Abstract
Spliced leader trans-splicing adds a short exon, the spliced leader (SL), to pre-mRNAs to generate 5’ ends of mRNAs. Addition of the SL in metazoa also adds a new cap to the mRNA, a trimethylguanosine (m32,2,7GpppN)(TMG) that replaces the typical eukaryotic monomethylguanosine (m7GpppN)(m7G) cap. Both trans-spliced (m32,2,7GpppN-SL-RNA) and not trans-spliced (m7GpppN-RNA) mRNAs are present in the same cells. Previous studies using cell-free systems to compare the overall translation of trans-spliced vs. non-trans-spliced RNAs led to different conclusions. Here we examine the contribution of m32,2,7GpppG-cap and SL sequence and other RNA elements to in vivo mRNA translation and stability in nematode embryos. Although 70–90% of all nematode mRNAs have a TMG-cap, the TMG cap does not support translation as well as an m7G-cap. However, when the TMG cap and SL are present together, they synergistically interact and translation is enhanced, indicating both trans-spliced elements are necessary to promote efficient translation. The SL by itself does not act as a cap-independent enhancer of translation. The poly(A)-tail synergistically interacts with the mRNA cap enhancing translation and plays a greater role in facilitating translation of TMG-SL mRNAs. In general, recipient mRNA sequences between the SL and AUG and the 3’ UTR do significantly contribute to the translation of trans-spliced mRNAs. Overall, the combination of TMG cap and SL contribute to mRNA translation and stability in a manner typical of a eukaryotic m7G-cap and 5’ UTRs, but they do not differentially enhance mRNA translation or stability compared to RNAs without the trans-spliced elements.
Keywords: Trimethylguanosine cap, spliced leader, translation, trans-splicing, nematode, Ascaris, mRNA stability
Introduction
Spliced leader (SL) trans-spliced mRNAs are formed by splicing a small exon, the spliced leader (16–52 nt depending on the species), to a splice acceptor site in an independently transcribed pre-mRNA [1–5]. SL addition thereby generates the mature 5’ end of the mRNA. This form of gene expression is present in diverse eukaryotes including some protozoa (Sarcomastigophora), several major invertebrate lineages (cnidarians, flatworms, rotifers, and nematodes), and in primitive chordates (tunicates) [6–12].
Trans-splicing in the Kinetoplastid protozoans serves to resolve polycistronic transcripts into mature monocistronic mRNAs by trans-splicing at the 5’ end of each protein coding cistron [13–16][for review see [17]]. Analysis of a subset of these polycistronic transcripts indicates, that unlike protein coding genes in most eukaryotes, these primary transcripts are uncapped [18]. Thus, trans-splicing, in addition to resolving these polycistronic transcripts into monocistronic mRNAs, also serves as a capping mechanism for some transcripts, and adds a novel cap 4 structure (m7G-ppp-N6,N6,2-O-trimethyladenosinep-2-O-methyladenosine-p-2-O-methylcytosine-p-N3,2–O-methyluridine) to the RNAs [19]. Polycistronic transcripts are also produced in C. elegans [1, 20, 21] and the organization of a number of trans-spliced genes in other nematodes, flatworms and tunicates suggests they are likely to be polycistronic [22–25]. Thus, a major function of trans-splicing in diverse eukaryotes is the resolution of polycistronic transcription units to form monocistronic, capped mRNAs.
The majority of trans-spliced transcripts (~50–80%) in nematodes are not derived from polycistronic transcripts. While the SL RNA substrate and a spliced leader are necessary for C. elegans embryogenesis [26, 27], the function of most trans-splicing in nematodes (and other metazoa) remains unknown. It has been proposed that trans-splicing in metazoa might be functionally correlated with unusual characteristics of the promoters, transcription, capping, or the 5’ untranslated regions (UTR) of trans-spliced genes or that it plays a role in mRNA export, localization, translation, or stability [1–3, 5, 28, 29].
The SL RNA in metazoa has a trimethylated (m32,2,7GpppN or trimethylguanosine = TMG) cap in which methyl groups are also present at the N2 position of guanine [9, 10, 24, 30, 31]. This cap is transferred to mRNAs with the spliced leader sequence producing m32,2,7GpppN – SL – mRNAs [32, 33]. Although 100% of the mRNA are trans-spliced in the Kinetoplastida, this is not the case in metazoa where from ~10% to 80% of mRNA are trans-spliced. Consequently, two populations of mRNAs co-exist in these organisms: 1) non-trans-spliced mRNAs with the m7GpppN cap structure and different sequences for the 5’ UTR of each mRNA and 2) trans-spliced mRNAs with the TMG cap and conserved spliced leader.
mRNA cap structure and the 5’ UTR play important roles in gene expression contributing to efficient mRNA translation and stability [34–36]. In previous studies using Ascaris cell-free translation systems [37, 38], one study concluded that mRNAs with trans-spliced elements were most efficiently translated whereas a second study observed non-trans-spliced mRNAs were slightly more efficiently translated. Here, we describe in vivo studies extending our understanding of nematode mRNA translation and further examine the hypothesis that TMG cap and conserved spliced leader sequence differentially contribute to mRNA translation and stability in early embryos of the nematode Ascaris. The current data indicate that the addition of trans-spliced elements to an RNA does not appear to significantly enhance the translation or stability of the mRNAs compared with non-trans-spliced 5’ UTRs. In addition, our in vivo studies demonstrate that a previously described Ascaris embryo cell-free translation system [37] faithfully reproduces many aspects of in vivo translation.
Materials and Methods
Isolation and Preparation of Ascaris Embryos
Adult female Ascaris were obtained from Carolina Biological Supply, the eggs isolated from uteri, stored, embryonated, and prepared for particle bombardment essentially as described [39].
Biolistics
Preparation of gold microcarriers and biolistics were performed as previously described [39] with the following modifications: 1) gold particles were spherical ~2.2 μm gold from Degussa (10KM) (dmc2 Metals Group, South Plainfield, NJ) and 2) instead of lyophilization of the RNA onto the gold carriers, an alcohol precipitation was used. In vitro transcribed RNAs were precipitated onto gold particles using 2.5 M ammonium acetate/ethanol precipitation using 0.1–5 μg of reporter RNA per 1 mg of gold particles. The RNA/gold pellet was washed with 200 μl of ice-cold ethanol, resuspended in ice-cold 100% ethanol (18 μl/mg gold) and spread onto macrocarriers and processed as previously described [39].
Binding of RNA onto the gold particles was evaluated by formaldehyde agarose gel analysis to determine the integrity of RNA bound to the beads, and RiboGreen (Molecular Probes, Carlsbad, CA) [40] was used to quantitate RNA loaded onto the particles for data normalization. Analysis of RNA bound to beads indicated little to no degradation of the RNA occurred prior to particle bombardment and that typically 80–85% of the precipitated RNA was bound onto the gold particles. Approximately 800,000 Ascaris embryos were evenly spread onto 35-mm Petri dishes and bombarded in a Bio-Rad Biolistic PDS-1000/HE Particle Delivery System at 15-in Hg of chamber vacuum, target distance of 3 cm (stage 1), and 1,350-psi particle acceleration pressure. After bombardment, 2–3 ml of nematode blastomere media [41] was added, and the embryos were incubated in an air shaker at 30°C at 110 rpm (to keep the embryos in suspension). Following incubation, embryos were collected, pelleted, and assayed for luciferase activity. Evaluation of gold particle delivery was carried out as previously described [39].
Transcription Template and RNA Preparation
PCR templates for in vitro transcription reactions were derived from constructs made in Promega plasmids pGL3 Basic and pRLnull or pGLUC Basic-1 (Targeting Systems, Santee, CA). Using PCR primers that added a 5’ T7 promoter and additional sequences to the 5’ or 3’ end of the template, transcription templates were generated that contained different 5’ UTRs, a luciferase open reading frame (Firefly, Renilla, or Gaussia) and 3’UTRs (derived from pGL3, pRLnull, pGluc-1, the Ascaris vacuolar ATP synthase 16 kDa proteolipid subunit RNA, and the Ascaris ribosomal protein L23 RNA) with or without a poly(A) tail of various lengths. RNAs were generated by in vitro transcription from these templates using a T7 MegaScript kit as described by the manufacturer (Ambion, Austin, TX) with either no cap or ApppG, m7GpppG, or m32,2,7GpppG cap at a ratio of 4:1 or 10:1 cap analog to GTP. The 5’ and 3’ UTRs of the synthesized RNAs are provided in Suppl. Figure 1. Transcription reactions were DNase I-treated, extracted with TRIzol (Invitrogen, Carlsbad, CA), and the RNAs precipitated twice, once with isopropanol and then with ammonium acetate/ethanol. Precipitated RNAs were further washed with 70% ethanol, dissolved in water, quantitated spectrophotometrically, and examined by agarose-formaldehyde denaturing gel electrophoresis. Capping efficiency and orientation on transcripts was carried out as described and indicated that similar capping efficiencies for the two caps and that 90% of the m7GpppG and 80% of the m32,2,7GpppG caps were added in the correct orientation [37]. These analyses demonstrated that differences in translation or stability observed were not a function of differences in capping efficiency or orientation on the transcripts.
Luciferase Assays
Following introduction of the RNA by particle bombardment, embryos were incubated in 35-mm Petri dishes and processed as previously described using Promega Renilla, Luciferase, or Dual Assay Systems with a Sirius Luminometer (Berthold Detection Systems, Germany) [39]. Although experimental data were similar with or without normalization to protein and RNA concentrations in the samples as previously described [39], in a number of experiments normalization was carried out by co-transfection of a firefly reporter RNA with either Renilla or Gaussia RNA and the use of the Dual Luciferase Assay.
Kinetic Determination of RNA Translation Efficiency and Functional Half-life
The kinetics of luciferase activity over time in the embryos was used to determine both in vivo mRNA translational efficiency and functional half-life [42]. Translational efficiency was determined from the slope of the rising curve derived from luciferase activity over time using the equation Y=Ymax*(1-exp[-K*X]). Functional half-life, or the length of time over which the mRNA is actively engaged in translation, was determined and defined as the time required following mRNA delivery to reach 50% of the final level of protein produced and calculated using the equation t1/2=0.69/K. Functional half-life measures the length of over time in which the mRNA is actively engaged in translation. In comparison, physical half-life only measures the physical longevity of the RNA without regard to translational competence and may not readily reflect changes in mRNA poly(A) tail length or mRNA cap removal which play important roles in mRNA translation.
Analysis of Physical RNA Half-life
RNA transcripts were biolistically introduced into Ascaris embryos and total RNA isolated from the embryos over time using Trizol [39]. For PCR analyses of transfected RNAs in the embryos, cDNA was synthesized using random hexamers and Superscript III as recommended by the manufacturer (Invitrogen) and semi-quantitative PCR carried out as described (9) or by Real-Time PCR using SYBR Green and a Roche LightCycler 480. For analysis of the decay of 32P-labeled transcripts, 32P-GTP labeled transcripts were synthesized using Promega’s Riboprobe in vitro transcription system. Decay of labeled RNAs introduced into embryos was analyzed and quantified by phosphorimager analysis of denaturing gel electrophoresis separations using Molecular Dynamics STORM 860 and ImageQuant software.
Results
Nematode translation is highly m7G cap-dependent
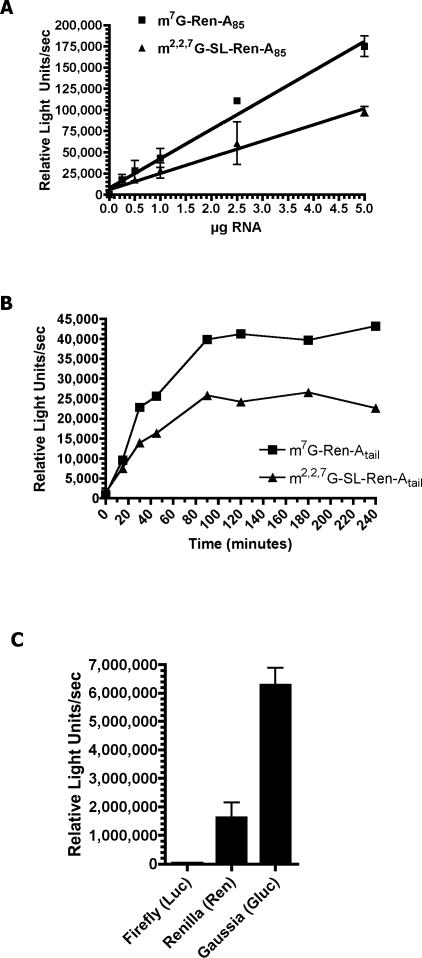
Three different luciferase reporters (Firefly, Renilla, and Guassia) were used to evaluate the contribution of the TMG cap and SL to RNA translation and stability in vivo. Reporter RNAs were synthesized in vitro with different caps (or no cap), 5’ UTRs, luciferase reporter open reading frames, 3’ UTRs, and poly(A) tails (or no tail) (see Suppl. Figure 1 for reporter characteristics). These RNAs were then biolistically introduced into ~800,000, 32–64 cell Ascaris embryos and luciferase activity assayed (Davis et al., 1999). The relative activities, time courses and dose responses were examined for all three reporters (Figure 1; Suppl. Figure 2; some data not shown). All three reporters demonstrated a linear dose response and were used at the lower end of the linear range (2.5 μg Firefly; 1 μg Renilla; 0.5 μg Gaussia). Maximal luciferase activity following introduction of the reporter RNA was typically reached at 60 min (Gaussia) or 90 min (Firefly and Renilla).
Figure 1. Luciferase RNA reporter dose response, activity over time, and comparison of reporter activities in Ascaris embryos.
A). Renilla (Ren) RNA reporter dose response. Varying amounts of the illustrated RNAs (0.25 to 5 μg) were precipitated onto gold particles, the particles introduced into 32–64 cell Ascaris embryos, the embryos incubated at 30 C for 90 minutes, and the embryos collected and assayed for luciferase activity. B) Renilla luciferase activity over time following introduction into Ascaris embryos. The 30 nt 5’ UTR for m7G-Ren is GGUCAUUCCGGUACUGUUAGGCUAGCCACCAUG and the 33 nt 5’ UTR for m2,2,7G-SL-Ren is GGUUUAAUUACCCAAGUUUGAGGGCUAGCCACCAUG, where orange is the SL sequence, grey Renilla sequence, and the green AUG the initiator methionine. C) Comparison of relative reporter luciferase activity after 90 min following introduction of 1 μg of reporter RNA. The reporter RNAs contained an m7GpppG cap and 75–85 nt poly(A) tail and are described in Suppl. Figure 1. All data illustrate representative experiments and show the mean and standard error derived from triplicate transfections for A and C and the mean for triplicate transfections in B.
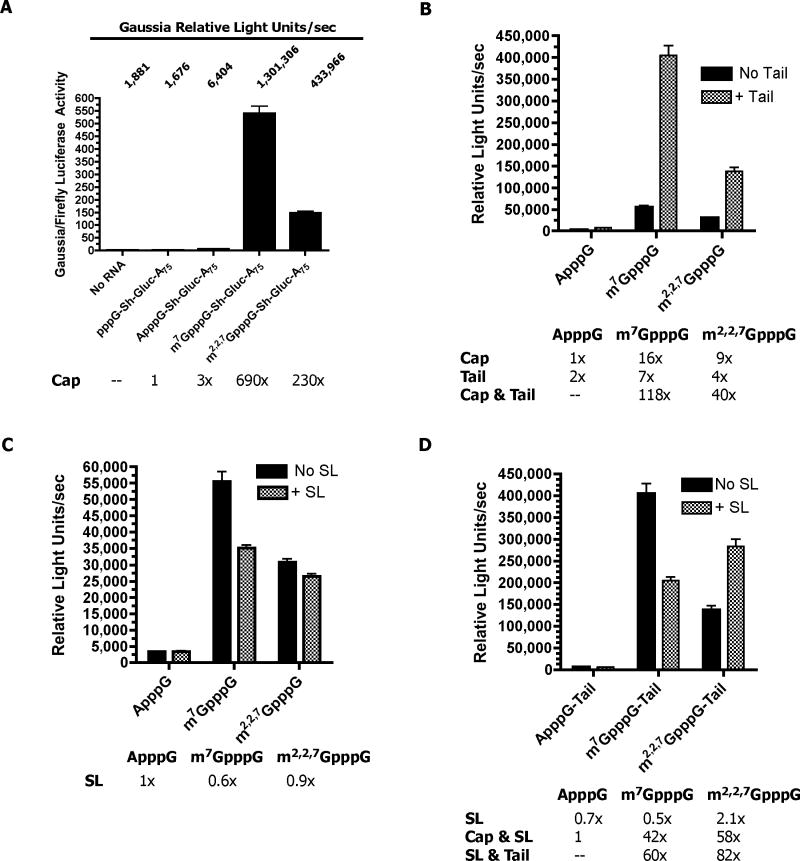
To provide a framework for the cap-dependence of nematode translation, we initially compared the translation of uncapped (pppG), GpppG-, and m7GpppG-capped RNAs. Luciferase activity derived from uncapped (pppG) RNAs was not detectable whereas significant levels of luciferase activity were derived from m7GpppG-capped RNAs (Figure 2A; Suppl. Figure 3). Translation of GpppG-capped RNAs was also observed (Suppl. Figure 3). However, GpppG-capped RNAs introduced into Ascaris cell-free extracts or embryos are rapidly N7-methylated [37](data not shown). Addition of the methyl donor inhibitor SAH (S-adenosyl-homocysteine) prevents N7-methylation and eliminates translation of GpppG-capped RNA in the cell- free extracts. Overall, these data indicate that the GpppG-capped RNAs are not the direct substrates for translation.
Figure 2. Translation in Ascaris embryos is methyl-guanosine cap-dependent, enhanced by a poly(A) tail, and both a spliced leader and poly(A) tail are required for efficient translation of TMG-capped RNAs.
A) Methyl-guanosine cap dependence of translation. The reporter RNA was Gaussia (Sh Gluc)(5′ UTR = GGUACCGAGCUCGGAUCCAGCCACCAUG) with a 75 nt poly(A) tail (see Suppl. Figure 1). (B) Poly(A) tail enhances translation of methyl-guanosine capped RNAs. The reporter RNA was Ren (5′ UTR = GGUCAUUCCGGUACUGUUAGGCUAGCCACCAUG)(see Suppl. Figure 1) with and without an 85 nt poly(A) tail. C) Effect of spliced leader on translation of different capped RNAs. The reporter RNAs were Ren (no SL) or SL Ren (+ SL) without a poly(A) tail (see Suppl. Figure 1). D) Effect of spliced leader and poly(A) tail on translation of different capped RNAs. The reporter RNAs were Ren (no SL)(5′ UTR = GGUCAUUCCGGUACUGUUAGGCUAGCCACCAUG) or SL Ren (+ SL)(5′ UTR = GGUUUAAUUACCCAAGUUUGAGGGCUAGCCACCAUG) with an 85 nt poly(A) tail (see Suppl. Figure 1). Gaussia (A) or Renilla (B-D) reporter RNAs were biolistically introduced into Ascaris embryos and the luciferase activity measured after 90 minutes. Data illustrate representative experiments and show the mean and standard error derived from triplicate transfections. The Gaussia activity was normalized to a co-transfected Firefly RNA using the Dual-Luciferase Assay.
In vivo methylation of GpppG-capped RNAs precluded our use of this cap as a baseline for guanine methylation-dependent translation. As an alternative, we examined ApppG-capped reporter RNAs, reasoning that the 5’ end of the mRNA would likely be more stable than pppG, as its 5’ –ppp- 5’ linkage would protect the 5’ end of the mRNA from degradation (Figure 2A). Translation of ApppG capped RNAs was detectable at a low level and therefore we used this capped RNA as a baseline for comparisons with other methylated, guanosine capped RNAs in subsequent experiments (Figure 2B–D).
TMG cap does not support translation as efficiently as an m7GpppG cap
We next examined the individual contributions of N7- and N2-methylation of the guanosine cap to translation. Nematode embryo translation is highly m7G-cap-dependent, as illustrated by the comparison between identical RNAs that differ only in an ApppG-vs. m7GpppG-cap (Figures 2A–B, Table 1). Overall, translation of TMG-capped RNA (2 additional methyl groups at N2) was 2–3 fold lower than m7G-capped RNA. The m7GpppG-cap dependence and lower overall translation of TMG-capped RNA were observed with all three luciferase reporters and different 5’ and 3’ UTRs (see Figure 3A–B; Suppl. Figure 3; data not shown).
Table 1. Effect of cap, poly(A) tail, and spliced leader sequence on translation of Renilla RNAs in Ascaris embryos.
The illustrated RNAs were biolistically introduced into Ascaris embryos and the luciferase activity measured after 2 hr. Relative light units/sec represent the mean and standard error of triplicate transfections from the experiments illustrated in Figure 2. Bold numbers within a column represent the level of increase over a baseline RNA shown as the number 1.
| Renilla Luciferase Activity (RLU/sec) | Effect of Methylated Capa | Effect of Poly(A)-Tailb | Effect of Methylated Cap AND Poly(A)-tailc | Effect of Spliced Leaderd | Effect of Methylated Cap AND Spliced Leadere | Effect of Spliced Leader AND Poly(A)-tailf | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| ApppG-Ren | 3,428 ± 47 | 1 | 1 | 1 | 1 | 1 | 1 |
| m7GpppG-Ren | 55,578 ± 2,993 | 16 | 1 | 1 | 1 | ||
| m2,2,7GpppG-Ren | 30,799 ± 850 | 9 | 1 | 1 | 1 | ||
|
|
|||||||
| ApppG-Ren-A85 | 6,945 ± 247 | 1 | 2 | 1 | 1 | ||
| m7GpppG-Ren-A85 | 404,639 ± 22,762 | 58 | 7 | 118 | 1 | ||
| m2,2,7GpppG-Ren-A85 | 137,325 ± 9,638 | 20 | 4 | 40 | 1 | ||
|
|
|
||||||
| ApppG-SL-Ren | 3417 ± 231 | 1 | 1 | 1 | 1 | 1 | |
| m7GpppG-SL-Ren | 31,450 ± 3,712 | 9 | 1 | 0.6 | 9 | ||
| m2,2,7GpppG-SL-Ren | 28,581 ±2,223 | 8 | 1 | 0.9 | 8 | ||
|
|
|
||||||
| ApppG-SL-Ren-A85 | 4,881 ±527 | 1 | 1 | 0.7 | 1 | 1 | |
| m7GpppG-SL-Ren-A85 | 203,995 ± 8,797 | 42 | 6 | 60 | 0.5 | 29 | 3.7 |
| m2,2,7GpppG-SL-Ren-A85 | 282,684±16,863 | 58 | 10 | 83 | 2.1 | 40 | 9.2 |
|
| |||||||
effect of methylated guanosine cap on different RNAs (baseline is the same RNA with an ApppG cap)
effect of poly(A)-tail on different capped RNAs (baseline is the same RNA without a tail)
effect of methylated guanosine cap and poly(A)-tail on different RNAs (baseline is an ApppG capped RNA without a tail)
effect of spliced leader on different RNAs (baseline is the same RNA without the SL)
effect of methylated cap and spliced leader on different RNAs (baseline is an ApppG capped RNA with same 3′ end)
effect of spliced leader and poly(A)-tail on different capped RNAs (baseline is same RNA without the SL or poly(A)-tail)
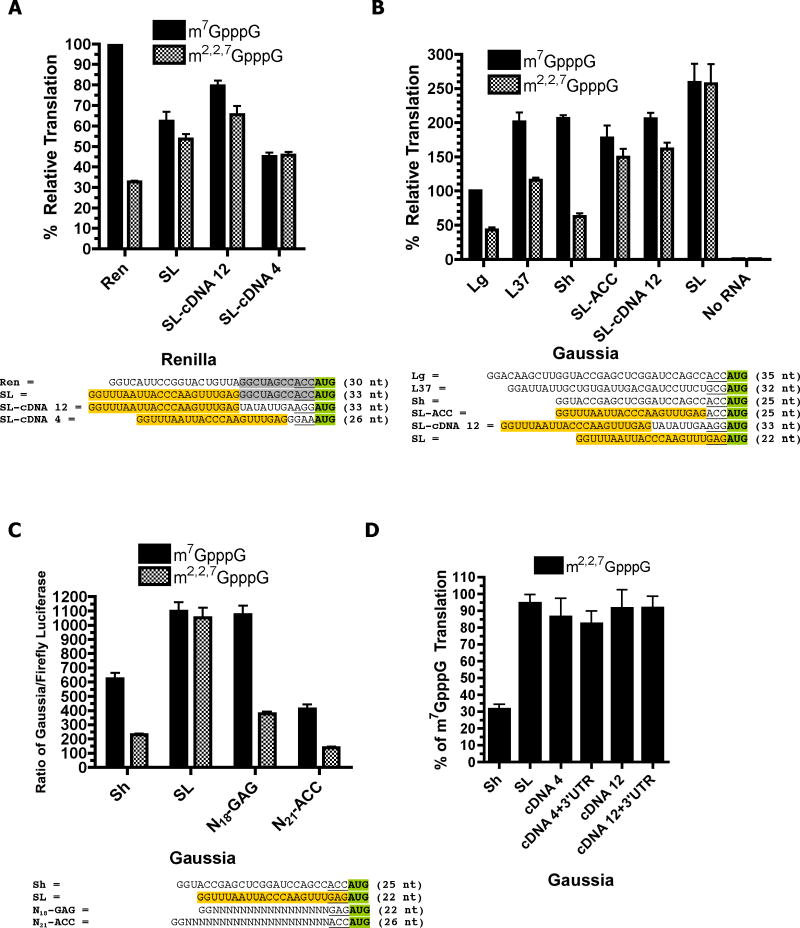
Figure 3. Contribution of m7GpppG vs m2,2,7GpppG cap, SL, and different 5’ and 3’ UTRs to translation in Ascaris embryos.
The illustrated RNAs (see Suppl. Figure 1 for additional information on the RNAs) and a Firefly control reporter RNA were biolistically co-transfected into 32–64 cell Ascaris embryos and luciferase activity measured after 60–90 minutes using a Dual-Luciferase assay. Activity was normalized to the co-transfected Firefly RNA activity. Data illustrate representative experiments and show the mean and standard error derived from a minimum of triplicate transfections. A). Comparison of Renilla, SL-Renilla and native trans-spliced mRNA 5’ UTRs on the translation of a Renilla RNA reporter. Normalized luciferase activity derived from the m7GpppG-Ren RNA was set at 100%. B). Effect of 5’ UTR length and native sequence on the translation of a Gaussia RNA reporter. Normalized luciferase activity derived from the m7GpppG-Lg Gaussia RNA was set at 100%. C). Effect of a Gaussia, SL, and random nucleotide 5’ UTRs and their initiation context on translation of a Gaussia RNA reporter. D). Effect of the native trans-spliced mRNA 5’ UTRs and their homologous 3’ UTRs on the translation of a Gaussia RNA reporter. Note that values represent the percentage of translation obtained the identical RNA except with an m7GpppG cap. The sequences for the 5’ UTRs are illustrated in A-C and the 3’ UTR sequences are in Suppl. Figure 1.
Poly(A) tail synergizes with methylated cap to enhance translation
Addition of a 85 nt poly(A) tail greatly increased translation of either m7GpppG- or m32,2,7GpppG-capped RNAs, whereas the tail had only a small effect on ApppG-capped RNA (Figure 2B and Table 1). The effect of the methylated, guanosine cap and poly(A) tail together on translation are synergistic (Table 2). Interestingly, this synergistic effect is greater for RNAs that have both the TMG cap and SL (the trans-spliced elements) compared to m7GpppG-capped RNAs.
Table 2. Ascaris embryo translation in vivo exhibits synergistic effects between the cap, poly(A) tail, and spliced leader sequence.
Data derived from Table 1 were used to calculate the synergistic effects as illustrated in Supplemental Tables 1–3 and the results summarized here. The set of RNAs evaluated to determine the synergism are illustrated.
| Synergistic Effect of Cap and Taila | Synergistic Effect of Cap and SLb | Synergistic Effect of SL and Tailc | |||
|---|---|---|---|---|---|
|
| |||||
| ApppG-Ren | ApppG-Ren-A85 | m7GpppG-Ren | |||
| m7GpppG-Ren | m7GpppG-Ren-A85 | m7GpppG-SL-Ren | |||
| ApppG-Ren-A85 | 6 | ApppG-SL-Ren-A85 | 0.5 | m7GpppG-Ren-A85 | 0.5 |
| m7GpppG-Ren-A85 | m7GpppG-SL-Ren-A85 | m7GpppG-SL-Ren-A85 | |||
|
| |||||
| ApppG-SL-Ren | ApppG-Ren-A85 | m2,2,7GpppG-Ren | |||
| m2,2,7GpppG-SL-Ren | m2,2,7GpppG-Ren-A85 | m2,2,7GpppG-SL-Ren | |||
| ApppG-SL-Ren-A85 | 9 | ApppG-SL-Ren-A85 | 2 | m2,2,7GpppG-Ren-A85 | 1.8 |
| m2,2,7GpppG-SL-Ren-A85 | m2,2,7GpppG-SL-Ren-A85 | m2,2,7GpppG-SL-Ren-A85 | |||
effect of methylated cap and tail/(effect of methylated cap + effect of A-tail) from Table 1 and Suppl. Table 1
effect of methylated cap and SL/(effect of methylated cap + effect of SL) from Table 1 and Suppl. Table 2
effect of SL and tail/(effect of SL + effect of A-tail) from Table 1 and Suppl. Table 3
SL facilitates translation of TMG-capped RNA
In order to determine the effect of the SL sequence itself on translation in vivo, we substituted the 5’ terminal 22 nt of the Renilla 5’ UTR with the SL sequence. The effect of this substitution was an ~50% decrease in overall translation of a m7GpppG-capped RNA (Figure 2C and Tables 1–2). However, translation of m32,2,7GpppG-capped RNAs increased ~ 2-fold when the spliced leader was in the 5’ UTR. This 2-fold enhancement requires a poly(A) tail, as the same SL substitution in the 5’ UTR of a non-polyadenylated, m32,2,7GpppG-capped RNA had little effect on translation (Compare Figures 2C–D, Tables 1–2). These data suggest the spliced leader has a positive, synergistic effect on the translation of TMG-capped RNAs, whereas when it is substituted into the 5’ UTR of m7GpppG-capped RNAs, the SL has a negative effect on translation (Figure 2D, Tables 1–2). Importantly, optimal translation of TMG-capped RNAs requires a downstream SL and a poly(A) tail. The data also indicate that the SL by itself does not act as a cap-independent enhancer of translation (Figures 2C–D). Similar results were observed using Firefly reporter RNAs (data not shown).
Sequences between the SL and AUG in recipient mRNAs do not enhance translation of TMG-SL RNAs
The 22 nt nematode SL is typically added 0–10 nucleotides upstream of the AUG in the recipient RNA [37]. To further examine the contribution of the SL and other downstream sequences between the SL and AUG in the 5’ UTR on translation, we evaluated native Ascaris 5’ UTRs of three trans-spliced mRNAs. RNA transfection efficiencies in these experiments were normalized by co-transfection of a control Firefly luciferase RNA. Consistent with the results in Figure 2C–D, overall translation of TMG-capped reporter RNAs is considerably less compared to identical RNAs with an m7G-cap unless the SL sequence is present at the 5’ end (Figure 3A–B). Furthermore, sequences located between the SL and the initiation codon in native trans-spliced 5’ UTRs (SL-cDNA 12 and SL-cDNA 4) do not appear to play a large role in influencing translation of TMG-capped RNAs as illustrated by the comparison of the level of translation of the SL alone compared to SL-cDNA 12 or SL-cDNA 4 (Figure 3A).
To more closely examine the specificity of SL enhancement of the translation of TMG-capped RNAs, we compared the effect of the SL with non-trans-spliced 5’ UTRs (L37, Lg, and Sh)(Figure 3B) and random sequences (N18-GAG and N21-ACC)(Figure 3C) on the translation of TMG-capped RNA. Enhanced translation of TMG-capped RNA is observed only in the presence of the SL in the 5’ UTR. Overall, these data and those using three different luciferase reporter RNA 5’ UTRs (Firefly, Renilla, and Gaussia) demonstrate that the SL enhancement of TMG-capped RNA translation is a specific property of the SL sequence.
5’ UTR length and context affect translation
Overall translation for several reporter RNAs is also affected by the length of the 5’ UTR as illustrated by a comparison of the two Guassia 5’ UTRs (Sh vs Lg = Short vs Long)(Figure 3B) and random sequence 5’ UTRs (data not shown). Reporters with shorter 5’ UTRs exhibited higher overall levels of translation compared to those with longer 5’ UTRs. The SL sequence immediately adjacent to an initiation codon (see SL in Figures 3B–C) constitutes a relatively efficient 5’ UTR and initiation context for translation when directly compared to a Gaussia 5’ UTR of similar length (Sh). By comparison, the first 22 nts of the Firefly and Renilla 5’ UTRs provide a more efficient translation context in our experiments, as substitution of the SL for the first 22 nts of these 5’ UTRs led to a reduction in overall translation (Figures 2C–D and Figure 3A). Thus, a Gaussia 5’ UTR of the same length as the SL is only ~2/3 as efficient in translation when the two are directly compared. The lower level of translation of the Gaussia 5’ UTR is likely due in part to a combination of both its sequence and the -1 to -3 AUG context difference (SL = GAG, Gaussia = ACC). As shown in Figure 3B–C (compare SL ACC, SL, GN21-ACC, and GN18-GAG), the SL GAG provides a more efficient -1 to -3 AUG context than ACC. In addition, the GAG context may contribute to the synergistic interaction between the SL and TMG cap. Thus, our comparisons of several different 5’ UTRs to the SL and its context demonstrate that, as observed in other systems, the length and sequence of the 5’ UTR contribute to translation efficiency in nematodes. However, in general the SL is not a significantly better component of the 5’ UTR than those sequences to which it was compared in the current study.
Native 3’ UTRs of trans-spliced mRNAs do not enhance translation of TMG-SL RNA
In experiments described above, the overall luciferase reporter translation in the context of the TMG cap and SL sequence was similar when different luciferase 3’ UTRs were present (Firefly, Renilla, and Gaussia 3’ UTRs). To determine whether the 3’ UTR of a trans-spliced mRNA contributes to the translation of TMG-SL mRNAs, we examined reporter RNAs with both the native 5’ and 3’ UTRs of two trans-spliced Ascaris mRNAs (cDNA 12, encoding vacuolar H+ ATPase 16 kd proteolipid subunit, and cDNA 4, encoding ribosomal protein L23)(Figure 3D). Reporter RNA activity was not significantly affected when both the native 5’ and 3’ UTR of these trans-spliced messages were present compared to only the trans-spliced 5’ UTR. Similar data were obtained with a Renilla and Firefly reporter (data not shown). These data demonstrate, that in the context of a luciferase reporter, the native 3’ UTRs did not cooperate with the SL or directly contribute to the in vivo translation of trans-spliced mRNAs.
TMG and SL do not play a differential role in 1–2 cell embryos
In the above studies, we examined the role of the TMG-cap and SL on translation in 32–64 cell embryos. To determine whether translation of trans-spliced mRNAs was the same in earlier embryos and/or in a discrete subset of embryo cells, we examined the translation of several mRNAs in 1–2 cell embryos. Translation of RNAs with TMG-cap alone compared to RNAs with an m7GpppG-cap was even lower in early embryos. Overall, similar data were obtained for 1–2 cell embryos as observed for the 32–64 cell embryos described above (Supplementary Figure 4).
TMG cap and SL together do not differentially affect RNA stability
The luciferase activity data presented above represent the sum of both RNA translation efficiency and stability. To differentiate the contributions of the TMG cap and SL on translation efficiency and functional RNA half-life, the kinetics of luciferase activity over time following RNA introduction into the embryos were determined (see Materials and Methods)[42]. This methodology facilitates analysis of the independent contributions of RNA elements to translation and stability. This point is illustrated for 3 different poly(A) tail lengths in Table 3 and Supplemental Figure 5. Increasing poly(A) tail length acts to enhance translation and to increase functional RNA half-life. Thus, the increased overall level of translation as a function of poly(A) tail length is a function of both an increase in mRNA translation efficiency and functional RNA half-life.
Table 3. Poly(A) tail affects both translation and stability in Ascaris embryos.
Data from the time course illustrated in Supplemental Figure 5 were used to calculate translation efficiency (Relative luciferase light units/sec/min), functional half-life (min), and maximum accumulation of luciferase (Relative luciferase light units/sec) as described in the Materials and Methods. Note that increasing poly(A) tail length on the m32,2,7GpppG-SL-Renilla RNA affects both the translation efficiency and half-life which together contribute to the overall level of relative expression. Similar results were obtained with an m7GpppG-Renilla RNA and Firefly RNAs. Data illustrate the mean of triplicate transfections.
| RNA | Translational Efficiency (RLU/sec/min) | Relative Rate of Translation | Functional half-life (min) | Relative mRNA Stability | Maximum Accumulation (RLU/sec) | Relative Level of Expression |
|---|---|---|---|---|---|---|
|
| ||||||
| m2,2.7GpppG-SL-Ren-A0 | 341 | 1.0 | 19 | 1.0 | 17,487 | 1.0 |
| m2,2,7GpppG-SL-Ren-A30 | 998 | 2.9 | 39 | 2.1 | 65,360 | 3.7 |
| m2,2,7GpppG-SL-Ren-A85 | 1291 | 3.8 | 57 | 3.0 | 120,630 | 6.9 |
|
| ||||||
As illustrated in previous experiments (Figure 2B), the SL or TMG cap added to a Renilla test RNA results in a reduction in translational efficiency (Table 4). The SL sequence substituted for sequences in the 5’ UTR of a Renilla test RNA causes a small decrease in mRNA stability. The reduction in both translation and stability by substitution of the SL by itself leads to an overall decrease in relative luciferase expression. Substitution of the m7G-cap with a TMG cap results in a significant decrease in translation efficiency, but does not affect functional mRNA half-life. Importantly, as shown in Table 4, the overall translational efficiencies and mRNA stabilities of non-trans-spliced (m7GpppG-Ren-A85) and trans-spliced test mRNAs (m2,2,7GpppG-SL-Ren-A85) are similar. Similar data were obtained with a Firefly reporter RNA and the functional half-lives of Gaussia reporter RNAs were not different (data not shown). In summary, the 1) luciferase activity data in previous sections are largely a function of overall translation efficiency and 2) RNAs that have both the TMG-cap and SL (representing trans-spliced RNA) do not have significantly different functional half-lives compared to similar RNAs with the m7GpppG cap (representing non-trans-spliced RNA).
Table 4. Trans-spliced and non-trans-spliced mRNAs have similar translation efficiencies and functional half-lives following introduction into Ascaris embryos.
The illustrated RNAs were introduced into Ascaris embryos and samples of embryos collected at time intervals and assayed for luciferase activity. The time course of luciferase activity was used to calculate translation efficiency (Relative luciferase light units/sec/min), functional half-life (min), and maximum accumulation of luciferase (Relative luciferase light units/sec) as described in Materials and Methods. Data illustrate a representative experiment and represent the mean of triplicate transfections.
| RNA | Translational Efficiency (RLU/sec/min) | Relative Rate of Translation | Functional half-life (min) | Relative mRNA Stability | Maximum Accumulation (RLU/sec) | Relative Level of Expression |
|---|---|---|---|---|---|---|
|
| ||||||
| m7GpppG-Ren-A85 | 979 | 1.0 | 56 | 1.0 | 158,479 | 1.0 |
| m7GpppG-SL-Ren-A85 | 820 | 0.8 | 43 | 0.8 | 62,420 | 0.4 |
| m2,2,7GpppG-Ren-A85 | 396 | 0.4 | 57 | 1.0 | 49,036 | 0.3 |
| m2,2,7GpppG-SL-Ren-A85 | 850 | 0.9 | 54 | 1.0 | 137,175 | 0.9 |
|
| ||||||
To further compare the half-lives of these mRNAs, we also examined the physical stability of m7G-capped (non-trans-spliced) and TMG-capped SL (representing trans-spliced) reporter RNAs in Ascaris embryos using PCR (semi-quantitative PCR and real-time PCR of Renilla and Gaussia RNAs) and the decay of 32-P-labeled Gaussia transcripts. The decay slopes for the two RNAs were similar (Figure 4 and data not shown). As shown in Figure 4, levels of RNA drop dramatically within the first 10 minutes following RNA transfection. Following biolistic introduction of the RNAs, media added to the embryos contained RNase A to limit evaluation of RNA present in the media or adhering to the surface of the embryos from our analysis. We attribute the dramatic drop in RNA in the first 10 minutes to the decay or degradation of RNAs that are not incorporated into Ascaris embryo cells. Similar data were obtained using real-time PCR (data not shown). In addition, differences in the half-lives of RNAs determined using the kinetics of Firefly luciferase in Table 4 and Gaussia physical half-life in Figure 4 are attributable to different luciferase RNAs (Renilla in Table 4 and Gaussia in Figure 4) and the methods used to measure RNA half-lives. The important observation derived from these experiments is that trans-spliced and non-trans-spliced RNAs have similar half-lives in vivo demonstrating that the mRNA stabilities of trans-spliced and non-trans-spliced reporter RNAs are similar.
Figure 4. Physical half-life of trans-spliced vs non-trans-spliced test mRNA in Ascaris embryos.
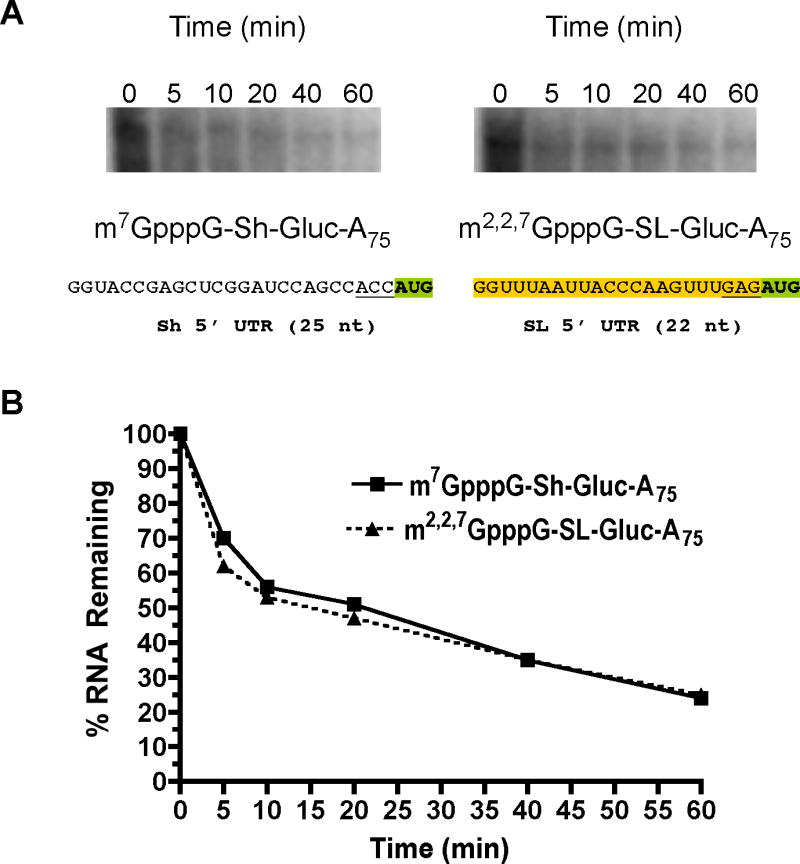
A). Decay of labeled Gaussia luciferase RNAs in embryos. 32P-labeled RNAs were introduced into embryos (see Suppl. Figure 1 for additional RNA information; orange shaded sequence is the SL), RNA extracted at intervals, the RNA resolved by electrophoresis, and visualized by phosphoimager analysis. B). Decay curves of 32P-labeled Gaussia RNAs in A. RNA levels illustrated in A were quantitated by phosphoimager analysis and plotted using levels observed at time zero as 100%. Similar data were obtained using transfection of unlabeled RNAs and analysis by real-time PCR (data not shown).
Real-time PCR analysis of luciferase RNAs over time following transfection enabled us to normalize luciferase activity to RNA levels in the embryos. These analyses (data not shown) led to similar data as those illustrated in Figures 2–3 and Tables 1 and 4. Thus, overall translation differences observed were not a function of different levels of RNA introduced or present in the embryos.
Discussion
Spliced leader trans-splicing adds a trimethylguanosine cap and conserved 5’ terminal sequence to metazoan mRNAs. Given the importance of the cap and 5’ UTR on mRNA translation, it has been proposed that spliced leader addition might function to enhance translation of recipient mRNAs. In the current in vivo study, we determined whether the TMG cap and SL acquired by SL trans-splicing differentially affected RNA translation and stability compared to m7GpppG-capped RNAs with several different 5’ UTRs. We also examined the contributions of different caps (ApppG, m7GpppG, and m2,2,7GpppG), the SL, trans-spliced RNA sequences between the SL and AUG, trans-spliced mRNA 3’ UTRs, and a poly(A) tail on in vivo mRNA translation in Ascaris embryos. Uncapped or ApppG-capped RNAs are very poorly translated (Figure 2A and Suppl. Figure 3). As observed for other eukaryotes, nematode mRNA translation in vivo is highly m7G-cap dependent (Figure 2, Table 1, Suppl. Figure 3), the cap synergizes with the poly(A) tail to enhance translation (Figure 2 and Tables 1 & 2), and the poly(A) tail is also a determinant of mRNA stability (Table 3). The TMG cap by itself does not support translation as well as an m7G-cap even though it is present on the majority of nematode mRNAs (Figures 2–3, Tables 1 & 4). However, when the TMG cap and SL are present together as is the case for trans-spliced RNAs, the SL synergistically enhances translation of TMG-capped, polyadenylated RNA (Figures 2–3, Suppl. Figure 4, and Tables 1 & 4). These in vivo studies extend previous studies carried out using Ascaris embryo cell-free systems [37, 38]. In addition, our in vivo studies demonstrate that several key characteristics of nematode translation are faithfully reproduced in the Ascaris cell-free system. These studies demonstrate the nematode translational machinery has evolved to translate the m32,2,7GpppG cap in the context of the SL, and both these trans-spliced elements are required to promote efficient translation of a recipient polyadenylated RNA..
In nematode cells, as well as those of other trans-splicing metazoa, trans-spliced and non-trans-spliced mRNAs are both present. Our in vivo data suggest the TMG cap and SL sequence play a similar role in mRNA translation and stability as the m7GpppG cap and 5’ UTR of non-trans-spliced mRNAs. On average, RNAs with the TMG-cap and SL are translated at relatively similar levels as non-trans-spliced m7G-capped RNAs (Figures 2–3). Thus, the trans-spliced elements themselves do not appear to confer any particular translational advantage to RNAs. However, both m32,2,7GpppG-cap and the spliced leader sequence must be present together to obtain translational efficiencies similar to those observed for m7GpppG-capped RNAs (Figures 2–3). Our data further suggest that RNAs with the TMG-cap and SL do not have different functional or physical half-lives compared to non-trans-spliced m7G-capped RNAs (Table 4 and Figure 4). Translation of mRNAs with the TMG-cap and SL is more dependent on the poly(A) tail (Figure 2C–D; Table 1–2), and consequently, trans-spliced RNAs may be more subject to translational regulation by poly(A) tail length alterations.
The effect on translation of the SL in the 5’ UTR was examined in a variety of contexts and compared to the 5’ UTRs from Firefly, Renilla, Gaussia, and a native, non-trans-spliced Ascaris RNA (Figures 2–3). The SL was less efficient in translation than the first 22 nts of the Firefly and Renilla 5’ UTRs, whereas the SL was equal to or 30–50% more efficient than a Gaussia (Sh) or non-trans-spliced native Ascaris 5’ UTR (L37) when the SL is placed immediately adjacent to the reporter AUG. The greater efficiency for the SL with the Gaussia reporter is attributable in part to 5’ UTR length (shorter 5’ UTRs are more efficient for Gaussia reporters and the L37 is longer) and to the different -1 to -3 AUG sequence. The -1 to -3 GAG of the SL is more efficient than the ACC present in several RNAs (see Figure 3B–C). Our interpretation of these data is that the spliced leader contributes a reasonably efficient 5’ UTR and initiation context for translation of mRNAs. Our data also suggest that the spliced leader sequence by itself does not enhance cap-independent translation. The SL therefore does not likely enhance translation by directly interacting with components of the translation machinery other than eIF4E binding to the cap [37].
Previous studies using lethal C. elegans mutants where the entire SL1 locus is deleted indicated that the reintroduction of mutant SLs could rescue lethality whereas several mutant SLs could not [26, 27]. The mutant SLs that could not rescue lethality may affect the synergistic interaction between the TMG cap and SL resulting in lower levels of trans-spliced RNA translation thus leading to lethality.
In vitro vs. in vivo analyses
Previously, Maroney et al (1995) noted that the m32,2,7GpppG-cap and spliced leader leader sequence interact cooperatively to increase translational efficiency in a cell-free system. We recently further characterized this interaction and demonstrated it was synergistic using a modified, competitive Ascaris cell-free translation system [37]. We have shown here that this interaction is physiologically relevant in vivo, that the synergism is dependent on the presence of a poly(A) tail, and that sequences between the SL and AUG and in the 3’ UTR of the trans-spliced mRNAs examined do not significantly contribute to translation of mRNAs with a TMG-cap and SL. Maroney et al. also observed that translation of the native trans-spliced vacuolar H+ ATPase mRNA (cDNA 12) with an m32,2,7GpppG-cap, SL, and poly(A) tail was ~ 2.5-fold more efficient than the same RNA with an m7GpppG-cap and no spliced leader sequence in the 5’ UTR. Both our current in vivo and previous in vitro data [37] differ from the data of Maroney et al. We did not observe a large enhancement in translation from addition of a m32,2,7GpppG cap and spliced leader to several luciferase reporters. Our experiments were also carried out with the complete native 5’ and 3’ UTRs of the same trans-spliced RNA (cDNA 12) studied by Maroney et al on a luciferase reporter. Similarly, addition of the complete native 5’ and 3’ UTRs of a second trans-spliced cDNA (cDNA 4) to a reporter RNA did not enhance translation compared to a non-transpliced reporter RNA (m7GpppG capped luciferase with various 5’ UTRs). The RNAs examined in vivo in the current study were also evaluated in our cell-free system [37], and produced results similar to that observed in vivo (data not shown). The simplest explanations for the differences observed are 1) the different translation conditions present in vivo and in our modified cell-free system [37] and 2) possible elements within the open reading frame of vacuolar ATP synthase 16 kDa proteolipid subunit RNA (cDNA 12) that contribute to translation [38].
Function of trans-splicing
The TMG cap and in some contexts the SL have negative effects on translation, while together they function synergistically to support translation. We interpret this result to suggest that the nematode translation machinery evolved to translate mRNAs with the TMG cap specifically in the context of the SL. If the TMG cap and SL do not confer any discrete properties to their recipient mRNAs such as enhanced translation or stability, why then is there SL trans-splicing of mRNAs that are not derived from polycistronic RNAs? One possible answer might be related to the primary transcripts of trans-spliced genes. We have recently shown that the spliced leader is typically added to nematode mRNAs at a relatively conserved distance from the mRNA AUG (usually less than 10 nucleotides). Notably, this conserved distance coincides with the optimal spacing for translation in a cell-free Ascaris system [37]. Thus, spliced leader addition in nematodes may serve to improve the translation initiation context for the primary transcripts of trans-spliced genes. The majority of trans-spliced test RNAs examined in the current study have this optimal spacing of SL to AUG. What function does trans-splicing serve in creating this spacing or improving the initiation context? If the initiation sites for primary transcripts of trans-spliced genes were heterogeneous, this might result in mRNAs with heterogeneous 5’ ends that could have upstream AUGs or short open reading frames in the 5’ UTR. These mRNAs would likely be poorly translated. SL addition might then function to trim these mRNAs to a uniform length that would be more optimal for translation initiation [37]. Alternatively, since trans-splicing trims a pre-mRNA to generate the mature 5’ UTR, there would be no selection pressure to eliminate mutations that could lead to upstream AUGs or open reading frames that would be deleterious to translation. Thus, once spliced leader trans-splicing is present it might need to be maintained to override deleterious effects of sequences in the 5’ UTR of pre-mRNAs [43].
Another potential function for nematode SL trans-splicing, albeit perhaps less likely, might be that primary transcripts for these genes are not efficiently capped. At this point there is no available information on the capping efficiency of primary transcripts for trans-spliced genes in nematodes. If capping efficiency were low, trans-splicing might serve a capping function as observed from some transcripts in the Kinetoplastida. Finally, it remains possible that the function(s) that SL trans-splicing originally served and/or evolved to address in nematodes other than processing of polycistronic transcripts is no longer required or needed. In this view, it could be that some trans-splicing remains, but no longer serves a discrete function.
SL and RNA decapping
We recently observed that addition of the SL sequence to test mRNAs leads to an ~10-fold reduction in nematode Dcp2 RNA decapping in vitro [44]. In addition, we previously observed that 5’ to 3’ RNA decay appears significantly less active than 3’ to 5’ RNA decay in an Ascaris embryo in vitro decay system [45]. In the current study, addition of the SL sequence to RNAs did not lead to significant increases in mRNA stability in vivo. One interpretation of these data is that Dcp2 decapping and 5’ to 3’ decay plays a minor role in general decay both in vivo and in vitro in nematode embryos. Dcp2 decapping of trans-spliced mRNAs might be a potential mechanism for regulated mRNA decay in nematodes.
Contribution of nuclear history
In the current studies, we introduced RNA directly into embryo cells bypassing the typical maturation of an mRNA in the nucleus and its export to the cytoplasm. This approach was necessary since it is currently not possible to design DNA constructs that can directly synthesize m32,2,7GpppG-capped mRNAs in vivo. Furthermore, the current strategy circumvents potential differences in promoter activity, splicing and polyadenylation (including tail length), and mRNA export that could compromise our ability to analyze the effect of individual RNA elements on translation and stability. However, we note that several lines of recent evidence suggests that the transcription and nuclear history of an mRNA can have a significant effect on splicing, export, and the cytoplasmic metabolism of the mRNA, including its translation [17, 46–55]. Thus, it remains possible that during cellular maturation of trans-spliced mRNAs that the m32,2,7GpppG-cap and spliced leader may play a greater role in mRNA translation and stability than observed here. Our introduction of the reporter RNAs into a variety of cells in the 32–64 cell embryos and subsequent assay of luciferase activity provides an average of the translation and stability of the introduced RNAs in a variety of different cell types. We have observed similar results in 1–2 and 2–4 cell embryos (Figure 4; data not shown). However, it remains possible that the m32,2,7GpppG-cap and spliced leader could play a discrete role in mRNA translation and/or stability in a subset of embryo cells. Furthermore, the presence of a number of eIF4E isoforms (5) expressed in different tissues in some nematodes such as C. elegans may play a role in the regulation of translation of trans-spliced mRNAs [56–59].
Conclusion
Overall, trans-splicing addition of the m32,2,7GpppG-cap and SL sequence to mRNAs does not result in mRNAs with significantly enhanced translation or stability in vivo when compared to non-trans-spliced test RNAs. The m32,2,7GpppG-cap and spliced leader sequence function best together and synergistically interact to increase translation in the presence of a poly(A) tail. It remains to be determined whether the nuclear maturation and export of trans-spliced mRNAs enables the m32,2,7GpppG-cap and spliced leader sequence to differentially affect mRNA translation and stability or if these elements play differential roles in a subset of nematode cells.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the technical help of David Ndegwa, Craig McFarland, Glenna Kunkel, Anthony Esposito, and Cassandra Friedmann. We thank Cassandra Friedmann for help with the analysis of translational efficiency and functional mRNA half-life. We thank David Bentley, Tom Blumenthal, and Lee Niswander and members of the Davis Lab for their constructive comments on the manuscript. This work was supported by funds from an Ellison Medical Foundation Grant ID-IA-0037, an NIH Grant AI49558, and CUNY-CSI and UCHSC startup funds to R.E.D. and a Howard Hughes Medical Institute Grant 55005604 and Polish Ministry of Science and Higher Education Grant 2 P04A 006 28 to E.D.
References
- 1.Blumenthal T. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 1995;11:132–6. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- 2.Davis RE. Spliced leader RNA trans-splicing in metazoa. Parasit Today. 1996;12:33–40. doi: 10.1016/0169-4758(96)80643-0. [DOI] [PubMed] [Google Scholar]
- 3.Hastings KE. SL trans-splicing: easy come or easy go? Trends Genet. 2005;21:240–7. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Nilsen TW. Trans-splicing of nematode premessenger RNA. Annu Rev Microbiol. 1993;47:413–40. doi: 10.1146/annurev.mi.47.100193.002213. [DOI] [PubMed] [Google Scholar]
- 5.Nilsen TW. Evolutionary origin of SL-addition trans-splicing: still an enigma. Trends Genet. 2001;17:678–80. doi: 10.1016/s0168-9525(01)02499-4. [DOI] [PubMed] [Google Scholar]
- 6.Boothroyd J, Cross G. Transcripts coding for different variant surface glycoproteins in Trypanosoma brucei have a short identical exon at their 5′ end. Gene. 1982;20:279–87. doi: 10.1016/0378-1119(82)90046-4. [DOI] [PubMed] [Google Scholar]
- 7.Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–61. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pouchkina-Stantcheva NN, Tunnacliffe A. Spliced-leader RNA mediated trans-splicing in phylum Rotifera. Mol Biol Evol. 2005 doi: 10.1093/molbev/msi139. [DOI] [PubMed] [Google Scholar]
- 9.Rajkovic A, et al. A spliced leader is present on a subset of mRNAs from the human parasite Schistosoma mansoni. Proc Natl Acad Sci USA. 1990;87:8879–83. doi: 10.1073/pnas.87.22.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stover NA, Steele RE. Trans-spliced leader addition to mRNAs in a cnidarian. Proc Natl Acad Sci USA. 2001;98:5693–8. doi: 10.1073/pnas.101049998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van der Ploeg L, et al. RNA splicing is required to make the messenger RNA for a variant surface antigen in trypanosomes. Nucl Acids Res. 1982;10:3591–604. doi: 10.1093/nar/10.12.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vandenberghe AE, Meedel TH, Hastings KE. mRNA 5′-leader trans-splicing in the chordates. Genes Dev. 2001;15:294–303. doi: 10.1101/gad.865401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imboden MA, et al. Transcription of the intergenic regions of the tubulin gene cluster of Trypanosoma brucei: evidence for a polycistronic transcription unit in a eukaryote. Nucleic Acids Res. 1987;15:7357–68. doi: 10.1093/nar/15.18.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson PJ, Kooter JM, Borst P. Inactivation of transcription by UV irradiation of T. brucei provides evidence for a multicistronic transcription unit including a VSG gene. Cell. 1987;51:273–81. doi: 10.1016/0092-8674(87)90154-1. [DOI] [PubMed] [Google Scholar]
- 15.Muhich ML, Boothroyd JC. Polycistronic transcripts in trypanosomes and their accumulation during heat shock: evidence for a precursor role in mRNA synthesis. Mol Cell Biol. 1988;8:3837–46. doi: 10.1128/mcb.8.9.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tschudi C, Ullu E. Polygene transcripts are precursors to calmodulin mRNAs in trypanosomes. EMBO J. 1988;7:455–63. doi: 10.1002/j.1460-2075.1988.tb02833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang XH, et al. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot Cell. 2003;2:830–40. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruderer T, Tu LC, Lee MG. The 5′ end structure of transcripts derived from the rRNA gene and the RNA polymerase I transcribed protein coding genes in Trypanosoma brucei. Mol Biochem Parasitol. 2003;129:69–77. doi: 10.1016/s0166-6851(03)00095-1. [DOI] [PubMed] [Google Scholar]
- 19.Bangs JD, et al. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J Biol Chem. 1992;267:9805–15. [PubMed] [Google Scholar]
- 20.Blumenthal T, Gleason KS. Caenorhabditis elegans operons: form and function. Nat Rev Genet. 2003;4:110–8. doi: 10.1038/nrg995. [DOI] [PubMed] [Google Scholar]
- 21.Spieth J, et al. Operons in C. elegans: polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell. 1993;73:521–32. doi: 10.1016/0092-8674(93)90139-h. [DOI] [PubMed] [Google Scholar]
- 22.Guiliano DB, Blaxter ML. Operon Conservation and the Evolution of trans-Splicing in the Phylum Nematoda. PLoS Genetics. 2006;2:e198. doi: 10.1371/journal.pgen.0020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis RE, Hodgson S. Gene linkage and steady state RNAs suggest trans-splicing may be associated with a polycistronic transcript in Schistosoma mansoni. Mol Biochem Parasitol. 1997;89:25–39. doi: 10.1016/s0166-6851(97)00097-2. [DOI] [PubMed] [Google Scholar]
- 24.Ganot P, et al. Spliced-leader RNA trans splicing in a chordate, Oikopleura dioica, with a compact genome. Mol Cell Biol. 2004;24:7795–805. doi: 10.1128/MCB.24.17.7795-7805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satou Y, et al. Genomic overview of mRNA 5′-leader trans-splicing in the ascidian Ciona intestinalis. Nucleic Acids Res. 2006;34:3378–88. doi: 10.1093/nar/gkl418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferguson KC, Heid PJ, Rothman JH. The SL1 trans-spliced leader RNA performs an essential embryonic function in Caenorhabditis elegans that can also be supplied by SL2 RNA. Genes Dev. 1996;10:1543–56. doi: 10.1101/gad.10.12.1543. [DOI] [PubMed] [Google Scholar]
- 27.Ferguson KC, Rothman JH. Alterations in the conserved SL1 trans-spliced leader of Caenorhabditis elegans demonstrate flexibility in length and sequence requirements in vivo. Mol Cell Biol. 1999;19:1892–900. doi: 10.1128/mcb.19.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonen L. Trans-splicing of pre-mRNA in plants, animals, and protists. FASEB J. 1993;7:40–6. doi: 10.1096/fasebj.7.1.8422973. [DOI] [PubMed] [Google Scholar]
- 29.Huang XY, Hirsh D. RNA trans-splicing. Genet Eng (N Y) 1992;14:211–29. doi: 10.1007/978-1-4615-3424-2_12. [DOI] [PubMed] [Google Scholar]
- 30.Maroney PA, Hannon GJ, Nilsen TW. Transcription and cap trimethylation of a nematode spliced leader RNA in a cell-free system. Proc Natl Acad Sci USA. 1990;87:709–13. doi: 10.1073/pnas.87.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas JD, Conrad RC, Blumenthal T. The C. elegans trans-spliced leader RNA is bound to Sm and has a trimethylguanosine cap. Cell. 1988;54:533–9. doi: 10.1016/0092-8674(88)90075-x. [DOI] [PubMed] [Google Scholar]
- 32.Liou RF, Blumenthal T. trans-spliced Caenorhabditis elegans mRNAs retain trimethylguanosine caps. Mol Cell Biol. 1990;10:1764–8. doi: 10.1128/mcb.10.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Doren K, Hirsh D. mRNAs that mature through trans-splicing in Caenorhabditis elegans have a trimethylguanosine cap at their 5′ termini. Mol Cell Biol. 1990;10:1769–72. doi: 10.1128/mcb.10.4.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv Virus Res. 2000;55:135–84. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cougot N, et al. ‘Cap-tabolism’. Trends Biochem Sci. 2004;29:436–44. doi: 10.1016/j.tibs.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 37.Lall S, et al. Contribution of trans-splicing, 5′-leader length, cap-poly(A) synergism, and initiation factors to nematode translation in an Ascaris suum embryo cell-free system. J Biol Chem. 2004;279:45573–85. doi: 10.1074/jbc.M407475200. [DOI] [PubMed] [Google Scholar]
- 38.Maroney PA, et al. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: a role for spliced leader addition in translational efficiency. RNA. 1995;1:714–23. [PMC free article] [PubMed] [Google Scholar]
- 39.Davis RE, et al. Transient expression of DNA and RNA in parasitic helminths by using particle bombardment. Proc Natl Acad Sci USA. 1999;96:8687–92. doi: 10.1073/pnas.96.15.8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones LJ, et al. RNA quantitation by fluorescence-based solution assay: RiboGreen reagent characterization. Anal Biochem. 1998;265:368–74. doi: 10.1006/abio.1998.2914. [DOI] [PubMed] [Google Scholar]
- 41.Shelton CA, Bowerman B. Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development. 1996;122:2043–50. doi: 10.1242/dev.122.7.2043. [DOI] [PubMed] [Google Scholar]
- 42.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–16. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 43.Blumenthal T. Trans-splicing and operons. In: T.C.e.R. Community, editor. WormBook. WormBook; 2005. [DOI] [PubMed] [Google Scholar]
- 44.Cohen LS, et al. Dcp2 Decaps m2,2,7GpppN-capped RNAs, and its activity is sequence and context dependent. Mol Cell Biol. 2005;25:8779–91. doi: 10.1128/MCB.25.20.8779-8791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen LS, et al. Nematode m7GpppG and m3(2,2,7)GpppG decapping: activities in Ascaris embryos and characterization of C. elegans scavenger DcpS. RNA. 2004;10:1609–24. doi: 10.1261/rna.7690504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 47.Hazelrigg T. Lost in translation gets an oskar. Dev Cell. 2004;6:611–3. doi: 10.1016/s1534-5807(04)00142-x. [DOI] [PubMed] [Google Scholar]
- 48.Huang Y, Steitz JA. SRprises along a messenger’s journey. Mol Cell. 2005;17:613–5. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 49.Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003;28:215–20. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 50.Lu S, Cullen BR. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA. 2003;9:618–30. doi: 10.1261/rna.5260303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 52.Matsumoto K, Wassarman KM, Wolffe AP. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998;17:2107–21. doi: 10.1093/emboj/17.7.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noe V, MacKenzie S, Ciudad CJ. An intron is required for dihydrofolate reductase protein stability. J Biol Chem. 2003 doi: 10.1074/jbc.M212746200. [DOI] [PubMed] [Google Scholar]
- 54.Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–17. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–22. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amiri A, et al. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development. 2001;128:3899–912. doi: 10.1242/dev.128.20.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dinkova TD, et al. Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Mol Cell Biol. 2005;25:100–13. doi: 10.1128/MCB.25.1.100-113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jankowska-Anyszka M, et al. Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in C.elegans can distinguish between mono- and trimethylated mRNA cap structures. J Biol Chem. 1998;273:10538–42. doi: 10.1074/jbc.273.17.10538. [DOI] [PubMed] [Google Scholar]
- 59.Keiper BD, et al. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J Biol Chem. 2000;275:10590–96. doi: 10.1074/jbc.275.14.10590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.