy. At least 6 grid squares were surveyed to ensure an appropriate sampling of the preparation.
RESULTS AND DISCUSSION
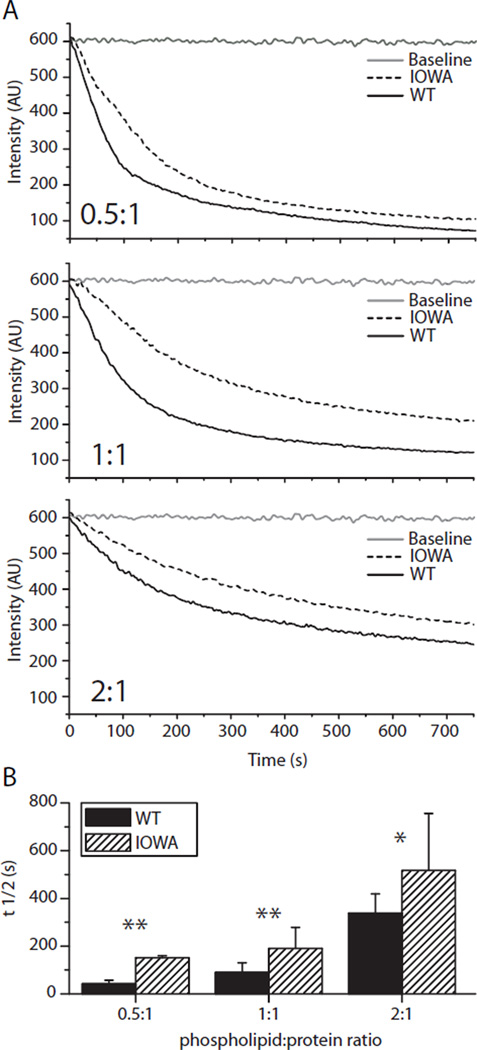
ApoA-I plasma concentration and function are important factors in cholesterol and phospholipid metabolism. Several naturally occurring mutations in the apoA-I gene induce in vivo fibril formation causing amyloidosis (9). The glycine to arginine mutation at position 26 in apoA-I in the Iowa kindred is associated with such hereditary systemic amyloidosis (11), where fibrils accumulate in kidney (9) and liver (20). The single mutation also leads to an increased turnover of the protein and lowered plasma levels (21, 22), and heterozygotes for apoA-IIOWA show a decreased plasma level of normal apoA-I. Since apoA-I is recycled between lipid-poor and lipid bound states, its lipid binding capacity would likely have a major influence on its in vivo half-life as well as its ability to form functional HDL. To investigate the effect of the G26R mutation on lipid-binding ability, wild-type apoA-I (apoA-IWT) and apoA-IIOWA were individually mixed with specified ratios of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) lipid vesicles and the lipid clearance was measured by light scattering (Figure 1). The apparent t1/2 values for all phospholipid:protein ratios tested were approximately twice as high for apoA-IIOWA compared to apoA-IWT, clearly indicating apoA-IIOWA bears a reduced lipid binding capacity.
FIGURE 1.
ApoA-IIOWA binds lipids but with lower efficiency than apoA-IWT. (A) Phospholipid (0.25 mg/ml DMPC) vesicle clearance efficiency as a function of time of wild-type and IOWA apoA-I is compared for the shown lipid:protein ratios. Dotted and solid black lines represent apoA-IIOWA and apoA-IWT, respectively. The baselines for lipid vesicles without protein are shown in gray. (B) T1/2 values for the different lipid:protein ratios were determined and plotted. Solid bars represent wild-type and hatched bars represent apoA-IIOWA. Mean values of three independent experiments are shown. Error bars indicate standard deviation. Asterisks denote statistically significant difference (*, p<0.05; **, p<0.01).
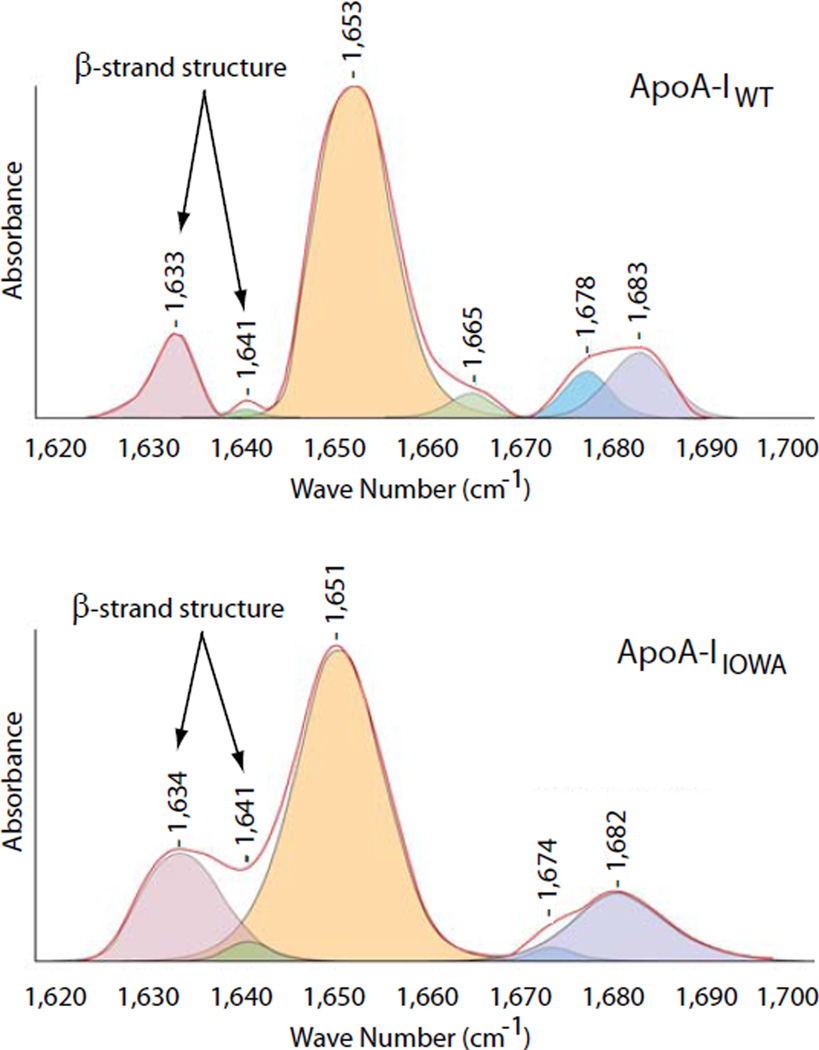
The amyloidogenic properties of apoA-IIOWA is shared with numerous proteins and protein fragments that give rise to specific pathological conditions such as Alzheimer’s disease, Creutzfeldt-Jacob disease, Parkinson’s disease, Huntington disease and type II diabetes mellitus (23). A mechanistic explanation of the amyloidogenic nature of such apolipoprotein variants is thus anticipated to provide important insight into the broad field of amyloid research. The common theme amongst proteins that cause these pathologies is that in the amyloidogenic state there is an increase in β-strand structure content, which leads to self-association and accumulation in vivo (24). A possible explanation for the amyloidogenic properties of apoA-IIOWA would thus be an increased β-strand structure content in the active conformer. Fourier transform infrared (FTIR) spectroscopy was used to quantify the β-strand content in lipid-free apoA-IIOWA (25). To examine the β-strand contribution to lipid-free apoA-IWT and apoA-IIOWA secondary structure, spectra were obtained in the amide I region (1620–1700 cm−1; Figure 2). The apoA-IWT protein displayed a β-strand structure content of 10%, a value that is in agreement with previously published data (5, 7, 8). In contrast, the percent β-strand structure of apoA-IIOWA is significantly higher and is calculated to be 22%. In terms of amino acid residues this corresponds to an increase in β-strand structure from 20 to 24 residues in apoA-IWT, to more than 50 residues in apoA-IIOWA.
FIGURE 2.
The apoA-IIOWA mutant has a two-fold increase in β-strand secondary structure content compared to the apoA-IWT protein. FTIR spectra (red traces) for apoA-IWT (top) and apoA-IIOWA (bottom) were recorded in the amide I region. The integrated peaks of the deconvoluted spectra were used for the determination of the secondary structure composition. Peaks corresponding to β-strand structures are indicated with arrows.
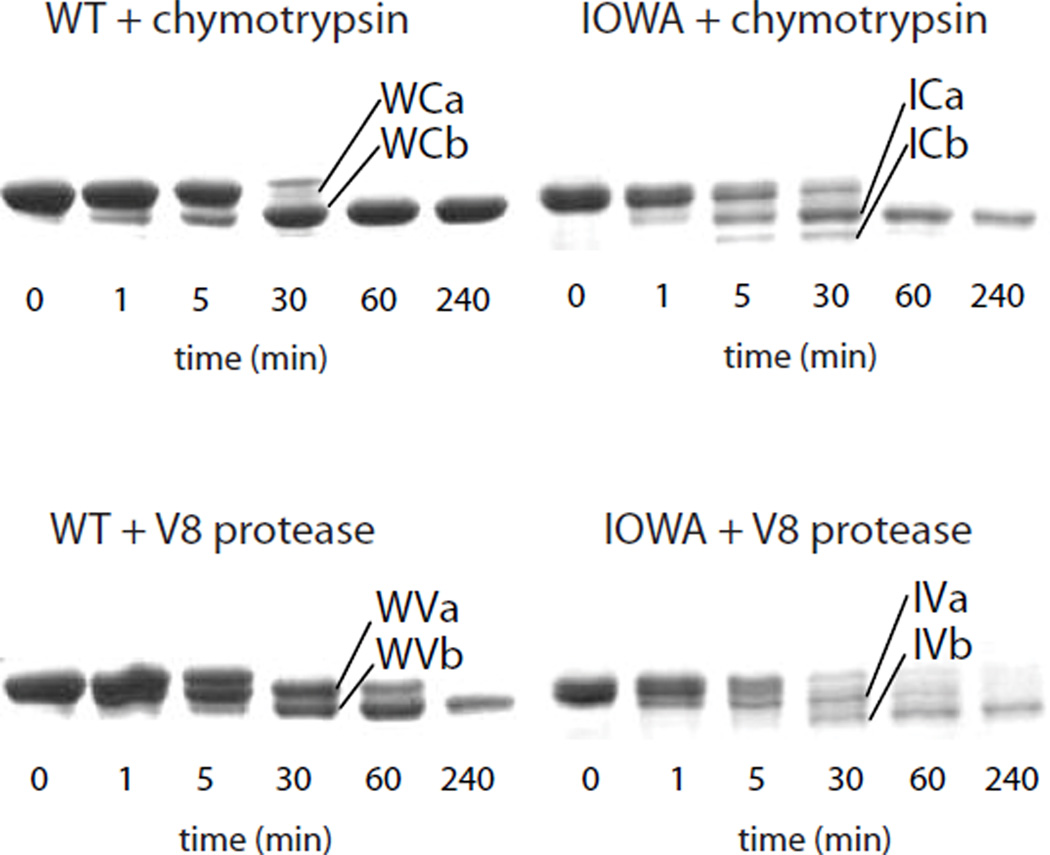
Limited proteolysis was used to assess the effect of increased β-sheet in the apoA-IIOWA on the protein's tertiary structure. Protease digestion of apoA-IWT using chymotrypsin, which is specific for large, non-polar side chains, leads to rapid production of fragments produced by C-terminal truncation (Figure 3, Table 1). As previously observed (16), the major product results from cleavage at Y192, with a lesser amount of cleavage occurring at F225. Protease digestion of apoA-IIOWA under the same conditions leads to the production of similar-sized fragments. Sequence analysis of these fragments revealed they are produced by cleavage in the N-terminus, at residues Y18 and F57. Digestion with V8 protease, which is specific for glutamic acid residues, yielded only C-terminal truncation in apoA-IWT, but generated both N- (at E34) and C-terminal truncations in apoA-IIOWA. Thus, the increase in β-sheet content determined by the FTIR secondary structure analysis coincides with increased protease sensitivity in the amino-terminus region. These results help explain previous in vivo findings (9) that show a preponderance of amino-terminal fragments of apoA-IIOWA in amyloid deposits.
FIGURE 3.
ApoA-IIOWA has increased sensitivity to proteolysis in the amino-terminus. ApoA-IWT (W) and apoaA-IIOWA (I) were each treated with chymotrypsin (C) or V8 protease (V) for the indicated times. The cleavage products were blotted onto PVDF membrane, excised and analyzed by Edman degradation (see Table 1).
Table 1.
Proteolytic Cleavage Fragments of apoA-IWT and apoA-IIOWA
| Band | Sequencea | Fragment IDb |
|---|---|---|
| WCa | MHHHH | M1-F229c |
| WCb | MHHHH | M1-Y192c |
| ICa | VDVLK | V19-D243 |
| ICb | SKLRE | S58-D243 |
| WVa | MHHHH | M1-E223c |
| WVb | MHHHH | M1-E212c |
| IVa | MHHHH | M1-E223 |
| GSALG | G35-D243 | |
| IVb | MHHHH | M1-E212 |
Determined by Edman analysis of blotted SDS-PAGE fragments;
Based on molecular mass estimate from SDS-PAGE;
this work and reference 15.
In order to elucidate the structural basis for apoA-IIOWA’s reduced lipid binding, increased amino-terminal protease sensitivity, higher turnover, amyloidogenic properties, and significantly increased β-strand structure content, we carried out EPR spectroscopy of site-directed spin-labels. We have recently shown that under physiological conditions, Gly26 in apoA-IWT is located at the end of a β-strand structure (residues 20 to 25) (4). We hypothesized that the role of Gly26 is to serve as a β-structure terminator, since glycine residues have both a low propensity for β-strand formation (26) and appear to be evolutionary preserved to prevent protein aggregation (27). The mutation of this residue in the apoA-IIOWA kindred would thus allow the β-strand of residues 20 to 25 to extend to position 26 and beyond, thereby increasing the likelihood for more global β-strand interactions.
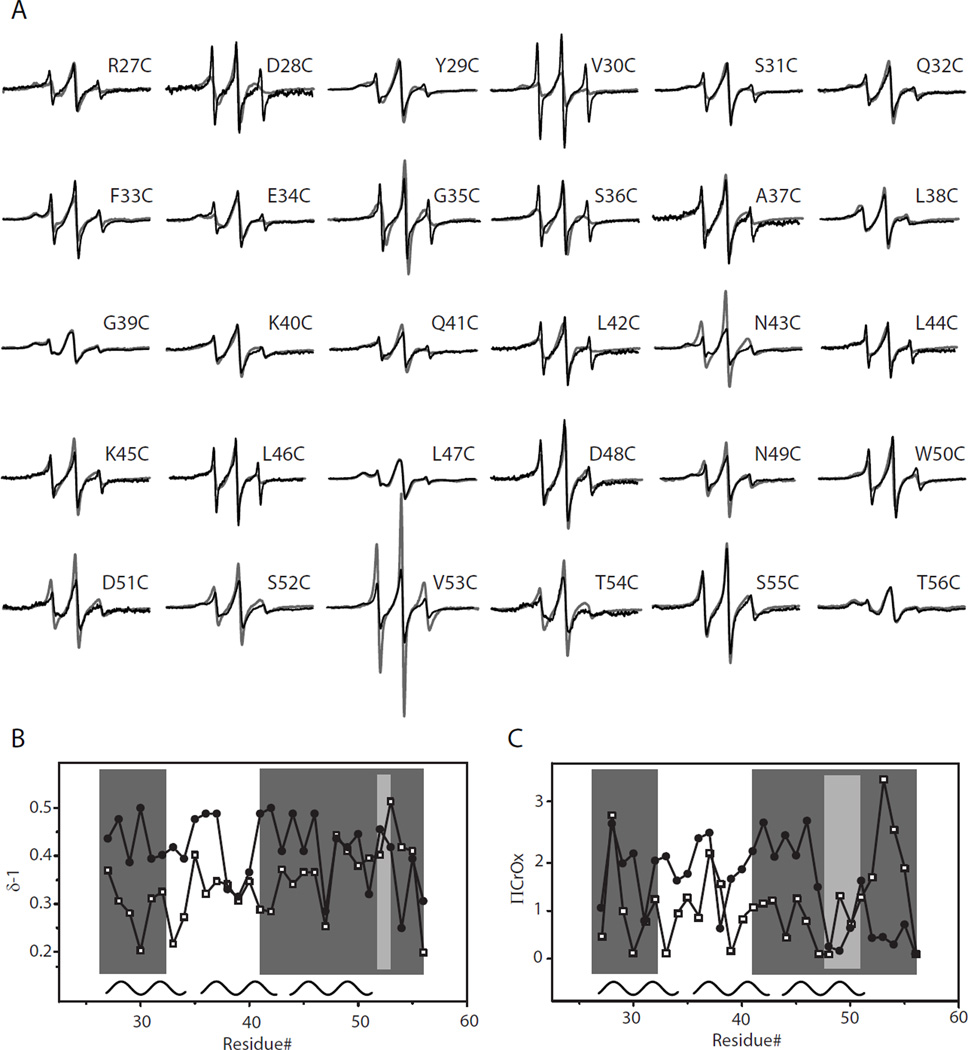
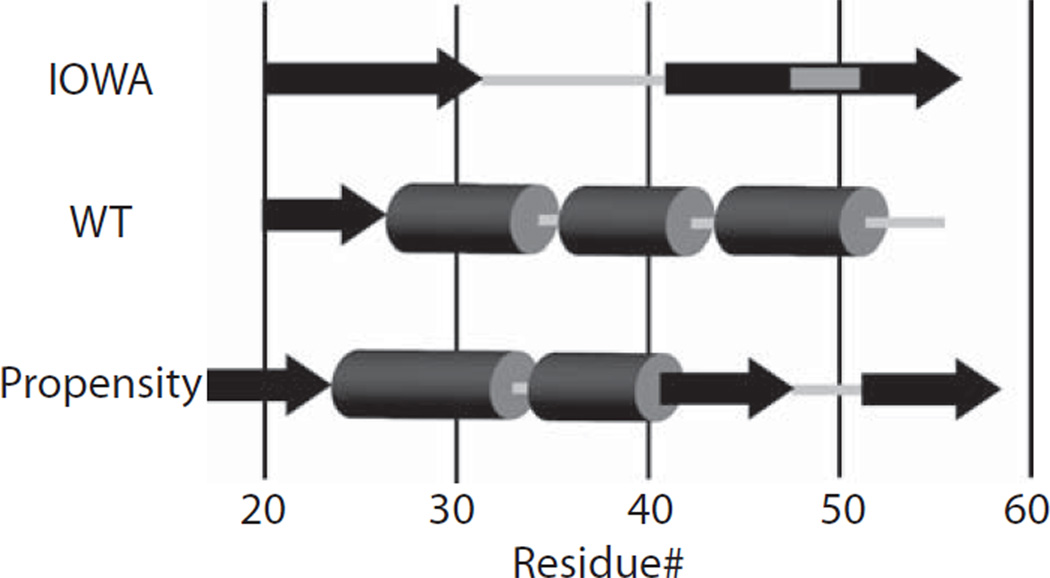
To investigate this possibility, we generated a series of cysteine substitution point mutations for residues 27 to 56, in the G26R background. These apoA-I variants bearing both the G26R mutation and a cysteine substitution were individually expressed in bacteria, purified, spin-labeled and analyzed by EPR. In figure 4A, the acquired spectra for apoA-IIOWA are compared to their apoA-IWT counterparts. Of the sites examined only four positions bear similar spectral line shapes (L38C, K40C, L47C and S55C; Figure 4A); the remainder of positions examined bear significantly different spectral line shapes. Structural differences were further analyzed by determining the inverse spectral line-width (δ−1; Figure 4B). This provides a measure of the degree of the side chain mobility of the nitroxide labeled residue and can be used to determine the secondary structure composition for a series of consecutive amino acids. From this analysis, residues 27 to 31, 41 to 52 and 53 to 56, show alternating higher and lower degrees of mobility, which is characteristic of β-strand structures (Figure 4B). In addition to side chain dynamics, the collision frequency between chromium oxalate (CrOx) and the nitroxide spin label was measured by power saturation EPR, allowing for the calculation of the accessibility parameter (ΠCrOx). Asymmetry in the accessibility of side chains provides an additional dimension of information for the mapping of secondary structure along the backbone fold. Consistent with the inverse spectral line-width, the plotted accessibility values (Figure 4C) display a periodicity of 2 within regions similar to those identified by the mobility parameter δ (residues 27 to 32, 41 to 47, and 50 to 56), providing further evidence for the presence of β-strand structure in this region. Interestingly, the experimentally determined secondary structure of apoA-IIOWA fits well with secondary structure predictive analysis of the N-terminal primary sequence (28) (Figure 5). This comparison implies that the G26R mutation allows the protein to adopt a regional structure, which is more probable based on the amino acid sequence, as found in other systems (29).
FIGURE 4.
EPR spectroscopy analysis reveals apoA-IIOWA specific β-structure patterns. Thirty residues (position 27 to 56) immediately downstream of the apoaA-IIOWA mutation (G26R) were individually mutated to cysteines, spin-labeled and scanned by EPR spectroscopy. (A) The resulting spectra of the mutant protein (black traces) displayed major differences compared to the corresponding spectra of apoA-IWT protein (gray traces). The secondary structure for this region was analyzed by plotting (B) the inverse central line-width (δ−1) and (C) the polar accessibility parameter (ΠCrOx) values for apoaA-IIOWA (filled circles) and apoaA-IWT (open squares). Regions of apoA-IIOWA displaying a periodicity of 2, consistent with β-structure, are shown in dark gray. The light gray indicates regions where this periodicity is interrupted. The helical regions of apoA-IWT are shown as black curves, with a periodicity of 3.6, at the bottom of panel (B) and (C).
FIGURE 5.
Schematic comparison of the secondary structure between apoA-IIOWA (this communication), apoA-IWT (4) and model based on primary sequence (28). β-structure is shown as black arrows whereas helical structures are indicated as dark gray cylinders. Light gray indicates non-structured regions.
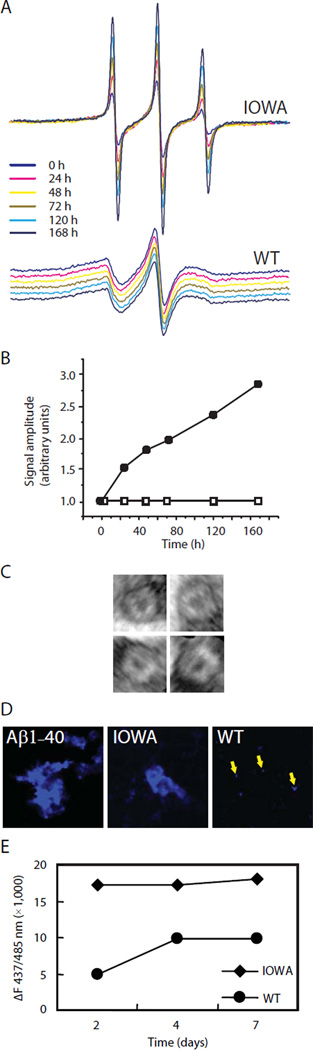
While fibrils formed in vivo have been found to contain full-length apoA-I (30), in vitro formation of apoA-I fibrils has only been reported for a peptide mimicking the N-terminal proteolytic fragment (13). To determine whether the G26R mutation induces a time-dependent change in the structure of full-length apoA-I we monitored spin-labeled apoA-IIOWA at 20–22 °C over a 7 day time course. The location (residue 29) of the spin-label was chosen, as it is the center of the first β-strand extension (residues 27 to 31) and thus a good reporter of early structural changes. Over the one week time course, the spectra of the spin-label at position 29 is dramatically altered (Figure 6A), with a threefold increase in amplitude (Figure 6B). The change in spectra reflects increased side chain mobility and thus a structural transition of the protein. In contrast, the spectra of the spin-label at position 29 in apoA-IWT background show no significant change under the same conditions (Figure 6AB).
FIGURE 6.
EPR, EM and fluorescent dye binding analyses of time-dependent structural change of apoA-IIOWA. (A) ApoA-IIOWA (upper panel) and apoA-IWT (lower panel) both labeled with nitroxide at their respective position 29 were incubated at 20–22 °C and the EPR spectral changes were monitored during 7 days. The spectra for apoA-IWT are baseline shifted to enable comparison. (B) The normalized amplitudes of the EPR spectra were plotted and showed a 3-fold increment for apoA-IIOWA (solid circles) during the time studied whereas the spectra for apoA-IWT (open squares) were unchanged. EM revealed the formation of (C) annular structures, approximately 20 nm in diameter, at the one-week time point. (D) Staining of apoA-IIOWA and apoA-IWT aggregates by the amyloidophilic dye FSB. Samples were incubated for 10 days and stained as described in Methods. Fibrils formed by Aβ1–40 were similarly stained as positive control. In the right panel arrows are used to indicate the presence of small aggregates formed by apoA-IWT. (E) ThT binding of apoA-IIOWA (filled diamonds) and apoA-IWT (filled circles). Samples (at 3.3 µM) were incubated at RT for indicated time and the fluorescence measured as described in Methods. Displayed fluorescence intensities are background-subtracted (given as ∆F).
To evaluate the extent of higher-order assembly, the protein sample was examined for fibril formation by electron microscopy (EM). Following a one-week incubation, annular (doughnut-shaped) protofibrils were observed for apoA-IIOWA (Figure 6C) but not for apoA-IWT. It should be noted that assemblies with annular morphology are common among amyloidogenic proteins, having been described for Parkinson’s disease-linked α-synuclein and Aβ mutants associated with Alzheimer’s disease (31). In these cases the fragments also display toxic properties that extend beyond amyloid formation [such as membrane permeabilization; (32, 33)].
To confirm the formation of amyloid structure and loss of solubility, samples were examined by the amyloidophilic dye, FSB (34). The fibrils formed by apoA-IIOWA stain with an intensity comparable to that of fibrils formed by Aβ peptides, a major component of the Alzheimer’s disease amyloid plaques. By contrast, only small FSB-reactive deposits are evident with apoA-IWT (Figure 6D). To observe the progression of β-strand formation in apoA-IWT and apoA-IIOWA, ThT binding (35) was measured for both samples over a period of 7 days. While both apoA-IWT and apoA-IIOWA display increasing amyloid structure with time, the apoA-IIOWA protein displays high ThT binding within 48 hours, and compared to apoA-IWT, a consistently higher amyloid signal throughout the one-week period (Figure 6E). Thus the appearance of β-strand structure is much faster in the presence of the Iowa mutation. Furthermore, while we do not see an appreciable amount of insoluble apoA-IWT accumulate over a 2-week time course, the apoA-IIOWA protein readily forms large-assemblies as evident by both EM and FSB staining.
Protein aggregation frequently arises when a region experiences a change in- or loss of- its native secondary structure (36). Due to their inherent conformational dynamics in the lipid-poor state, apoliporoteins display a high susceptibility of to form or associate with amyloids in vivo. The severe consequences of the Iowa mutation likely arise from the combination of losing the contribution of the native Gly residue in terminating β-strand propagation and the promotion of β structure when an Arg is introduced adjacent to succeeding residue of like charge and size, Arg27. The resulting downstream secondary structure conversion induces tertiary structure rearrangements. Together these changes decrease lipid binding function, increase protease susceptibility, and promote amyloid assembly. Because wild type apoA-I is also implicated in amyloid pathologies, apoA-IIOWA provides a valuable model for understanding the structural basis of mis-folding pathways in apoA-I and other apolipoproteins. While other regions of apoA-I may also be vulnerable to pro-amyloidogenic mis-folding, positions that occupy transitions between different backbone folds, such as Gly26, should in particular be considered as targets for factors that stabilize or destabilize the protein.
ACKNOWLEDGEMENT
We acknowledge Drs J.H. Crowe and L.M. Hayes for assistance in FTIR analyses.
Footnotes
This work was supported by National Institutes of Health grants HL77268, HL78615 and HL073826-01, and by the American Heart Association Scientist Development Grant 0235222N. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR-12088-01 from the National Center for Research Resources, National Institutes of Health. J.O.L. was a recipient of fellowships from the Sweden-America Foundation, the Bengt Lundqvist Foundation, and the Foundation Blanceflor Boncompagni-Ludovisi, neé Bildt.
The abbreviations used are: apoA-I, apolipoprotein A-I; CrOx, chromium oxalate; DMPC, 1,2-dimyristoyl-sn-glycero-3-phosphocholine; EPR, electron paramagnetic resonance; FTIR, Fourier transform infrared; FSB, (E,E)-1-Fluoro-2,5-bis(3–hydroxycarbonyl-4-hydroxy)styrylbenzene; HDL, high density lipoprotein
REFERENCES
- 1.Lewis GF. Determinants of plasma HDL concentrations and reverse cholesterol transport. Curr Opin Cardiol. 2006;21:345–352. doi: 10.1097/01.hco.0000231405.76930.a0. [DOI] [PubMed] [Google Scholar]
- 2.Oda MN, Forte TM, Ryan RO, Voss JC. The C-terminal domain of apolipoprotein A-I contains a lipid-sensitive conformational trigger. Nat Struct Biol. 2003;10:455–460. doi: 10.1038/nsb931. [DOI] [PubMed] [Google Scholar]
- 3.Martin DD, Budamagunta MS, Ryan RO, Voss JC, Oda MN. Apolipoprotein A-I assumes a "looped belt" conformation on reconstituted high density lipoprotein. J Biol Chem. 2006;281:20418–20426. doi: 10.1074/jbc.M602077200. [DOI] [PubMed] [Google Scholar]
- 4.Lagerstedt JO, Budamagunta MS, Oda MN, Voss JC. EPR spectroscopy of site-directed spin labels reveals the structural heterogeneity in the N-terminal domain of apo-AI in solution. J Biol Chem. 2007;282:9143–9149. doi: 10.1074/jbc.M608717200. [DOI] [PubMed] [Google Scholar]
- 5.Davidson WS, Silva RA. Apolipoprotein structural organization in high density lipoproteins: belts, bundles, hinges and hairpins. Curr Opin Lipidol. 2005;16:295–300. doi: 10.1097/01.mol.0000169349.38321.ad. [DOI] [PubMed] [Google Scholar]
- 6.Ajees AA, Anantharamaiah GM, Mishra VK, Hussain MM, Murthy HM. Crystal structure of human apolipoprotein A-I: insights into its protective effect against cardiovascular diseases. Proc Natl Acad Sci U S A. 2006;103:2126–2131. doi: 10.1073/pnas.0506877103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Brubaker G, Peng DQ, Somerlot B, Abdollahian DJ, Smith JD. Apolipoprotein A-I lysine modification: effects on helical content, lipid binding and cholesterol acceptor activity. Biochim Biophys Acta. 2006;1761:64–72. doi: 10.1016/j.bbalip.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Silva RA, Hilliard GM, Fang J, Macha S, Davidson WS. A three-dimensional molecular model of lipid-free apolipoprotein A-I determined by cross-linking/mass spectrometry and sequence threading. Biochemistry. 2005;44:2759–2769. doi: 10.1021/bi047717+. [DOI] [PubMed] [Google Scholar]
- 9.Joy T, Wang J, Hahn A, Hegele RA. APOA1 related amyloidosis: a case report and literature review. Clin Biochem. 2003;36:641–645. doi: 10.1016/s0009-9120(03)00110-3. [DOI] [PubMed] [Google Scholar]
- 10.Van Allen MW, Frohlich JA, Davis JR. Inherited predisposition to generalized amyloidosis. Clinical and pathological study of a family with neuropathy, nephropathy, and peptic ulcer. Neurology. 1969;19:10–25. doi: 10.1212/wnl.19.1.10. [DOI] [PubMed] [Google Scholar]
- 11.Nichols WC, Dwulet FE, Liepnieks J, Benson MD. Variant apolipoprotein AI as a major constituent of a human hereditary amyloid. Biochem Biophys Res Commun. 1988;156:762–768. doi: 10.1016/s0006-291x(88)80909-4. [DOI] [PubMed] [Google Scholar]
- 12.Nichols WC, Gregg RE, Brewer HB, Jr, Benson MD. A mutation in apolipoprotein A-I in the Iowa type of familial amyloidotic polyneuropathy. Genomics. 1990;8:318–323. doi: 10.1016/0888-7543(90)90288-6. [DOI] [PubMed] [Google Scholar]
- 13.Andreola A, Bellotti V, Giorgetti S, Mangione P, Obici L, Stoppini M, Torres J, Monzani E, Merlini G, Sunde M. Conformational switching and fibrillogenesis in the amyloidogenic fragment of apolipoprotein a-I. J Biol Chem. 2003;278:2444–2451. doi: 10.1074/jbc.M204801200. [DOI] [PubMed] [Google Scholar]
- 14.Berliner LJ, Grunwald J, Hankovszky HO, Hideg K. A novel reversible thiol-specific spin label: papain active site labeling and inhibition. Analytical biochemistry. 1982;119:450–455. doi: 10.1016/0003-2697(82)90612-1. [DOI] [PubMed] [Google Scholar]
- 15.MacDonald RC, MacDonald RI, Menco BP, Takeshita K, Subbarao NK, Hu LR. Small-volume extrusion apparatus for preparation of large, unilamellar vesicles. Biochim Biophys Acta. 1991;1061:297–303. doi: 10.1016/0005-2736(91)90295-j. [DOI] [PubMed] [Google Scholar]
- 16.Roberts LM, Ray MJ, Shih TW, Hayden E, Reader MM, Brouillette CG. Structural analysis of apolipoprotein A-I: limited proteolysis of methionine-reduced and -oxidized lipid-free and lipid-bound human apo A-I. Biochemistry. 1997;36:7615–7624. doi: 10.1021/bi962952g. [DOI] [PubMed] [Google Scholar]
- 17.Susi H, Byler DM. Resolution-enhanced Fourier transform infrared spectroscopy of enzymes. Methods Enzymol. 1986;130:290–311. doi: 10.1016/0076-6879(86)30015-6. [DOI] [PubMed] [Google Scholar]
- 18.Tamm LK, Tatulian SA. Infrared spectroscopy of proteins and peptides in lipid bilayers. Q Rev Biophys. 1997;30:365–429. doi: 10.1017/s0033583597003375. [DOI] [PubMed] [Google Scholar]
- 19.Oh KJ, Altenbach C, Collier RJ, Hubbell WL. Site-directed spin labeling of proteins. Applications to diphtheria toxin. Methods Mol Biol. 2000;145:147–169. doi: 10.1385/1-59259-052-7:147. [DOI] [PubMed] [Google Scholar]
- 20.Gillmore JD, Stangou AJ, Tennent GA, Booth DR, O'Grady J, Rela M, Heaton ND, Wall CA, Keogh JA, Hawkins PN. Clinical and biochemical outcome of hepatorenal transplantation for hereditary systemic amyloidosis associated with apolipoprotein AI Gly26Arg. Transplantation. 2001;71:986–992. doi: 10.1097/00007890-200104150-00026. [DOI] [PubMed] [Google Scholar]
- 21.Rader DJ, Gregg RE, Meng MS, Schaefer JR, Zech LA, Benson MD, Brewer HB., Jr In vivo metabolism of a mutant apolipoprotein, apoA-IIowa, associated with hypoalphalipoproteinemia and hereditary systemic amyloidosis. J Lipid Res. 1992;33:755–763. [PubMed] [Google Scholar]
- 22.Genschel J, Haas R, Propsting MJ, Schmidt HH. Apolipoprotein A-I induced amyloidosis. FEBS Lett. 1998;430:145–149. doi: 10.1016/s0014-5793(98)00668-1. [DOI] [PubMed] [Google Scholar]
- 23.Uversky VN, Fink AL. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim Biophys Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Sunde M, Blake CC. From the globular to the fibrous state: protein structure and structural conversion in amyloid formation. Q Rev Biophys. 1998;31:1–39. doi: 10.1017/s0033583598003400. [DOI] [PubMed] [Google Scholar]
- 25.Oberg KA, Ruysschaert JM, Goormaghtigh E. The optimization of protein secondary structure determination with infrared and circular dichroism spectra. Eur J Biochem. 2004;271:2937–2948. doi: 10.1111/j.1432-1033.2004.04220.x. [DOI] [PubMed] [Google Scholar]
- 26.Williams RW, Chang A, Juretic D, Loughran S. Secondary structure predictions and medium range interactions. Biochim Biophys Acta. 1987;916:200–204. doi: 10.1016/0167-4838(87)90109-9. [DOI] [PubMed] [Google Scholar]
- 27.Parrini C, Taddei N, Ramazzotti M, Degl'Innocenti D, Ramponi G, Dobson CM, Chiti F. Glycine residues appear to be evolutionarily conserved for their ability to inhibit aggregation. Structure (Camb) 2005;13:1143–1151. doi: 10.1016/j.str.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 28.Nolte RT, Atkinson D. Conformational analysis of apolipoprotein A-I and E-3 based on primary sequence and circular dichroism. Biophys J. 1992;63:1221–1239. doi: 10.1016/S0006-3495(92)81698-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiti F, Taddei N, Baroni F, Capanni C, Stefani M, Ramponi G, Dobson CM. Kinetic partitioning of protein folding and aggregation. Nat Struct Biol. 2002;9:137–143. doi: 10.1038/nsb752. [DOI] [PubMed] [Google Scholar]
- 30.de Sousa MM, Vital C, Ostler D, Fernandes R, Pouget-Abadie J, Carles D, Saraiva MJ. Apolipoprotein AI and transthyretin as components of amyloid fibrils in a kindred with apoAI Leu178His amyloidosis. Am J Pathol. 2000;156:1911–1917. doi: 10.1016/S0002-9440(10)65064-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 32.Lashuel HA. Membrane permeabilization: a common mechanism in protein-misfolding diseases. Sci Aging Knowledge Environ. 2005;2005:pe28. doi: 10.1126/sageke.2005.38.pe28. [DOI] [PubMed] [Google Scholar]
- 33.Volles MJ, Lansbury PT., Jr Zeroing in on the pathogenic form of alpha-synuclein and its mechanism of neurotoxicity in Parkinson's disease. Biochemistry. 2003;42:7871–7878. doi: 10.1021/bi030086j. [DOI] [PubMed] [Google Scholar]
- 34.Sato K, Higuchi M, Iwata N, Saido TC, Sasamoto K. Fluoro-substituted and 13C-labeled styrylbenzene derivatives for detecting brain amyloid plaques. European journal of medicinal chemistry. 2004;39:573–578. doi: 10.1016/j.ejmech.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 35.Naiki H, Higuchi K, Hosokawa M, Takeda T. Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Analytical biochemistry. 1989;177:244–249. doi: 10.1016/0003-2697(89)90046-8. [DOI] [PubMed] [Google Scholar]
- 36.Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol. 1998;8:101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]