Abstract
The paraventricular nucleus of the hypothalamus (PVH) plays a central role in regulating the hypothalamic-pituitary-adrenal (HPA) axis. Medial parvocellular neurons of the PVH (mpPVH) integrate sensory and humoral inputs to maintain homeostasis. Humeral inputs include glucocorticoids secreted by the adrenals, which down-regulate HPA activation. A primary glucocorticoid target is the population of mpPVH neurons that synthesize and secrete corticotropin-releasing factors, the most potent of which is corticotropin-releasing hormone (CRH). Although CRH gene (crh) expression is known to be down-regulated by glucocorticoids, the mechanisms by which this process occurs are still poorly understood. To begin this study we postulated that glucocorticoid repression of crh involves HDAC recruitment to the region of the crh proximal promoter. To evaluate this hypothesis, we treated hypothalamic cells that express CRH with the HDAC inhibitor trichostatin A (TSA). As predicted, treatment with TSA led to increased CRH mRNA levels and crh promoter activity. Although co-treatment with Dex (10−7 M) reduced the TSA effect on mRNA levels, it failed to reduce promoter activity; however co-transfection of HDAC1 but not 3 restored Dex inhibition. A distinction between HDAC1 and 3 was also apparent with respect to crh promoter occupancy. Dex led to increased HDAC1 but not HDAC3 occupancy. In vivo studies revealed that CRH-immunoreactive (-ir) neurons contained HDAC1- and HDAC3-ir. Collectively, these data point to a role for HDAC1 in the physiologic regulation of crh.
Keywords: corticotropin releasing factor, gene expression, histone deacetylase 1, stress
1.1 Introduction
The ability to meet successfully the physiologic demands of stress is paramount for survival. Mammals have developed multiple physiologic systems to meet such demands, one being the hypothalamic-pituitary-adrenal axis (HPA axis). This system receives inputs from numerous sites in the central nervous system (CNS), and it integrates them with signals from the hormonal milieu. This enormous amount of information is integrated by a small population of neuroendocrine motor neurons of the HPA axis, which synthesize and secrete corticotropin releasing factors, the most potent being the 41 amino acid corticotropin-releasing hormone (CRH)[1].
After CRH is secreted into the portal circulation of the median eminence, it is carried to the anterior pituitary where it binds to CRH receptors expressed by corticotropes. This CRH binding triggers the synthesis of the adrenocorticotrophic hormone (ACTH) and its secretion into the peripheral circulation. ACTH then acts by binding cells in the zona fasiculata of the adrenal cortex, which synthesize and secrete glucocorticoids into the peripheral circulation. These steroids then act in the liver to mobilize glucose, a necessary phenomenon in meeting the demands of a stressful situation.
Glucocorticoids modulate processes in virtually every cell in the body. Many of these effects are adaptive. For example, glucocorticoids are lympholytic, an adaptive property in the face of an autoimmune response, or lymphocytic neoplasms; however, in excess glucocorticoids lead to adverse consequences. In addition to being diabetogenic and immunosuppressive, they may lead to osteopenia and frank psychosis[2] ; thus, their levels must be tightly controlled.
Much of this control is mediated by a classic endocrine negative feedback system in which the end product of axis activation down-regulates axis activity; thus, glucocorticoids down-regulate the synthesis of corticotrope ACTH and modulate the activity of many components of the CNS circuitry that trigger HPA axis activation. The net result is down-regulation of CRH synthesis and secretion. One locus at which the glucocorticoids act is the paraventricular nucleus of the hypothalamus (PVH). Specifically medial parvocellular neurons are key integrators hormonal and neuronal input and mediate CRH outflow. Glucocorticoids have been shown in numerous studies to down regulate CRH gene (crh). Although substantial in-roads have been made in understanding the mechanisms by which down-regulation of CRH expression occurs, numerous steps in the process remain to be elucidated [3].
The prototypic mechanism by which glucocorticoids exert their effect involves binding to their receptor (GR). As is the case for all steroid receptors and numerous members of the nuclear receptor superfamily, these proteins are ligand-activated transcription factors. Ligand-bound GRs bind to cognate response elements in DNA, GREs, and then activate gene expression. Although much is known about mechanisms of GR-activated gene expression, less is known about mechanisms of gene repression, particularly of genes involved in the mammalian stress response.
At the level of promoter regulation, GRs may repress gene expression by binding to composite elements such as those found in the proliferin gene. Additionally, they may repress through alternate pathways in which GRs regulate gene expression by modulating the activity of other transcription factors such as AP-1 [4]. With respect to crh regulation, a negative GRE has been demonstrated to be involved in down regulation of CRH gene (crh) expression [5]. This nGRE is a composite element; GR and AP-1 family members both bind this site. In the case of modulating other transcription factor activity, down-regulation of crh expression traces to a cAMP regulatory element (CRE) [6].
Regardless of which element is utilized, regulation by the GR requires interaction with various coregulators [7]. In the case of repression though the AP-1 site, the CRE binding protein (CREB)-binding protein (CBP), a histone acetyl transferase (HAT), has been functionally implicated, the concept being that in the presence of high levels of CBP, the GR is sequestered from the promoter [8]. This mechanism, however, is controversial.
Another mechanism for repression could be to alter the balance of HATs and HDACs at the promoter. A reduced concentration or diminished activity of CBP or other HATs could lead to decreased acetylation. Alternatively, decreased acetylation could be effected by an increased concentration of HDACs or an increase in their activity; thus, increased HDAC occupancy of the crh promoter could lead to decreased histone acetylation, which is generally associated with decreased gene expression.
Numerous HDACs have been described, the prototypic ones being HDACs 1, 2, and 3. HDACs 1 and 2 are structurally more similar to each other than they are to HDAC3 [9]; so, HDACs 1 and 3 were chosen to afford a greater chance of detecting a functional difference between the three prototypes. HDACs 1 and 3 interact directly or indirectly with nuclear receptors. For example, HDAC1 has been functionally associated with the GR and in a complex with GR and Sin3A [10]. HDAC3 is part of a repressive complex that includes unliganded thyroid hormone receptor and the nuclear receptor co-repressor (NCoR) [11]. Thus, it was likely that HDACs 1 & 3 would functionally interact at the crh promoter, and might display a degree of specificity in doing so.
To determine whether or not the molecular data generated could have physiologic relevance we combined in vitro and in vivo approaches. For in vitro studies, we took advantage of the CRH-expressing hypothalamic cell line, IVB, which has been demonstrated to express CRH and functional GRs [12]. For in vivo studies, we monitored changes in the colocalization of GR and HDAC immunoreactivity (ir) with CRH-ir in rats that had been adrenalectomized and treated with vehicle (ADX) or Dex (ADX+Dex). As a measure of deacetylase function, we measured levels of histone 3 and 4 (H3 and H4) acetylation, both in vitro and in vivo.
2.1 Materials and Methods
2.1.1 Reagents and their sources, buffers, and methods
These reagents have been previously described: Luciferase assay [13], mRNA isolation, RT-PCR, ChIP analysis, and ICC [14].
2.1.2 Animals and Animal Handling
Animal surgery, experimental protocols, and monitoring were approved by the Animal Care and Use Committee at Colorado State University and carried out in accordance with NIH and AAALAC guidelines. Young adult Sprague Dawley male rats (60–90 days old) were obtained from Charles Rivers Laboratories (Wilmington, MA). Animals were double housed within the Colorado State University vivarium and maintained on a 12-h dark 12-h light schedule (lights on at 0700 h) with ad libitum access to food and water. Animals were acclimated for 4 days prior to surgery. Rats were adrenalectomized (ADX) and maintained on 0.9% NaCl, while shams received tap water. A cohort of ADX animals were injected daily with 30 ug/kg Dex (s.c.) for 4 days; Controls received vehicle (safflower oil) This regimen has been previously described by Suzuki and Handa [15] where it was shown to alter gene expression within the hypothalamic PVN. Three to four animals were included in each cohort. On the final day of treatment, animals were perfused transcardially with saline followed by 4% buffered paraformaldehyde. Brains were post-fixed for 24 hours and cryoprotected in 30% sucrose, cryostat-sectioned at 35 µm and stored in cryopreservative solution at −20°C until processed [16].
2.1.3 Cell Culture
Hypothalamic IVB Cells were cultured and maintained in phenol red-free Dulbecco’s Modified Eagle’s / F12 (Sigma) supplemented with 10% newborn calf serum; 100 units/ml penicillin/streptomycin (100 units/ml, Gibco), 0.1 mM non-essential amino acids 1mM sodium pyruvate and 2mM L-glutamine.
2.1.4 Immunocytochemistry (ICC)
IVB cells (1 × 105 cells/ 0.5 ml/ well) were plated and maintained in Lab-Tek™ II Chamber Slides (Nalge Nunc International, USA) for 48 h in a medium supplemented with charcoal stripped serum. They were treated for 2hrs with 10−7M Dex or ethanolic vehicle (veh) before fixation in 4% paraformaldehyde. Cells were permeablized with 0.1 % Triton X-100 and blocked in normal goat serum to reduce nonspecific binding of the antibody. They were then incubated with the primary antibody for 48 h at 4 °C. Anti-GR monoclonal antibody (mAb, 1:200 dilution) was purchased from Pierce/ Thermo Scientific, Rockford, IL; anti-HDAC1 mAb (1:200 dilution) from Abcam (Cambridge, MA); anti- HDAC3 mAb (1:200 dilution) from Cell Signaling (Danvers, MA); pan Ac-H3 polyclonal Antibody (pAb, 1:200 dilution) from Millipore (Temecula, California); and pan Ac-H4 pAb (1:1000) from Active Motif (Carlsbad, CA). After having been washed with PBS, cells were incubated with 1:1000 dilution of either Alexa Fluor 594-goat anti rabbit IgG or Alexa Fluor 488-goat anti mouse (Molecular Probes; Invitrogen) for 1h. Cells were then mounted in FlourSave (Calbiochem) mounting media; images were captured with a CCD camera and digitized images were arranged using Adobe Photoshop (Adobe, Mountain View, CA).
2.1.5 mRNA analysis
IVB cells were grown to 80–90% confluency in media containing stripped serum for 48 hrs prior to lysis. They were then treated with Dex 10−7M for 2 hrs. Total RNA was isolated using TRIzol (Invitrogen) followed by isopropanol precipitation.
2.1.6 Reverse transcription of total RNA, and PCR amplification
Two hundred and fifty U of MultiScribe™ reverse transcriptase (RT) in 10 uL RT buffer (composition proprietary) with 1mM dNTP, and 5 uM random primers (High Capacity cDNA Archive Kit; Applied Biosystems, Foster City, CA) were used to transcribe total RNA (5 ug). cDNA synthesis was performed at 25°C for 10 minutes followed by 37°C for 2 hours. Primer sequences for CRH and cycling parameters have been previously described [14]
2.1.7 Transient Transfections
Cells were grown for 48 hrs. in medium containing stripped serum prior to transfection. IVB cells (1.5×106) were transfected with 5 µg of the reporter construct along with HDAC1 or HDAC3 expression vector as indicated in legends. Transfection was performed using Lipofectamine (Invitrogen), and transfection efficiency was monitored by cotransfection of a beta galactosidase reporter construct, as described [17]. Data were obtained from 3 or more independent experiments using different cell passages. Each transfection was performed in triplicate
2.1.8 Chromatin immunoprecipitation followed by real time PCR (ChIP)
For ChIP assay, cells were grown to 80 to 90% confluency in media containing stripped serum for 48 hrs. Cells were treated for 2h with α-amanitin to synchronize the cell cycle and then treated with Dex (10−7M) for 2 hours. All methodological procedures were performed as described [14].
2.1.9 Real-time PCR quantification for ChIP analysis
For real-time PCR, 5 µl aliquots of each ChIP DNA sample were used in triplicate for qPCR analysis of the CRH promoter. Primer sequences, cycling parameters, the threshold cycle (Ct), and generation of the standard curve have been described [14]. pFoxLuc DNA was added to chromatin samples and served as a control for DNA recovered by ChIP. PCR amplification of pFoxLuc DNA recovered by ChIP served to correct for sample loss throughout the assay. Calculation methodology has been reported [18, 19].
2.1.10 Immunohistochemistry (IHC)
CRH-, HDAC1- or HDAC3-, and Ac-H3- or Ac-H4- immunoreactivities were co-localized using a dual-immunofluorescent procedure. All steps were carried out on free-floating sections at room temperature except for primary antibody incubation, which was performed at 4°C. Four series of sections were taken through the anterior hypothalamic area, and each series was processed for CRH immunoreactivity and either HDAC or either acetylated histone. Sections were washed in 0.1 M phosphate buffer with 0.9% saline (PBS) to remove cryoprotectant. The sections were next incubated in PBS containing 4% normal goat serum (Sigma, G6767) and 0.1% Triton X-100 (Sigma, X-100; PBSTX) for 1 h. Sections were then incubated in rabbit polyclonal antibody against CRH (1:10,000; CRH antibody generously provided by W. Vale, Salk Institute) for 48 hr in PBSTX at 4°C. Following incubation, sections were washed and then placed in a solution of PBSTX with biotinylated goat anti-rabbit IgG (1:1000; Vector Laboratories, BA-1000) for 1h. Sections were washed and incubated for 1h in avidin-biotin complex (1:1000; Vector Laboratories, PK-6101), then washed and incubated in Cy3 strepavidin (1:250; Jackson ImmunoResearch Laboratories Inc, 111-166-003). After repeated washing, sections were incubated for 48 hours at 4°C in PBSTX containing a rabbit polyclonal antibody against either rabbit anti-HDAC1 (1:500; Santa Cruz Biotechnology, sc-7872), anti-HDAC3 (1:100; Santa Cruz Biotechnology, sc-11417), anti-Ac-H3 (1:200; Upstate Cell Signaling Solutions, 06-599) or anti-Ac-H4 (1:10,000; Upstate Cell Signaling Solutions, 06-866). After incubation, sections were washed and then placed in PBSTX containing Alexa 488-conjugated Affinipure goat anti-rabbit IgG (1:250; Molecular Probes, A11008) for 1 h. Following washes in PBS, each series was mounted on slides and coverslipped with Flourosave. Omission of each primary antibody eliminated immunoreactivity for the corresponding antigen.
2.1.11 Cell visualization and quantification
Sections were imaged under a LSM 710 laser scanning confocal microscope (Carl Zeiss Microscopy, Jena, Germany). The number and location of CRH immunoreactive (-ir) cells and the percentage of CRH-ir neurons that colocalized either with HDAC-ir or with pan-acetylated-ir histone were determined by an investigator blind to treatment conditions. Cells were considered doubled-labeled only if a ring of CRH-ir cytoplasm completely surrounded an immunoreactive nucleus through multiple planes of focus. From each animal, the parvocellular region of the PVH was atlas matched [20]. The total number of CRH-ir, HDAC-ir or pan acetylated histone-ir cells, the number of CRH-ir cells and the number of CRH-ir cells colocalized with either HDAC or either acetylated histone were recorded bilaterally through the largest extent of the parvocellular region of PVH in two adjacent sections, resulting in a single number for each animal.
2.1.12 Statistical analyses
One-way ANOVA was used for colocalization and ChIP analysis. For IHC data, Fisher’s PLSD post hoc test was used. Data was analyzed using the computer program StatView 5.0.1 (SAS Institute Inc., Cary, NC). Statistical significance was set at p≤0.05. ChIP data was analyzed with the Mann-Whitney U test using the Number Cruncher Statistical System program (NCSS, Kaysville, UT).
3.1 Results
3.1.1 The IVB cell line displays predicted responses to glucocorticoid exposure
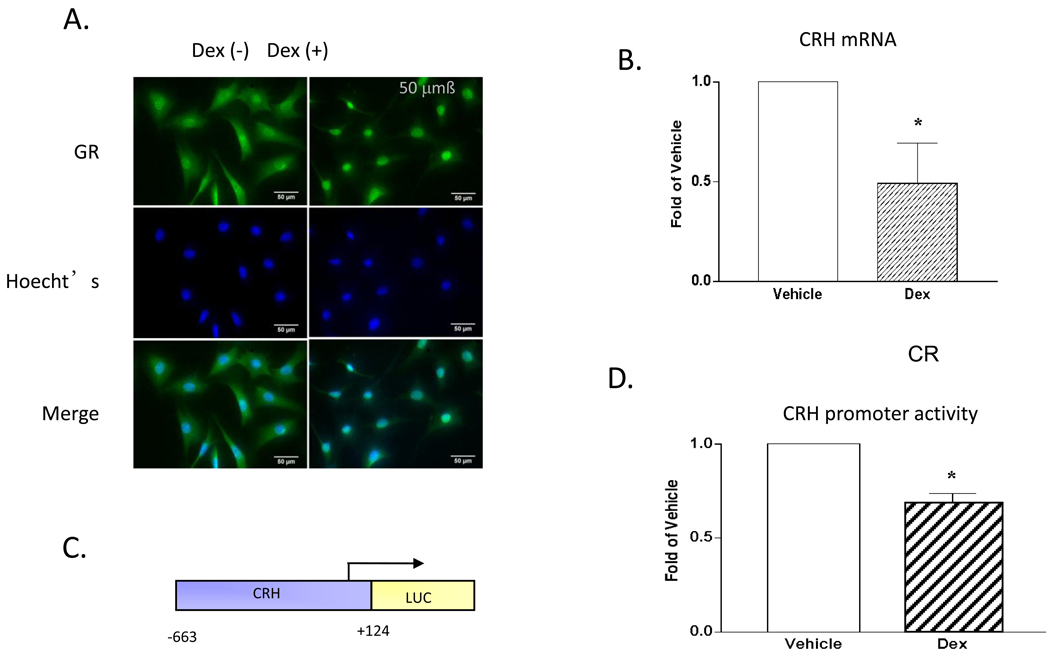
Dex treatment led to GR-ir translocation from the cytoplasm to the nucleus, as expected (Fig 1A). Dex also reduced basal CRH mRNA levels (Fig 1B) and crh promoter activity (Figs 1C and D). Consistent with previous reports (e.g. [12, 21, 22]), the IVB line is suitable for studying GR regulated crh expression.
Figure 1. IVB cells express GR-ir and support Dex-mediated reduction of CRH mRNA levels and crh promoter activity.
Dex (10−7M) treatment for 2h leads to translocation of the GR-ir to the nucleus and reduction of CRH mRNA. Treatment with Dex (10−7M) for 18 hrs leads to reduction of crh promoter activity. (A) GR ICC: top - ICC for GR; middle Hoecht’s; bottom - merged images. (B) CRH mRNA (C) crh promoter: luciferase construct used in all transfection experiments. The numbers indicate the bases from the transcription start site, indicated by the arrow. (D) Promoter activity: Cells were treated with vehicle (EtOH) or Dex (10−7M). After correcting for efficiency of transfection, fold change was calculated from relative light units of luciferase activity. Error bars represent SEM. (*) Dex treatment differs from vehicle: (B) p<0.02, (D) p<0.001. (LUC) luciferase, (Dex) dexamethasone.
3.1.2 HDAC1 permits Dex reduction of TSA-stimulated crh promoter activity
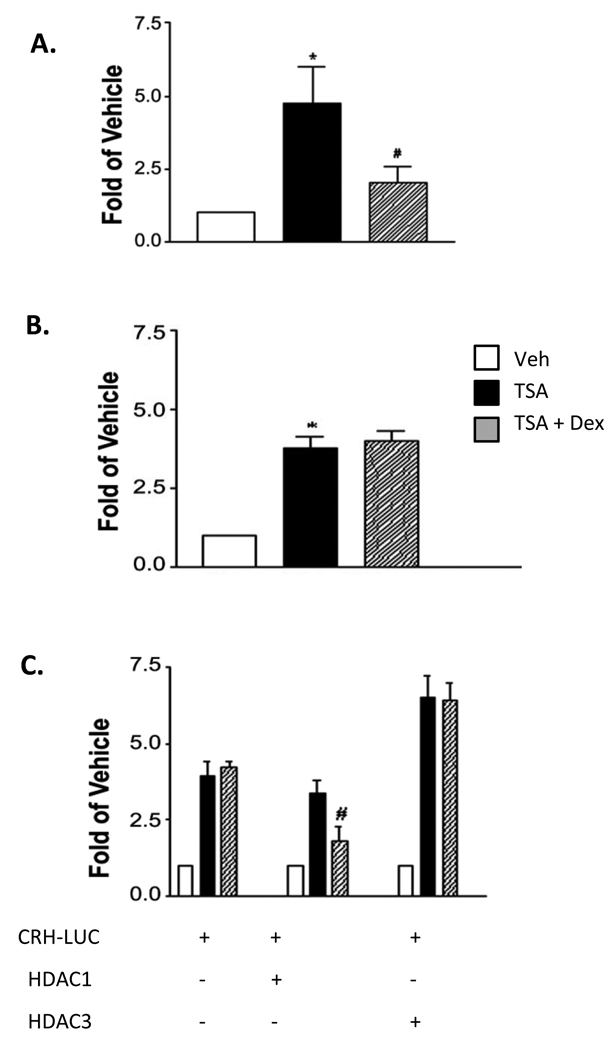
To investigate the role of HDACs in Dex-mediated crh repression, IVB cells were treated with the HDAC inhibitor TSA ± Dex, and CRH mRNA levels were measured. At a concentration that elicited approximately one-half maximal stimulation of promoter activity (10−8M; data not shown), TSA increased CRH mRNA levels 4.7-fold. This effect was reversed with co-administration of Dex (10−7M, Fig 2A). crh promoter activity also increased in response to TSA, by 3.8-fold, but Dex did not reduce this effect (Fig 2B). In the presence of over-expressed HDAC1, however, Dex did inhibit of TSA-induced promoter activation, by 47%. This effect was HDAC-specific, as over-expression of HDAC3 had no effect (Figure 2C). Taken together, these results suggest that HDAC1 plays a role in Dex inhibition of crh promoter activity and displays specificity in doing so.
Figure 2. TSA-induced crh expression is attenuated by HDAC1 but not HDAC3.
(A) TSA increases CRH mRNA levels. These levels are reduced by the co-administration of Dex. IVB cells were treated with TSA (10−8M) with or without Dex (10−7M) for two hours prior to mRNA isolation and quantification by real-time PCR. (B) Dex fails to suppress the TSA-mediated increase of crh promoter activity. IVBs transfected with the crh-LUC reporter and treated with TSA (10−8M) ± Dex (10−7M) for 18 hrs. (C) Over-expression of HDAC1 permits Dex reduction of TSA-enhanced transcription. IVBs were co-transfected with the crh reporter construct as in B, and with either 1 ug of HDAC 1 or 3 expression vector. Treatments: TSA (10−8M) ± Dex (10−7M) for 18 hours prior to luciferase assay. Error bars represent SEM. (*) TSA differs from vehicle: (A) p<0.02, (B) p<0.002 (#) TSA + Dex differs from TSA (A) p<0.05, (C) p<0.04.
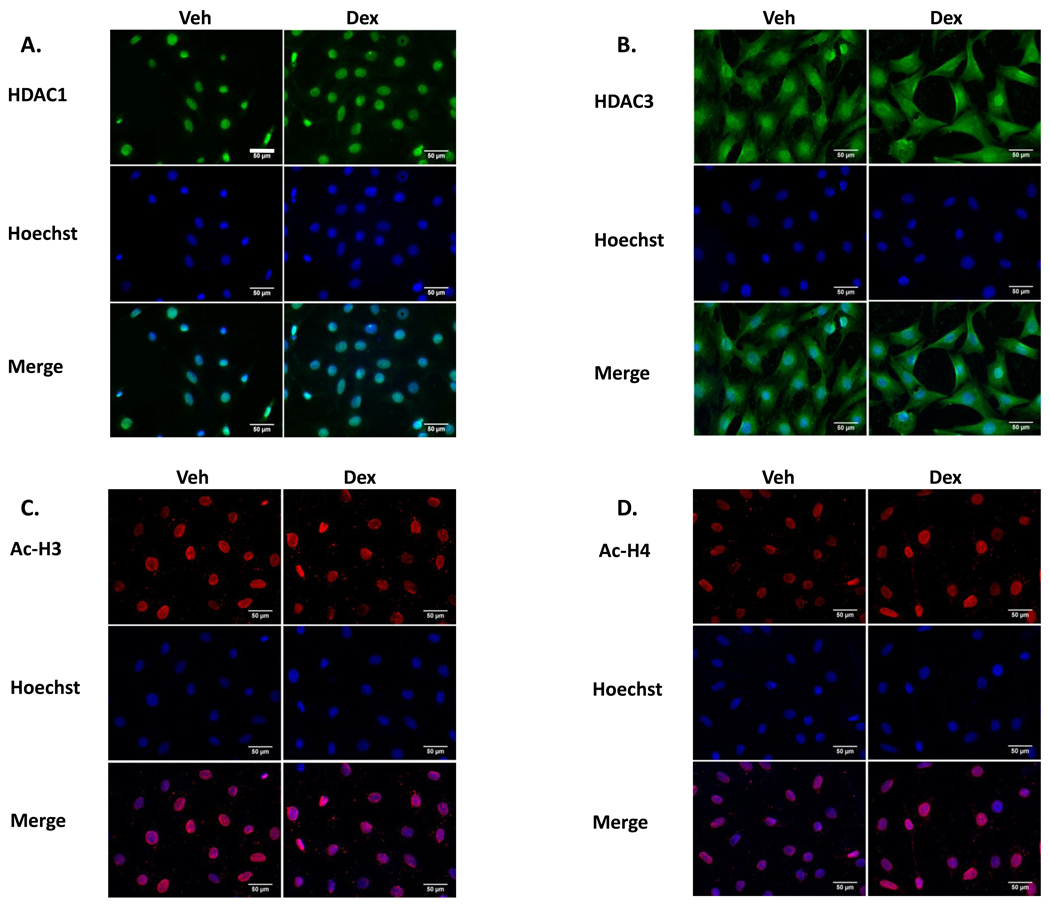
3.1.3 Dex fails to alter nuclear localization of HDACs 1- and 3-ir in IVB cells
As has been previously shown, HDAC1-ir is predominantly nuclear, whereas HDAC3 is both nuclear and cytoplasmic [23]. Subcellular localization of HDAC1- or HDAC3- ir was unaltered by Dex. (Fig 3A & B). Similarly, Ac-H3- or 4– ir were present exclusively in the nucleus, regardless of Dex treatment (Fig 3C & D).
Figure 3. The compartmental localization of HDACs 1 and 3 is unchanged by Dex.
(A,B) Top row: HDAC1-ir is Nuclear and HDAC3-ir is nuclear and cytoplasmic. (C,D) Top row: pan Ac H3 and H4-ir are nuclear. (A–D) Middle row: Hoechst stain; lower row: merged ICC and Hoechst images. All Bars = 50 µm. The bar in A, top row, left is thicker, for easier visualization.
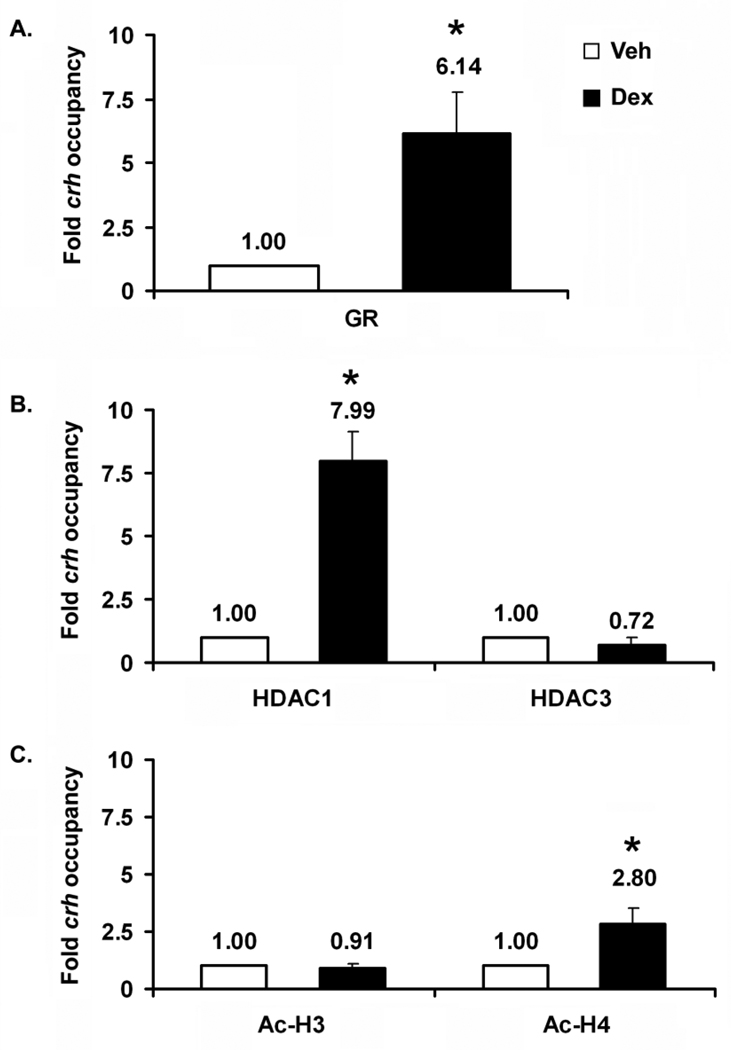
3.1.4 Dex alters GR and HDAC1 crh promoter occupancy and levels of H4 acetylation
To determine whether Dex treatment elicited changes in crh promoter occupancy by GR and HDACs 1 and 3, we performed ChIP analysis. GR occupancy increased over 6-fold after 2 hrs of Dex treatment (Fig 4A). Dex treatment led to an 8-fold increase in HDAC1 occupancy but had no effect on HDAC3 occupancy (Fig 4B). Dex treatment also led to increased levels of Ac- H4 but not pan-acetylated H3 (Fig 4C). Lastly, no GR occupancy was detected in far upstream region, between nucleotides −5113 through −1867 (not shown); thus, Dex modulates HDAC1 occupancy of the proximal crh promoter and displays specificity in doing so. That levels of pan-Ac-H4 but not pan-Ac-H3 are altered, suggests that changes in H4 but not 3 acetylation play a role in GR inhibition of crh promoter activity. Although an increase in acetylation is counter-intuitive with respect to gene repression, there is at least one instance in which acetylation is found at a repressed gene (see discussion, 4.1.3.2).
Figure 4. In the region of the proximal crh promoter, Dex increases crh occupancy by GR and HDAC1, and leads to increased Ac-H4.
ChIP analyses were performed to determine (A) GR occupancy, (B) HDAC1 and 3 occupancy, and (C) degree of H4 and H3 pan-acetylation. As in the case of the ICC experiments (Figs 1 and 3), cells were treated for 2 hrs. Error bars represent SEM. n=3–5 for each ChIP analysis. Vehicle is taken as one (left bars). (*) Dex treatment is significantly different than vehicle for GR (p<0.003) and HDAC1 (p<0.014) occupancy, and for the degree of H4 acetylation (p<0.006).
3.1.5 CRH-ir co-localizes with HDAC1- & 3-ir in the mpPVH
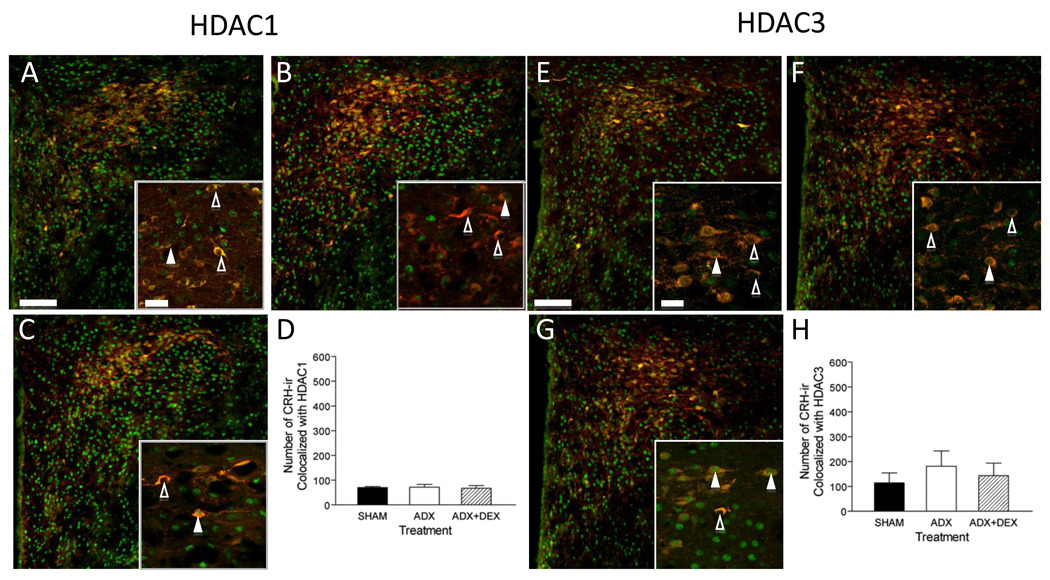
To study the effects of Dex in vivo, rats were subjected to adrenalectomy (ADX) with or without Dex treatment. Numerous cells displayed HDACs and histone acetylation marks. Furthermore, immunoreactivity for both HDACs and Ac-H3 and -H4 co-localized with CRH-ir, suggesting physiologic relevance of histone acetylation to crh regulation (Figs. 5 & 6).
Figure 5. HDACs 1 and 3-ir are present in CRH-ir PVH parvocellular neurons.
Hypothalamic sections from Sham ADX (A, E), ADX (B, F), and ADX + Dex (C, G) treated rats were examined for CRH-ir (red) and HDAC1- and 3-ir (green). (D,H) There is no significant difference between treatments for either combined HDAC1- and CRH-ir (D) or combined HDAC3- and CRH-ir (H). Bars: Low magnification =100µm; high magnification (inset) = 20µm. (△) no co-colocalization, (▲) co-localization.
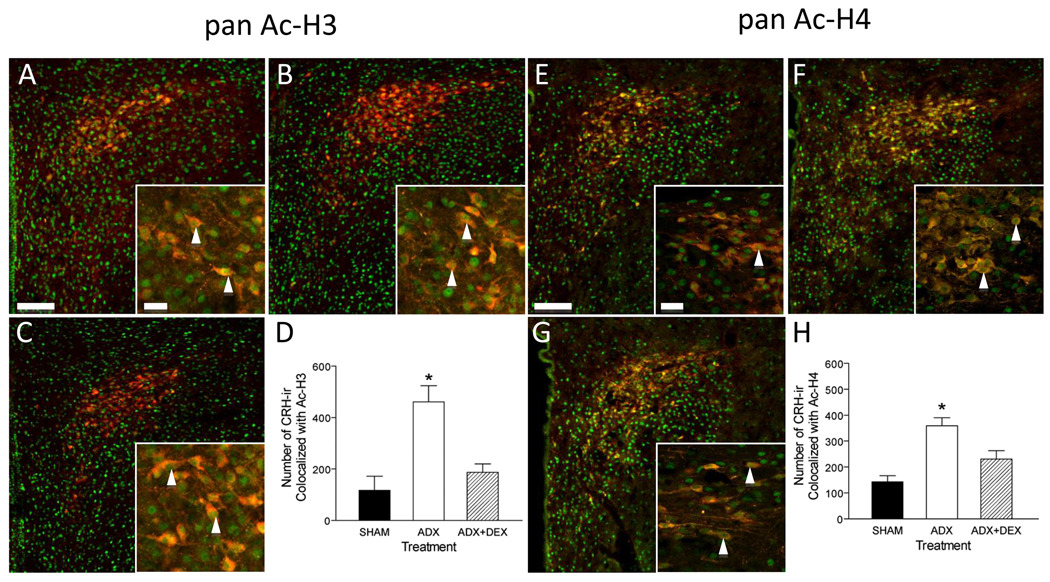
Figure 6. Dex replacement leads to increased numbers of pan-acetylated H3- and H4- ir in CRH-ir PVH parvocellular neurons.
Hypothalamic sections from Sham ADX (A, E), ADX (B, F), and ADX + Dex (C, G) treated rats were examined for CRH-ir (red) and pan-Ac H3- or 4-ir (green). (D,H) The number of CRH-ir cells that display either Ac-H3- or Ac-H4- ir increases after ADX. This increase is attenuated in the presence of Dex. Bars: Low magnification =100µm; high magnification (inset) = 20µm. (*) Significantly different from SHAM and ADX+DEX p<0.05. (▲) co-localization.
As predicted from numerous studies, ADX led to an increased number of CRH-ir cells (here 2–3 fold), which was attenuated by treatment with Dex (Figs. 5 & 6, B & F and data not shown). The number of neurons that displayed dual immunoreactivity for CRH-ir and either HDAC1 or 3 was determined across treatments. There were no significant differences between treatments for either HDAC1 and HDAC3 (Fig 4D and H). In distinction, the number of Ac-H3 and –H4 –ir cells that displayed CRH-ir increased after ADX (Fig 6 D and H) and were reduced by Dex administration.
4.1 Discussion
4.1.1 Summary
The findings presented indicate that HDAC1 plays a role in glucocorticoid-mediated down regulation of crh expression. Over-expression of HDAC1 restores Dex inhibition of TSA-stimulated crh promoter activity, and Dex leads to increased HDAC1 occupancy in the region of the crh proximal promoter. These findings are specific in that neither happens in the case of HDAC3. Specificity is also present in the degree to which H3 and H4 acetylation is present in the proximal region of the promoter. There is no response to Dex in the degree of H3 acetylation, whereas the degree of H4 acetylation increases. In addition, the in vivo findings reveal co-localization of CRH-ir with both HDACs and with both pan-AcH3 and -AcH4; thus, the in vitro findings likely bear on in vivo regulation of crh expression.
4.1.2 Comparative analysis of in vitro findings with the in vivo findings
One element necessary for a meaningful in vitro/in vivo correlation is that GRs are present in CRH-ir neurons that display HDAC1 and 3 and AcH3/4 immunoreactivity. GR-ir is present in 100% of CRH-ir mpPVH neurons [24]; thus, by extrapolation, every CRH-ir neuron positive for HDAC1 or 3 has the potential for an HDAC1 or 3 and functional interaction with GR.
Two sets of in vivo data appear to be at odds with the in vitro data. In the first case, the number of HDAC1-ir neurons that colocalize with CRH-ir in vivo is invariant by glucocorticoid status (Fig 5D and H), even though by all in vitro measures HDAC1 appears to play a role in modulating crh expression. These in vitro and in vivo findings, however, are not mutually exclusive. Even though the cellular concentration of HDAC1 may be unchanged in vivo, its engagement with the promoter could increase in the presence of Dex. In the second case, the number of CRH-ir cells that co-express either Ac-H3 and Ac-H4 both vary by glucocorticoid status. In each case, ADX leads to increased numbers of CRH-ir cells that co-localize with AcH3- or AcH4-ir, and addition of Dex attenuates the number of cells displaying the colocalization (Fig 6D and H). This finding is distinct from the in vitro data, in which only pan-AcH4 is responsive to glucocorticoid status. The in vivo data reflect the chromatin state of an entire nucleus thus, the chromatin of individual sets of genes that are differentially acetylated in response to altered glucocorticoid levels could well be obscured by extensive histone acetylation.
4.1.3 Mechanisms of GR repression
4.1.3.1 Transcription factors involved
Early studies of crh regulation demonstrated that Dex could repress stimulated crh promoter activity via the proximal CRE at approximately −220 [6]. Involvement of this CRE in crh regulation has been found repeatedly [6, 21, 25–27]. For example, Liu et al. (2006) showed that forskolin treatment led to a shift from CREB occupancy to ICER occupancy[21], indicating that the involvement of a given CREB family member is a point of regulation. In addition to this CRE, an nGRE is present at −278 to −249 [5, 28]. This nGRE is a composite element and, like the composite element in the proliferin gene promoter [4], it binds to hetero-dimers of AP-1 family members and GR. Such composite elements are known to mediate GR repression, as in the case of the proliferin gene [4].
4.1.3.2 Acetylation
In general, states of acetylation are associated with activated gene expression and deacetylation with repressed gene expression; thus, the finding of an increased level of HCAC1 occupancy in tandem with increased H4 pan-acetylation (Fig 4), appears incongruous. It is unlikely, however, that an open chromatin state permits access to activators alone; thus, one could imagine a scenario in which local transition from a hypoacetylated state to an acetylated state would permit access to a co-repressor. This mechanism would be in keeping with the ChIP data.
An alternate scenario would be that acetylation of a quiescent promoter would be necessary for later recruitment of a different histone acetylase in response to an altered cell state. This scenario has been described in the case of the PHO5 gene in yeast whose regulation is critical for appropriate phosphate metabolism. In the case of PHO5, acetylation by the nucleosome acetyltransferase of H4 (NuA4) is required for an appropriate response to altered phosphate levels; thus, NuA4 prepares PHO5 for engagement of another HAT, Spt-ADA-Gcn5-acetyl transferase (SAGA), which opens the chromatin for transcription in response to a change in phosphate levels (see [29] for review).
The PHO5 sequence of events points to the importance of considering transcriptional regulation in a temporal context. It has been shown in many cases that promoter occupancy is a sequence of ordered and cyclic events; regulatory proteins engage and disengage in a state of equilibrium. Germane here is the elegant work performed by Gordon Hager and his colleagues. For example, Straveva et al. showed that pulsatile glucocorticoid levels lead to pulsatile gene expression at the MMTV promoter. This pattern mimics the naturally occurring ultradian rhythm of corticosterone. In contrast, constant exposure fails to elicit similar oscillations [30]. Cyclic occupancy has been demonstrated for ERα as well. A comprehensive study by Metivier et al. showed that in the presence of estradiol (E2) at the pS2 promoter ERα and numerous coregulators also display ordered cyclic occupancy [31]. Interestingly, he later showed that apo-ERα also displays ordered cyclic occupancy of the same promoter [32]. As for the crh promoter, exposure to E2 leads to phasic occupancy of ERα and ERβ, which are coordinated with pCREB, CBP, and SRC-1 in a specific manner. The patterns of ERα and β occupancy, in turn, correspond with the degree of Ac-H3 and Ac-H4, suggesting that assembly of ERα and ERβ complexes are functional [14]. Such analysis has not yet been reported for GR occupancy of the crh promoter. Given the profile of GR at MMTV, ERα at pS2 and ERα and β at the crh promoter, in all likelihood GR regulates ordered cyclic occupancy of coregulators and associated histone modification enzyme at crh as well. Whether this is driven by glucocorticoid pulses or is an intrinsic property of GR:crh interactions remains to be determined.
4.1.4 Acetylation in relation to methylation
Taken together, our studies suggest that in the sub-population of parvocellular neurons, that contain CRH and HDAC1, glucocorticoids negatively regulate crh expression at the level of chromatin via a mechanism that involves acetylation. Specifically the mechanism includes increased GR and HDAC1occupancy and resultant changes in H4 acetylation.
Although histone acetylation is arguably the first chromatin modification to have been studied extensively, there are numerous other chromatin modifications that play a role in gene expression. These include methylation, both of histones and of DNA, phosphorylation and ubiquitination [33]. Of these, methylation of CpG islands has been demonstrated to play a role in crh expression. Strikingly, the inability to appropriately transfer information from methylated CpGs [34] and the degree of methylation in the proximal crh promoter [35]are both tightly correlated with behavior associated with abnormal regulation of CRH expression.
One reason for an inability to transfer information from CpGs in the crh promoter has been shown to be a result of mutated methylated CpG-2 binding protein (MeCP-2). Mutations in MeCP2 underlie Rett Syndrome, a cognitive disorder with behavioral and emotional manifestations. The emotional manifestations include anxiety and low mood. In one study, these manifestations were found to be present in 76 and 70 per cent of the cases, respectively [36]. Anxiety and low mood are relevant here in that they both are associated with abnormal CRH regulation. Increased CRH in extra-hypothalamic sites is associated with anxiety. Increased CRH in the PVH is associated with depression. Using a mouse model of Rett Syndrome, one in which the mice carry an Mecp2 mutation (Mecp2308/Y), McGill et al found that anxiety was increased in the Mecp2308/Y mice as measured by the elevated-plus maze. Additionally, activation of the HPA axis was excessive in response to restraint stress. In situ hybridization in unperturbed mice revealed that CRH mRNA levels are greater in the PVH and the amygdala of mutant mice compared to wild type. In their ChIP analysis of the crh promoter McGill et al. found that in wild type mice MeCP2 bound to CpG sequences in the proximal promoter. This was not the case for mice bearing the Mecp2308/Y mutation [34]. Thus mutation of MeCP2 has profound influences on crh expression, which in turn lead to features associated with anxiety and depression.
To determine the extent to which CpG methylation plays a role in crh regulation, Elliott et al studied the degree of CpG methylation present at the crh proximal promoter [35]. Increased CRH mRNA and decreased methylation were tightly correlated with exposure to chronic social stress in a subset of mice. This subset displayed social avoidance behavior and were considered defeated. Furthermore, treatment of defeated mice with imipramine restored both CRH mRNA levels to levels of unstressed mice and increased the degree of CpG methylation.
Two major points may be made with respect to the relevance of the McGill and Elliott studies to the studies reported here. First, they both targeted a region of the promoter included in the in vitro studies presented, the region that includes the CRE. The McGill study and ours also included the nGRE. Second, acetylation and DNA methylation have been linked in mechanisms of gene repression, see Dobosy for review [37]. For example, HDAC1 is present in the MeCP2/Sin3A repressive complex. Taken together, the data suggest that functional interactions between HDAC1 and MeCP2 are involved in repression of crh gene expression.
4.1.5 Significance
The HPA axis is the neuroendocrine underpinning of the response to stress. Axis dysregulation has been long associated with depression, and the maladaptive step is thought to be the inability to appropriately down-regulate aspects of CRH signaling. From the discussion above, it is highly likely that a repressive complex participates in down-regulating crh expression, in particular the MeCP2/HDAC1/Sin3A complex. The order of events that leads to a functional repressive complex at the crh proximal promoter has yet to be elucidated. Regardless, the HDAC1 component of this complex may now be considered a novel target for re-setting HPA axis activity to a homeostatic state.
Acknowledgments
This work was funded by NIH R01 MH82900 (RMU)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vale W, Spiess J, Rivier C, Rivier J. Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science. 1981;213:1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- 2.Goodman & Gilman's The Pharmacological Basis of Therapeutics. 11e ed2011. [Google Scholar]
- 3.de Kloet ER, Joels M, Holsboer F. Stress and the brain: From adaptation to disease. Nature Reviews Neuroscience. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- 4.Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 5.Malkoski SP, Handanos CM, Dorin RI. Localization of a negative glucocorticoid response element of the human corticotropin releasing hormone gene. Mol Cell Endocrinol. 1997;127:189–199. doi: 10.1016/s0303-7207(96)04004-x. [DOI] [PubMed] [Google Scholar]
- 6.Guardiola-Diaz HM, Kolinske JS, Gates LH, Seasholtz AF. Negative glucorticoid regulation of cyclic adenosine 3', 5'-monophosphate-stimulated corticotropin-releasing hormone-reporter expression in AtT-20 cells. Mol Endocrinol. 1996;10:317–329. doi: 10.1210/mend.10.3.8833660. [DOI] [PubMed] [Google Scholar]
- 7.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 8.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, Rosenfeld MG. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 9.Wang DF, Helquist P, Wiech NL, Wiest O. Toward selective histone deacetylase inhibitor design: homology modeling, docking studies, and molecular dynamics simulations of human class I histone deacetylases. J Med Chem. 2005;48:6936–6947. doi: 10.1021/jm0505011. [DOI] [PubMed] [Google Scholar]
- 10.Jee YK, Gilmour J, Kelly A, Bowen H, Richards D, Soh C, Smith P, Hawrylowicz C, Cousins D, Lee T, Lavender P. Repression of interleukin-5 transcription by the glucocorticoid receptor targets GATA3 signaling and involves histone deacetylase recruitment. J Biol Chem. 2005;280:23243–23250. doi: 10.1074/jbc.M503659200. [DOI] [PubMed] [Google Scholar]
- 11.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasckow J, Mulchahey JJ, Aguilera G, Pisarska M, Nikodemova M, Chen HC, Herman JP, Murphy EK, Liu Y, Rizvi TA, Dautzenberg FM, Sheriff S. Corticotropin-releasing hormone (CRH) expression and protein kinase A mediated CRH receptor signalling in an immortalized hypothalamic cell line. J Neuroendocrinol. 2003;15:521–529. doi: 10.1046/j.1365-2826.2003.01026.x. [DOI] [PubMed] [Google Scholar]
- 13.Miller WJ, Suzuki S, Miller LK, Handa R, Uht RM. Estrogen receptor (ER)beta isoforms rather than ERalpha regulate corticotropin-releasing hormone promoter activity through an alternate pathway. Journal of Neuroscience. 2004;24:10628–10635. doi: 10.1523/JNEUROSCI.5540-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lalmansingh AS, Uht RM. Estradiol regulates corticotropin-releasing hormone gene (crh) expression in a rapid and phasic manner that parallels estrogen receptor-alpha and -beta recruitment to a 3',5'-cyclic adenosine 5'-monophosphate regulatory region of the proximal crh promoter. Endocrinology. 2008;149:346–357. doi: 10.1210/en.2007-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki S, Handa RJ. Regulation of estrogen receptor-beta expression in the female rat hypothalamus: differential effects of dexamethasone and estradiol. Endocrinology. 2004;145:3658–3670. doi: 10.1210/en.2003-1688. [DOI] [PubMed] [Google Scholar]
- 16.Watson RE, Jr, Wiegand SJ, Clough RW, Hoffman GE. Use of cryoprotectant to maintain long-term peptide immunoreactivity and tissue morphology. Peptides. 1986;7:155–159. doi: 10.1016/0196-9781(86)90076-8. [DOI] [PubMed] [Google Scholar]
- 17.Uht RM, Anderson CM, Webb P, Kushner PJ. Transcriptional activities of estrogen and glucocorticoid receptors are functionally integrated at the AP-1 response element. Endocrinology. 1997;138:2900–2908. doi: 10.1210/endo.138.7.5244. [DOI] [PubMed] [Google Scholar]
- 18.Chakrabarti SK, James JC, Mirmira RG. Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1. Importance of chromatin structure in directing promoter binding. Journal of Biological Chemistry. 2002;277:13286–13293. doi: 10.1074/jbc.M111857200. [DOI] [PubMed] [Google Scholar]
- 19.Chakrabarti SK, Francis J, Ziesmann SM, Garmey JC, Mirmira RG. Covalent histone modifications underlie the developmental regulation of insulin gene transcription in pancreatic beta cells. J Biol Chem. 2003;278:23617–23623. doi: 10.1074/jbc.M303423200. [DOI] [PubMed] [Google Scholar]
- 20.Swanson L. Brain Maps: Structure of the Rat Brain. Amsterdam: The Netherlands: Elsevier Science Publishers B.V.; 1992. [Google Scholar]
- 21.Liu Y, Kalintchenko N, Sassone-Corsi P, Aguilera G. Inhibition of corticotrophin-releasing hormone transcription by inducible cAMP-early repressor in the hypothalamic cell line, 4B. J Neuroendocrinol. 2006;18:42–49. doi: 10.1111/j.1365-2826.2005.01383.x. [DOI] [PubMed] [Google Scholar]
- 22.Nikodemova M, Kasckow J, Liu H, Manganiello V, Aguilera G. Cyclic adenosine 3',5'-monophosphate regulation of corticotropin-releasing hormone promoter activity in AtT-20 cells and in a transformed hypothalamic cell line. Endocrinology. 2003;144:1292–1300. doi: 10.1210/en.2002-220990. [DOI] [PubMed] [Google Scholar]
- 23.Yang WM, Tsai SC, Wen YD, Fejer G, Seto E. Functional domains of histone deacetylase-3. J Biol Chem. 2002;277:9447–9454. doi: 10.1074/jbc.M105993200. [DOI] [PubMed] [Google Scholar]
- 24.Uht RM, McKelvy JF, Harrison RW, Bohn MC. Demonstration of glucocorticoid receptor-like immunoreactivity in glucocorticoid-sensitive vasopressin and corticotropin-releasing factor neurons in the hypothalamic paraventricular nucleus. J. Neurosci. Res. 1988;19:405–411. doi: 10.1002/jnr.490190404. [DOI] [PubMed] [Google Scholar]
- 25.Thompson RC, Seasholtz AF, Herbert E. Rat corticotropin-releasing hormone gene: sequence and tissue-specific expression. Mol Endocrinol. 1987;1:363–370. doi: 10.1210/mend-1-5-363. [DOI] [PubMed] [Google Scholar]
- 26.Thompson RC, Seasholtz AF, Douglass JO, Herbert E. The rat corticotropin-releasing hormone gene. Ann N Y Acad Sci. 1987;512:1–11. doi: 10.1111/j.1749-6632.1987.tb24947.x. [DOI] [PubMed] [Google Scholar]
- 27.Guardiola-Diaz HM, Boswell C, Seasholtz AF. The cAMP-responsive element in the corticotropin-releasing hormone gene mediates transcriptional regulation by depolarization. J Biol Chem. 1994;269:14784–14791. [PubMed] [Google Scholar]
- 28.Malkoski SP, Dorin RI. Composite glucocorticoid regulation at a functionally defined negative glucocorticoid response element of the human corticotropin-releasing hormone gene. Mol Endocrinol. 1999;13:1629–1644. doi: 10.1210/mend.13.10.0351. [DOI] [PubMed] [Google Scholar]
- 29.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 30.Stavreva DA, Wiench M, John S, Conway-Campbell BL, McKenna MA, Pooley JR, Johnson TA, Voss TC, Lightman SL, Hager GL. Ultradian hormone stimulation induces glucocorticoid receptor-mediated pulses of gene transcription. Nat Cell Biol. 2009;11:1093–1102. doi: 10.1038/ncb1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 32.Metivier R, Penot G, Carmouche RP, Hubner MR, Reid G, Denger S, Manu D, Brand H, Kos M, Benes V, Gannon F. Transcriptional complexes engaged by apo-estrogen receptor-alpha isoforms have divergent outcomes. EMBO J. 2004;23:3653–3666. doi: 10.1038/sj.emboj.7600377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 34.McGill BE, Bundle SF, Yaylaoglu MB, Carson JP, Thaller C, Zoghbi HY. Enhanced anxiety and stress-induced corticosterone release are associated with increased Crh expression in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A. 2006;103:18267–18272. doi: 10.1073/pnas.0608702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat Neurosci. 2010;13:1351–1353. doi: 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- 36.Sansom D, Krishnan VH, Corbett J, Kerr A. Emotional and behavioural aspects of Rett syndrome. Dev Med Child Neurol. 1993;35:340–345. doi: 10.1111/j.1469-8749.1993.tb11646.x. [DOI] [PubMed] [Google Scholar]
- 37.Dobosy JR, Selker EU. Emerging connections between DNA methylation and histone acetylation. Cell Mol Life Sci. 2001;58:721–727. doi: 10.1007/PL00000895. [DOI] [PMC free article] [PubMed] [Google Scholar]