Abstract
Variations in diabetic phenotypes are caused by complex interactions of genetic effects, environmental factors, and the interplay between the two. We tease apart these complex interactions by examining genome-wide genetic and epigenetic effects on diabetes-related traits among different sex, diet, and sex-by-diet cohorts in a Mus musculus model. We conducted a genome-wide scan for quantitative trait loci affecting serum glucose and insulin levels and response to glucose stress in an F16 Advanced Intercross Line of the LG/J and SM/J intercross (Wustl:LG, SM-G16). Half of each sibship was fed a high-fat diet and half was fed a relatively low-fat diet. Context-dependent genetic (additive and dominance) and epigenetic (parent-of-origin imprinting) effects were characterized by partitioning animals into sex, diet, and sex-by-diet cohorts. We find that different cohorts often have unique genetic effects at the same loci, and that genetic signals can be masked or erroneously assigned to specific cohorts if they are not considered individually. Our data demonstrate that the effects of genes on complex trait variation are highly context dependent, and that the same genomic sequence can affect traits differently depending on an individual’s sex and/or dietary environment. Our results have important implications for studies of complex traits in humans.
Keywords: quantitative trait loci, imprinting, context-dependency, mouse models, diabetes
The prevalence of type-2 diabetes mellitus (T2D) has increased steadily over the past two decades, currently afflicting ≈ 8% of the adult population in the United States (National_Diabetes_Information_Clearinghouse 2005). Hypotheses have been proposed to explain this epidemic in terms of changing environmental factors, for example the thrifty gene and the sedentary lifestyle hypotheses, which suggest that increased caloric intake combined with decreased caloric output is the major cause (Neel 1962). However, there is clearly an underlying genetic component: T2D is highly heritable, ranging from ~50–90% in monogenetic twin studies, and disease incidence varies with an individual’s percent ethnic ancestry (Clee and Attie 2007; Permutt et al. 2005). Linkage analysis and positional cloning have led to identification of several genes with large effect sizes associated with T2D, generally through studies of relevant phenotypes such as pancreatic β-cell function (e.g. the ATP-binding cassette, ABCC8, (Huopio et al. 2003)), insulin resistance (e.g. insulin-degrading enzyme, IDE, (Farris et al. 2003)), and obesity (e.g. peroxisome proliferator-activated receptor γ, PPARG (Altshuler et al. 2000)). More recently, genome-wide association studies (GWAS) attempt to identify new variants with small effect sizes through hypothesis-free testing (Wolfs et al. 2009).
The National Human Genome Research Institute compiled a hand-curated catalog of published human GWAS (www.genome.gov/gwastudies) (Hindorff et al. 2009), and as of this writing, there are 30 unique genes associated with T2D. However, these genes account for a very small percentage, ≈ 5–10%, of the overall heritable variation of T2D in humans (Salanti et al. 2009). This percentage could in fact be an inflation because gene-by-environmental covariation and non-assortative mating are not accounted for in GWAS (Falconer and Mackay 1996). This is partly because these meta-analyses were designed with the common-disease/ common-variant hypothesis in mind. As such, they are effective in identifying associations between common phenotypes and common allelic variants in homogeneous populations, but they are not very effective in identifying such associations in heterogeneous populations, such as humans (McCarthy et al. 2008). T2D is a common disease, but the alleles underlying the phenotype are many and relatively rare, and most will not pass the stringent multiple testing criteria necessary to claim association, hence the “missing heritability” that is attracting so much attention in the wake of GWAS popularity (McCarthy and Zeggini 2009). In addition to lack of statistical power to detect genes of small effect size, other suggested culprits underlying the low proportion of heritable variation explained include cryptic sub-population structure, epistatic interactions among loci, and genomic structural polymorphisms such as copy number variants (Manolio et al. 2009). We suggest an additional factor contributes to this missing heritability in GWAS studies: not accounting for the context-dependency of the genetic effects of the trait.
Here we present results of a study investigating the context dependency of genetic effects on glucose and insulin phenotypes in response to a high- and low-fat diet in an F16 Advanced Intercross Line (AIL) formed from the LG/J and SM/J inbred mouse strains (Wustl:LG, SM-G16). Variation in complex traits in LG/J x SM/J is due to many genes of small effect interacting with each other and with the environment, and quantitative trait loci (QTL) have previously been mapped for obesity, serum chemistry and growth-related phenotypes (Cheverud et al. 2004; Cheverud et al. 1996; Ehrich et al. 2005; Fawcett et al. 2009; Fawcett et al. 2008; Kenney-Hunt et al. 2006). In addition to reporting additive and dominance effects, in this study we also present for the first time genome-wide parent-of-origin imprinting effects. Genomic imprinting can be generally defined as the unequal expression of maternally and paternally derived copies of a gene, and its effects have been shown to play a role in development of T2D and its co-morbidities, such as obesity (Rampersaud et al. 2008; Weinstein et al. 2009; Xie et al. 2008).
RESEARCH DESIGN AND METHODS
Mouse population
The mice used in this study are from the F16 generation of the LG/J x SM/J AIL (Wustl:LG, SM-G16). The LG/J mice were generated as part of an experiment selecting for large body size at 60 days (Goodale 1941). The SM/J mice were generated as part of a separate selection experiment for small body size at 60 days (MacArthur 1944). At this age, there is ~20g difference in body size between these two strains (Kramer et al. 1998). Animals from each strain have been inbred by brother-sister mating for over 150 generations.
The LG/J x SM/J AIL is managed as a pseudo-randomly mated line starting from the F2 generation. Initially, 10 male SM/J mice were crossed with 10 female LG/J mice obtained from the Jackson Laboratories. Animals are randomly mated except that brother-sister mating is not allowed. As a further check on inbreeding, one male and one female are chosen, when possible, from each family as breeders for the next generation. Eliminating variation in familial contributions to the next generation doubles the effective population size of a colony relative to its census size and is an effective method of reducing the rate of inbreeding (Templeton 2006). The average number of breeding pairs in the production of the AIL is 75, giving a census size of 150 and an effective size of approximately 300.
For this study, 71 pairs of F15 animals were double mated, resulting in an experimental F16 sample of 1,002 animals in 76 litters, with an average of 6.8 animals per sibship. Pups were housed with their mothers until being weaned at 3 weeks of age and then separated into sex-specific cages of four or five animals per cage. The animal facility operates on a 12-hour light/dark cycle with a constant temperature of 21°C. Male (n=500) and female (n=502) animals from each litter were each partitioned and fed low-fat (247 males; 254 females), and high-fat (253 males, 248 females) diets. The diets were selected to be isocaloric and as similar as possible in nutrient composition, except for fat (supporting information, Table S1). Calories from fat are 15% and 43% in the low- (catalog TD88137, Harlan Teklad, Madison, WI) and high- (catalog D12284, Research Diets, New Brunswick, NJ) fat diets, respectively. All animals were fed ad libitum.
Phenotypic data
Animals were weighed weekly for 20 weeks. A subset of animals (217 females, 113 fed the low-fat diet and 104 fed the high-fat diet; 213 males, 103 fed the low-fat diet and 110 fed the high-fat diet) were subject to an intra-peritoneal glucose tolerance test (IPGTT) at 10 and 20 weeks of age as described in Ehrich et al. (2005). Briefly, a 4-hour fasting glucose level was measured followed by an intra-peritoneal injection of 0.01ml of 10% glucose solution per gram of body weight. Measurements taken over the course of 2 hours were used to calculate the area under the curve (AUC), an overall measure of glucose tolerance. At 20 weeks of age, animals were necropsied as described in Ehrich et al. (2005). Briefly, animals were anesthesized by intra-peritoneal injection of sodium pentobarbital after a 4 hour fast. Serum samples were obtained via cardiac puncture and processed to measure serum glucose and insulin levels. Genetic mapping of fat pad weights (reproductive, renal, inguinal and mesenteric) and organ weights (heart, liver, kidneys and spleen) is reported in (Cheverud et al. 2010a) and (Lawson et al. 2010a).
Genotypic Data
DNA was extracted from liver tissue using the QIAGEN kit. We selected 1536 single nucleotide polymorphisms (SNPs) from the CTC/Oxford SNP survey (www.well.ox.ac.uk/mouse/INBREDS/) for scoring with the Illumina Golden Gate Assay. SNP typing was performed at the Washington University Genome Sequencing Center, and 1402 autosomal SNPs were reliably scored and used for this analysis (supporting information, Table S2). A genetic map was created for these SNPs, based on their physical order as given in the mouse genome database (mm9; NCBI build 37), and recombination fractions between the markers were estimated using the package R/qtl (Broman and Saunak 2009). Due to the accumulation of recombination over the generations, the F16 population described here has approximately 8 times the recombination of the F2 generation.
Using SNP data from families, ordered genotypes were reconstructed at each marker for all F16 animals using the Integer Linear Programming algorithm as implemented in PedPhase 2.1 (Li and Jiang 2005). Additive (Xa) and dominance (Xd) genotypic scores were assigned at each marker: Xa = 1, 0, −1 and Xd = 0, 1, 0 for the LG/LG, LG/SM and SM/LG, and SM/SM genotypes, respectively. The ‘LG’ refers to an allele derived from the LG/J strain and ‘SM’ refers to an allele derived from the SM/J strain. Additionally, we assigned imprinting genotypic scores (Xi) to distinguish between the two reciprocal heterozygotes, LG/SM and SM/LG, where the first allele is inherited from the father and the second from the mother. For the four ordered genotypes, LG/LG, LG/SM, SM/LG, and SM/SM, Xi = 0, +1, −1, 0, respectively (Wolf et al. 2008). Additional genotypes were imputed at 1cM intervals among the SNPs using the equations of Haley and Knott (Haley and Knott 1992) with the addition of newly derived equations for imputing imprinting genotypic scores (supporting information, Table S3).
QTL analysis
Single locus analyses were performed using maximum likelihood in the Mixed Procedure in SAS 9.2. Our full mapping model included: sex, diet, the direct effects of the genomic locations (Xa, Xd, Xi), and their two- and three-way interactions with sex and diet as fixed effects. The full model explains variation in trait (Y) using the linear equation:
where μ is the population mean and e is the residual. Family and its interactions with sex and diet, including the three-way interactions, were included as random effects in the model. The addition of these random effects corrects for inflation of LOD scores caused by family structure within the colony. The −2 ln(likelihood) of this model was compared to a null model including only sex, diet and sex-by-diet interaction terms using a chi-square test with 12 degrees of freedom. Probabilities were transformed into LOD = −log10(Pr). The regression coefficients are the additive [a = (GLG/LG)−(GSM/SM))/2], dominance [d = ((GLG/SM+GSM/LG)−(GLG/LG−GSM/SM))/2] and imprinting [i = (GLG/SM−GSM/LG)/2] genotypic values, where G refers to the mean phenotype of all individuals sharing the subscripted genotype, and their interactions with sex (s) and/or with diet (d).
The number of independent tests was calculated using the Li and Ji method based on the eigenvalues of the correlation matrix of marker additive genotype scores (Li and Ji 2005). This was then used to calculate Bonferroni adjusted significance thresholds, 1−(1−α)1/M, where M is the number of independent tests. A significance threshold was calculated at the genome-wide level (LOD ≥3.90) as well as separately for each autosome (supporting information, Table S4). With chromosome-wise significance we expect 1 false positive chromosomal result per trait in the study. These results overwhelm this expectation in that there are 6- to 10-times the number of significant chromosomal results for each trait than expected by chance under the null model of no QTL. QTL support regions were determined using a standard one LOD drop from the peak of the QTL.
RESULTS
QTL Results
We find 70 trait-specific QTL mapping to 64 locations across the genome. Of these 70 QTL, 17 are significant at the genome-wide level and 53 are significant at the chromosome-wise level. The most commonly mapped trait is glucose tolerance at 20 weeks with 25 QTL. Glucose tolerance at 10 weeks has 17 QTL, followed by serum insulin level at necropsy with 15 QTL, serum glucose level at necropsy with 6 QTL, basal glucose level at 10 weeks with 4 QTL, and basal glucose level at 20 weeks with 3 QTL. Fifty-nine percent of these QTL have significant additive effects, 54% have significant dominance effects and 59% have significant imprinting effects. However, only ~9% have additive, dominance and imprinting as main effects (n=6) without interactions. The majority of these QTL have significant interactions with sex and/or with diet (Table 1; 91%, n=64).
TABLE 1.
Breakdown of genome-wide QTL fine-mapped in this study
| Chr | QTL | Trait* | LOD | POS (Mb) | Proximal CI | Distal CI | Interaction¶
|
Total Genes | Glucose and/or Insulin Affecting Genes | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ADD | DOM | IMP | |||||||||
| 1 | Ddiab1a | AUC_20wks | 3.32 | 67.99 | 66.31 | 68.56 | sd | s | 0 | 9 | Acadl |
| Ddiab1b | bGLC_20wks | 3.79 | 77.73 | 76.25 | 78.28 | d | sd | 0 | 2 | ||
| Ddiab1c | AUC_10wks | 3.35 | 190.62 | 189.77 | 191.29 | 0 | sd | 0 | 4 | ||
|
| |||||||||||
| 2 | Ddiab2a | AUC_10wks | 4.06 | 59.26 | 58.53 | 61.58 | sd | s | 1 | 15 | |
| Ddiab2b | INS | 4.13 | 69.20 | 67.96 | 70.35 | sd | 0 | d | 23 | G6pc2 | |
| Ddiab2c | GLC | 3.30 | 73.81 | 71.73 | 76.29 | 1 | 0 | 0 | 39 | Rapgef4 | |
| Ddiab2d | GLC | 3.38 | 99.06 | 93.29 | 99.82 | 1 | 0 | s | 14 | ||
| Ddiab2e | INS | 3.60 | 103.71 | 102.74 | 106.63 | 0 | d | 0 | 34 | ||
| Ddiab2f | AUC_20wks | 4.18 | 145.71 | 145.01 | 147.25 | 0 | 0 | s | 13 | Insm1, Nkx2-2 | |
|
| |||||||||||
| 3 | Ddiab3a | AUC_20wks | 3.55 | 15.21 | 14.27 | 16.50 | 0 | s | d | 12 | |
| Ddiab3b | AUC_10wks | 5.61 | 61.07 | 59.44 | 66.07 | 1 | 0 | 0 | 32 | Sucnr1 | |
| Ddiab3c | INS | 3.57 | 135.46 | 133.52 | 135.82 | 0 | sd | sd | 27 | Fabp2 | |
|
| |||||||||||
| 4 | Ddiab4a | AUC_20wks | 3.58 | 55.34 | 55.34 | 60.38 | 0 | 0 | sd | 40 | |
| Ddiab4b | AUC_20wks | 3.68 | 122.63 | 120.73 | 123.57 | sd | 0 | 0 | 26 | ||
| Ddiab4c | AUC_20wks | 3.80 | 151.63 | 150.49 | 152.94 | 0 | 0 | sd | 23 | ||
|
| |||||||||||
| 5 | Ddiab5a | AUC_10wks | 11.89 | 141.54 | 139.55 | 143.37 | 0 | 0 | sd | 46 | Gper |
|
| |||||||||||
| 6 | Ddiab6a | AUC_20wks | 3.24 | 48.94 | 48.02 | 51.57 | 0 | sd | 0 | 44 | Igf2bp3 |
| Ddiab6b | bGLC_20wks | 2.86 | 78.00 | 77.21 | 79.74 | 0 | sd | sd | 7 | ||
| Ddiab6c | AUC_20wks | 7.32 | 88.54 | 84.28 | 91.81 | sd | sd | sd | 112 | Alms1, Gfpt1, Tpra1, Klf15 | |
| Ddiab6d | INS | 8.85 | 107.02 | 100.75 | 115.43 | sd | sd | 0 | 65 | Edem, Ghrl, Pparg | |
| nGLC | 3.19 | 107.02 | sd | 1 | 0 | ||||||
| AUC_2 0wks | 3.43 | 107.21 | sd | 0 | d | ||||||
|
| |||||||||||
| 7 | Ddiab7a | INS | 2.75 | 26.18 | 25.25 | 26.60 | 0 | sd | 0 | 53 | Gsk3a, Lipe |
| Ddiab7b | AUC_10wks | 3.01 | 52.12 | 48.49 | 58.32 | 1 | d | 0 | 222 | Nr1h2, Akt1s1, Trpm4, Gys1, Fgf21, Kcnj11, Abcc8 | |
| AUC_20wks | 3.65 | 56.01 | 0 | sd | 0 | ||||||
| Ddiab7c | bGLC_10wks | 2.87 | 69.70 | 65.96 | 73.76 | 1 | 0 | s | 39 | Atp10a | |
| INS | 2.76 | 72.38 | 1 | 0 | d | ||||||
| Ddiab7d | AUC_20wks | 4.90 | 79.00 | 78.13 | 80.14 | 0 | sd | 0 | 1 | ||
|
| |||||||||||
| 8 | Ddiab8a | AUC_10wks | 4.26 | 29.36 | 21.55 | 39.08 | sd | 0 | sd | 132 | Ikbkb, Adrb3 |
| Ddiab8b | AUC_10wks | 4.11 | 71.23 | 64.98 | 75.50 | sd | 0 | sd | 148 | Cpe, Lpl, Npy1r, Pik3r2, Rab3a, Slc27a1 | |
|
| |||||||||||
| 9 | Ddiab9a | AUC_10wks | 3.66 | 15.64 | 15.12 | 18.02 | 1 | 0 | sd | 8 | |
| Ddiab9b | AUC_20wks | 3.02 | 54.42 | 52.12 | 56.50 | 1 | 0 | sd | 45 | ||
|
| |||||||||||
| 10 | Ddiab10a | AUC_20wks | 2.96 | 28.07 | 24.01 | 32.45 | s | 0 | 0 | 31 | Enpp1, Med23 |
| Ddiab10b | INS | 2.70 | 105.44 | 104.95 | 107.74 | d | sd | 0 | 13 | ||
|
| |||||||||||
| 11 | Ddiab11a | INS | 4.98 | 7.32 | 6.46 | 7.85 | sd | 0 | sd | 10 | Igfbp1, Igfbp3 |
| Ddiab11b | INS | 8.15 | 12.52 | 11.51 | 13.77 | sd | 1 | sd | 5 | Grb10 | |
| Ddiab11c | AUC_20wks | 2.88 | 80.69 | 80.01 | 81.60 | 0 | sd | 0 | 12 | ||
| Ddiab11d | AUC_20wks | 3.15 | 105.13 | 104.43 | 106.39 | 0 | 0 | sd | 31 | Gh | |
| Ddiab11e | AUC_20wks | 3.03 | 108.38 | 107.61 | 109.26 | 0 | 0 | 1 | 10 | ||
|
| |||||||||||
| 12 | Ddiab12a | nGLC | 2.98 | 52.49 | 49.63 | 55.02 | 0 | 0 | 1 | 16 | |
| Ddiab12b | nGLC | 2.93 | 55.70 | 55.12 | 57.08 | 0 | 1 | 1 | 23 | Insm2 | |
| Ddiab12c | AUC_20wks | 3.50 | 72.09 | 71.01 | 72.87 | sd | 0 | 0 | 14 | ||
| Ddiab12d | AUC_20wks | 2.68 | 77.40 | 76.99 | 79.31 | sd | d | 0 | 18 | ||
| Ddiab12e | AUC_20wks | 2.78 | 103.77 | 103.00 | 104.13 | sd | 0 | 0 | 16 | ||
| Ddiab12f | AUC_10wks | 2.82 | 121.12 | 118.32 | 123.92 | 0 | 0 | sd | 10 | Ptprn2 | |
|
| |||||||||||
| 13 | Ddiab13a | AUC_20wks | 3.06 | 29.67 | 21.92 | 33.36 | sd | 0 | 0 | 152 | |
| Ddiab13b | bGLC_10wks | 2.70 | 40.51 | 39.41 | 41.83 | 0 | s | d | 12 | ||
| Ddiab13c | bGLC_10wks | 2.73 | 61.03 | 57.90 | 62.99 | d | d | d | 52 | ||
| Ddiab13d | INS | 4.56 | 68.92 | 67.81 | 73.33 | 0 | d | sd | 20 | ||
| Ddiab13e | AUC_20wks | 3.24 | 87.10 | 86.46 | 88.39 | sd | s | 0 | 0 | ||
| Ddiab13f | AUC_20wks | 3.83 | 96.15 | 93.17 | 97.71 | sd | s | 0 | 37 | ||
| Ddiab13g | bGLC_10wks | 3.07 | 105.42 | 104.62 | 107.09 | 0 | s | s | 15 | ||
| Ddiab13h | AUC_10wks | 4.75 | 114.70 | 112.87 | 119.12 | sd | 0 | sd | 30 | ||
| AUC_20wks | 2.91 | 116.61 | sd | s | sd | ||||||
|
| |||||||||||
| 14 | Ddiab14a | AUC_10wks | 4.02 | 23.78 | 23.24 | 29.48 | sd | 0 | sd | 31 | |
| Ddiab14b | INS | 3.09 | 31.66 | 28.31 | 34.68 | sd | sd | sd | 66 | ||
|
| |||||||||||
| 15 | Ddiab15a | AUC_10wks | 3.43 | 21.94 | 21.94 | 23.61 | 0 | sd | sd | 2 | |
| Ddiab15b | AUC_10wks | 3.63 | 31.47 | 30.83 | 32.63 | 0 | 0 | sd | 11 | ||
| Ddiab15c | INS | 3.47 | 34.26 | 31.47 | 35.03 | d | d | 0 | 20 | ||
| Ddiab15d | AUC_10wks | 5.30 | 66.84 | 66.00 | 68.85 | d | sd | 0 | 12 | ||
|
| |||||||||||
| 16 | Ddiab16a | AUC_10wks | 2.85 | 5.28 | 3.99 | 8.33 | 0 | 1 | sd | 32 | |
| Ddiab16b | INS | 2.58 | 50.30 | 49.00 | 50.37 | 0 | 0 | sd | 5 | ||
| Ddiab16c | INS | 2.60 | 51.06 | 50.44 | 58.54 | sd | sd | sd | 29 | ||
| AUC_20wks | 3.40 | 51.26 | 50.44 | 58.54 | 0 | sd | 0 | ||||
| Ddiab16d | INS | 3.44 | 56.02 | 53.44 | 59.52 | sd | sd | sd | 63 | ||
|
| |||||||||||
| 17 | Ddiab17a | AUC_20wks | 2.56 | 10.92 | 10.40 | 12.31 | sd | 0 | 0 | 3 | |
| Ddiab17b | AUC_10wks | 2.85 | 25.48 | 23.81 | 25.90 | 0 | 0 | sd | 104 | Pdpk1, Sstr5 | |
|
| |||||||||||
| 18 | Ddiab18a | AUC_10wks | 5.15 | 33.96 | 31.33 | 35.20 | sd | sd | 0 | 43 | Egr1 |
| Ddiab18b | bGLC_20wks | 3.19 | 57.67 | 56.90 | 58.74 | s | 0 | 0 | 12 | ||
|
| |||||||||||
| 19 | Ddiab19a | nGLC | 2.67 | 18.58 | 17.08 | 20.01 | 0 | sd | sd | 10 | |
AUC_10wks: area under the curve at 10 weeks; AUC_20wks: area under the curve at 20 weeks; bGLC_10wks: basal glucose level at 10 weeks; bGLC_20wks: basal glucose level at 20 weeks; GLC: serum glucose level at necropsy; INS: serum insulin level at necropsy. Bold LOD scores indicate QTL significant at the genome-wide threshold, others are significant at their respective chromosome-wise thresholds.
ADD: additive genotypic effect; DOM: dominance genotypic effect; IMP: imprinting genotypic effect; s: sex interaction; d: diet interaction; sd: sex-by-diet interaction; 1: main effect with no interaction; 0: no effect.
On average, for QTL with additive effects among the sex- and diet-based cohorts, animals that are LG homozygotes at the significant locus have higher serum glucose levels and lower serum insulin levels, but respond better to a glucose challenge than animals that are SM homozygotes. For QTL with dominance effects among the sex- and diet-based cohorts, the LG allele is dominant to the SM allele 50% of the time for glucose tolerance, 60% of the time for serum glucose levels (combined results from serum glucose levels measured at necropsy and basal glucose levels measured at 10 and 20 weeks), and 40% of the time for serum insulin levels. For QTL with imprinting effects among the sex- and diet-based cohorts, 66% of imprinting values are positive for glucose tolerance at 10 weeks, indicating that most often animals with the LG/SM genotype, inheriting their LG allele from their father and their SM allele from their mother, have a higher AUC and thus a relatively poorer response to a glucose challenge. At 20 weeks, 39% of imprinting values are positive for glucose tolerance. Sixty-nine percent of imprinting values are positive for serum glucose levels, and 43% are positive for serum insulin levels. Genotypic values for all 70 significant trait-specific QTL for the full F16 population as well as for each sex- and diet cohort are provided in supporting information, Table S5.
The average QTL interval is ~4 MB and contains 34 genes. Many of these intervals contain genes associated with variation in glucose and insulin levels (Table 1). Of note are Pparg, encoding peroxisome proliferator-activated receptor gamma, and Kcnj11, encoding the potassium inwardly rectifying channel J11. These two genes are well studied not only in mouse models of T2D, but also in human studies where variations associated with T2D-susceptibility have replicated across populations (McCarthy and Zeggini 2009). Our results suggest natural variants at these loci may be responsible for variation in diabetes-state in mice as well.
Context Dependency
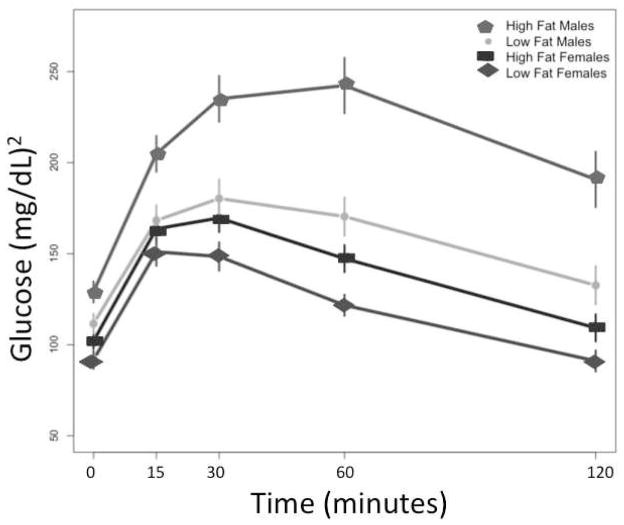
Phenotypic variation in response to high- and low-fat diets between the LG/J and SM/J strains for glucose and insulin levels, as well as for T2D co-morbidities such as obesity, and their heritabilities, have been reported for LG/J x SM/J parent strains (Ehrich et al. 2003). This cross has proven an excellent model system for identifying QTL associated with variation in these traits (Cheverud et al. 2004; Cheverud et al. 2009; Ehrich et al. 2005; Fawcett et al. 2009; Fawcett et al. 2008). Phenotypic variation among sex-by-diet cohorts in the F16 is illustrated here by response to glucose stress (Figure 1, IPGTT). Males have higher basal glucose levels and a weaker overall response to a glucose challenge. However, males fed a high-fat diet stand out among the cohorts with glucose levels that continue to elevate for a longer time period, and then decline at a slower rate than in any other sex-by-diet cohort. This phenotypic variation among the cohorts hints at the underlying genomic complexity of glucose and insulin traits, and we are able to dissect the genetic underpinnings using our mapping results.
FIGURE 1.
Phenotypic variation in response to glucose stress, as measured by an intra-peritoneal injection of 0.01ml of 10% glucose solution per gram of body weight (IPGTT). Measurements taken over the course of 2 hours were used to calculate the area under the curve (AUC), an overall measure of glucose tolerance, among each sex-by-diet cohort. High-fat fed male glucose levels continue to elevate for a longer time period, and then decline at a slower rate, than any other sex-by-diet cohort in this population.
Of the QTL showing significant interactions with sex, 78% affect males. Of the QTL showing significant interactions with diet, 63% affect animals fed a high-fat diet. We find most significant interactions affect individual sex-by-diet cohorts, and 72% of these QTL are found in males fed a high-fat diet. We find that 23 of these QTL show genotypic effects in multiple cohorts, but in different ways, and that these effects are not always seen in the full population. In the accounts that follow, the separate cohort effects described are supported by significant interaction tests.
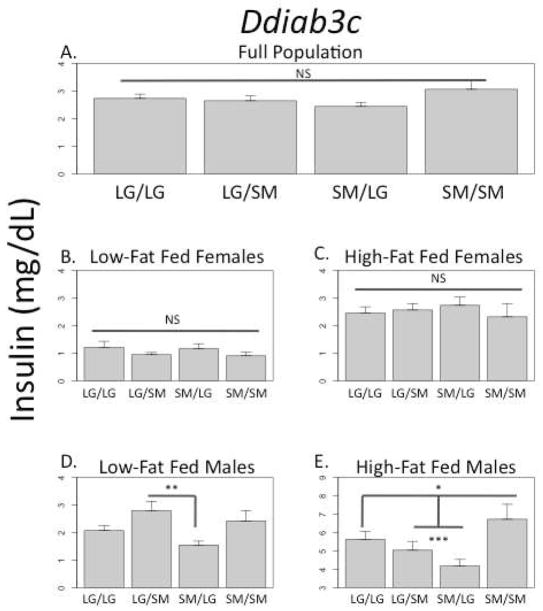
For example, a QTL on chromosome 3 (Figure2, Ddiab3c, INS) is associated with variation in serum insulin levels. In males fed a low-fat diet, there is a significant bipolar dominance imprinting effect (no additive or dominance effect). The imprinting value is positive, meaning heterozygous animals in this cohort have higher insulin levels if they inherit their LG allele from their fathers and their SM allele from their mothers. At this same QTL, there are significant negative additive effects in males fed a high-fat diet, whereby animals homozygous for the SM allele in this cohort have higher insulin levels. Additionally, high-fat fed males show highly significant under-dominance, whereby heterozygous animals, whether LG/SM or SM/LG, have lower insulin levels than animals homozygous with either LG or SM alleles in this cohort. Although the two reciprocal heterozygotes do not show the same magnitude of under-dominance, there is not a significant imprinting effect in the high-fat fed males at this locus. There are no significant effects in females fed either high- or low-fat diets, and the QTL does not register as significant in the full population. The absence of significant genetic effects in the females washes out the significant effects found in the males when all animals are considered together.
FIGURE 2.
QTL Ddiab3c associated with serum insulin level at necropsy (INS). There are no significant genetic effects in the full population, in low-fat fed females, and high-fat fed females (A–C). In low-fat fed males there is a significant bipolar dominance imprinting effect (D). In high-fat fed males there are significant negative additive effects and significant under-dominance effects, but no significant imprinting effect (E). The lack of significant genotypic effects in females at this locus washes out the sex-by-diet effects found in males when all individuals are pooled together as a full population. The different scales among the cohorts reflect their different mean phenotypic scores for this trait. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001
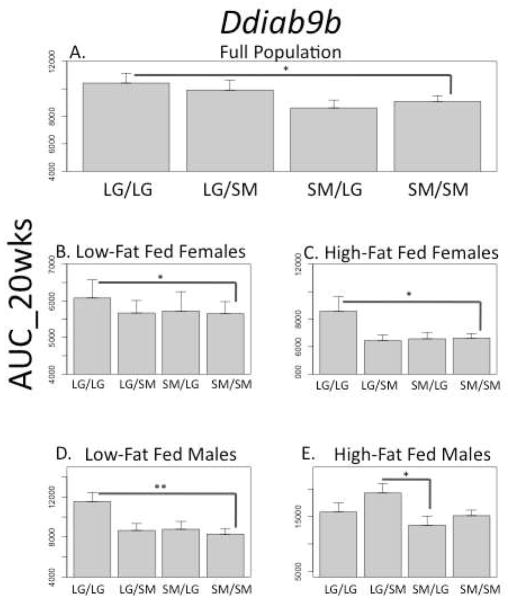
A similar example is seen at a QTL on chromosome 9 associated with glucose tolerance at 20 weeks, as measured by the IPGTT (Figure 3, Ddiab9b, AUC_20wks). There are significant positive additive effects in the full population, whereby animals homozygous for the LG allele at this locus have a poorer response to a glucose challenge. When this effect is examined in individual cohorts, it is seen in all but the high-fat fed males. In this cohort, there is a significant bipolar dominance imprinting effect (no additive or dominance effect). The imprinting value is positive, meaning heterozygous animals inheriting their LG allele from their father and their SM allele from their mother have a poorer response to a glucose challenge. Genomic imprinting is not seen in any other cohort at this locus, and it does not register as a significant effect in the full population.
FIGURE 3.
QTL Ddiab9b associated with glucose tolerance at 20 weeks as measured by the IPGTT. There are significant positive additive effects in the full population, as well as in all cohorts (A–D) except the high-fat fed males. In males fed a high-fat diet, there is a significant bipolar dominance imprinting effect (E). The different scales among the cohorts reflect their different mean phenotypic scores for this trait. *p ≤ 0.05; **p ≤ 0.01
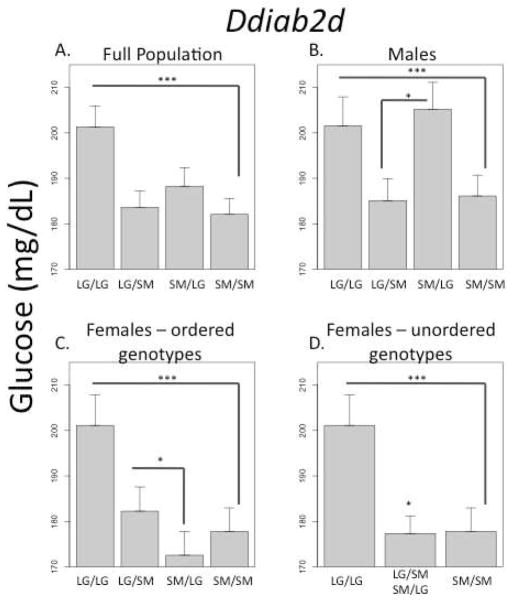
At a QTL on chromosome 2 there are significant positive additive effects in the full population, whereby animals that are homozygous for the LG allele have higher serum glucose levels at necropsy (Figure 4, Ddiab2d, GLC). Significant imprinting effects are found in both males and females, however the imprinting values are of opposite signs: In females, the imprinting value is positive, meaning heterozygous females inheriting their LG allele from their father and their SM allele from their mother have higher glucose levels. In males, the imprinting value is negative, meaning heterozygous males inheriting their SM allele from their father and their LG allele from their mother have higher glucose levels. Imprinting is not significant in the full population because the opposite values in the male and female cohorts cancel each other out. The imprinting effect in males is clearly maternal expression. The imprinting effect in females reflects paternal expression, however the effect is less clear than that seen in the males because females also have significant under-dominance effects at this same locus. If the paternally and maternally inherited alleles are not differentiated, all heterozygote females appear to have lower glucose levels than expected from an additive model (Fig. 4d).
FIGURE 4.
QTL Ddiab2d associated serum glucose level at necropsy (GLC). There are highly significant positive additive effects in the full population (A), which is reflected in all cohorts (B–C). Both males and females have significant imprinting effects, but the imprinting values are of the opposite signs: in males the imprinting value is negative and the effect is maternal expression. In females the imprinting value is positive and the effect is paternal expression. Additionally, females show significant under-dominance, which affects the magnitude of the imprinting effects seen in this cohort. The dominance effects are seen when the two reciprocal heterozygotes are combined (D). *p ≤ 0.05; ***p ≤ 0.001
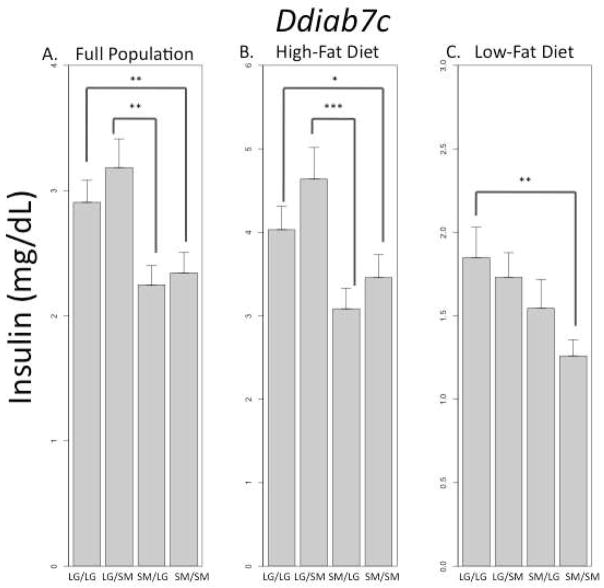
A converse example is seen at a QTL on chromosome 7 associated with variation in serum insulin levels (Figure 5, Ddiab7c, INS). Significant positive additive effects and positive imprinting values are found in the full population, meaning animals homozygous for the LG allele and animals heterozygous that inherit their LG allele from their father and their SM allele from their mother have higher serum insulin levels. However, when individual cohorts are examined, the imprinting effects are only significant in animals fed a high-fat diet. Animals fed a low-fat diet show only additive effects at this locus, but because the imprinting value in the high-fat fed animals is so highly significant (p < 0.001), the effect registers as significant in the full population (p < 0.01). The genomic effects in the high-fat fed cohort are a complex combination of a strong bipolar dominance imprinting effect with a relatively weaker additive effect, and the bipolar dominance imprint effect is the overriding pattern.
FIGURE 5.
QTL Ddiab7c associated serum insulin level at necropsy (INS). Significant positive additive effects and positive imprinting values are found in the full population, across all cohorts (A). However, the imprinting effects are only found in ½ the population. The highly significant bipolar dominance imprinting effects found in animals fed a high-fat diet register in the full population even though there is no significant imprinting effect in animals fed a low-fat diet (B–C). *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001
DISCUSSION
An intriguing result of this study is the ubiquity of genomic parent-of-origin imprinting effects. As of this writing, more than 80 imprinted genes have been identified in both humans and mice, and it is estimated that ≈30% of genes imprinted in one species are imprinted in the other (Williamson et al. 2009). It is becoming apparent that imprinting is an important aspect of the genetic architecture of many complex traits, including T2D (Rampersaud et al. 2008; Weinstein et al. 2009; Xie et al. 2008), and the epigenetic patterns identified are complex. Bioinformatic tools have been developed to identify imprinting signatures, such as methylation and histone modification, and genome-wide scans indicate that several hundred genes are likely to be imprinted across the genome (Luedi et al. 2005; Mantey et al. 2005). We find that imprinting is not only a dynamic contributor to variation in glucose and insulin traits in this population, but also an effect that is highly context dependent. These results support previous findings that imprinting patterns vary among genotypes and environments (Hager et al. 2009; Wolf et al. 2008).
Our results illustrate that context dependency is an important consideration when dissecting the genetic architecture of a complex trait such as T2D. We show that genotype interacts with environment in important ways, and that these interactions are not always consistent among genotypes and across environments within the same population. Further, the results presented here complement those found in this same population examining variation in obesity-related traits and serum lipid levels (Cheverud et al. 2010b; Lawson et al. 2010b). While most effects are seen in high-fat fed males for T2D-related traits, most effects are seen in high-fat fed females for obesity and serum lipids levels. However, in general, the majority of genetic effects are seen in multiple cohorts in different ways across all traits.
From a clinical perspective, this result is intuitive: it is well known that T2D penetrances vary within and between human sub-populations, and that, in general, women are less prone than men of the same body mass (Cornier et al. 2008). While general lifestyle dietary and activity modifications have proven therapeutic, individual response to such treatment varies (Ordovas and Shen 2008). From a research perspective, this result implies that meta-analyses such as GWAS miss an important aspect of the genetic architecture underlying variation in T2D. While some human studies have successfully examined gene-by-environmental interactions (Junyent et al. 2009; Kabagambe et al. 2009), these interactions are typically regarded as nuisance factors in analyses, despite the fact that they may underlie the increasing prevalence of T2D. Identifying context-dependent genetic effects is challenging in human studies because it is generally not feasible to control and/or to record an individual’s diet over time.
Mouse models are especially appropriate for addressing issues of context dependency because the animals studied are of known genomic background with measurable phenotypic differences in a controlled environment. Other studies using mouse models have found that context dependency underlies variation in complex traits such as cholesterol metabolism (Kitami et al. 2008), blood serum levels (Svenson et al. 2007), adiposity (Taylor et al. 1999; York et al. 1996), bone density (Ackert-Bicknell et al. 2008), and hepatic carcinoma (Hill-Baskin et al. 2009). Further, experimental mouse populations contain no rare alleles as polymorphisms are at 50% throughout the genome. This not only increases the power to detect QTL, and eventually quantitative trait genes (QTG) or quantitative trait nucleotides (QTN) having small effects (Mackay et al. 2009), but also allows for detailed analysis of other aspects of genetic architecture such as epistasis. Indeed, recent work has implicated epistasis in human insulin resistance (Baratta et al. 2003), and rodent models indicate that epistasis is an important aspect of the genetic architecture of complex traits (Shao et al. 2008). Although our study did not examine epistasis, it is likely that gene-by-gene interactions also contribute to variation in these traits. Mouse results such as those presented here are directly applicable to human studies because loci identified in the mouse can be translated to the homologous region in human, and then can be further used to elucidate context-dependent genetic effects.
Supplementary Material
Acknowledgments
This work was supported by NIDDK R01 DK055736 to James M. Cheverud and Clay F. Semenkovich, by NHLBI T32-HL091823, program director Dr D.C. Rao, to Heather A. Lawson, and by the Biotechnology and Biological Sciences Research Council BBSRC-BB/C/516936 to Jason B. Wolf. The authors would like to thank Dr Seth Crosby and the Washington University Genome Sequencing Center for their help in SNP genotyping.
LITERATURE CITED
- Ackert-Bicknell CL, Demissie S, Marin de Evsikova C, Hsu YH, DeMambro VE, Karasik D, Cupples LA, Ordovas JM, Tucker KL, Cho K, Canalis E, Paigen B, Churchill GA, Forejt J, Beamer WG, Ferrari S, Bouxsein ML, Kiel DP, Rosen CJ. PPARG by dietary fat interaction influences bone mass in mice and humans. J Bone Miner Res. 2008;23:1398–1408. doi: 10.1359/JBMR.080419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- Baratta R, Di Paola R, Spampinato D, Fini G, Marucci A, Coco A, Vigneri R, Frittitta L, Trischitta V. Evidence for genetic epistasis in human insulin resistance: the combined effect of PC-1 (K121Q) and PPARgamma2 (P12A) polymorphisms. J Mol Med. 2003;81:718–723. doi: 10.1007/s00109-003-0466-3. [DOI] [PubMed] [Google Scholar]
- Broman KW, Saunak S. A Guide to QTL Mapping with R/qtl. New York: Springer; 2009. [Google Scholar]
- Cheverud JM, Ehrich TH, Hrbek T, Kenney JP, Pletscher LS, Semenkovich CF. Quantitative trait loci for obesity- and diabetes-related traits and their dietary responses to high-fat feeding in LGXSM recombinant inbred mouse strains. Diabetes. 2004;53:3328–3336. doi: 10.2337/diabetes.53.12.3328. [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Fawcett GL, Jarvis JP, Norgard EA, Pavlicev M, Pletscher LS, Polonsky KS, Ye H, Bell GI, Semenkovich CF. Calpain-10 is a component of the obesity-related quantitative trait locus, Adip1. J Lipid Res. 2009 doi: 10.1194/jlr.M900128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Lawson HA, Fawcett G, Wang B, Pletscher LS, Fox A, Maxwell TJ, Ehrich TH, Kenney-Hunt J, Wolf J, Semenkovich CF. Diet-Dependent Genetic and Genomic Imprinting Effects on Obesity in Mice. Obesity (Silver Spring) 2010a doi: 10.1038/oby.2010.141. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Lawson HA, Fawcett GL, Wang B, Pletscher LS, ARF, Maxwell TJ, Ehrich TH, Kenney-Hunt JP, Wolf JB, Semenkovich CF. Diet-Dependent Genetic and Genomic Imprinting Effects on Obesity in Mice. Obesity (Silver Spring) 2010b doi: 10.1038/oby.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Routman EJ, Duarte FA, van Swinderen B, Cothran K, Perel C. Quantitative trait loci for murine growth. Genetics. 1996;142:1305–1319. doi: 10.1093/genetics/142.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clee SM, Attie AD. The genetic landscape of type 2 diabetes in mice. Endocr Rev. 2007;28:48–83. doi: 10.1210/er.2006-0035. [DOI] [PubMed] [Google Scholar]
- Cornier MA, Dabelea D, Hernandez TL, Lindstrom RC, Steig AJ, Stob NR, Van Pelt RE, Wang H, Eckel RH. The metabolic syndrome. Endocr Rev. 2008;29:777–822. doi: 10.1210/er.2008-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich TH, Hrbek T, Kenney-Hunt JP, Pletscher LS, Wang B, Semenkovich CF, Cheverud JM. Fine-mapping gene-by-diet interactions on chromosome 13 in a LG/J x SM/J murine model of obesity. Diabetes. 2005;54:1863–1872. doi: 10.2337/diabetes.54.6.1863. [DOI] [PubMed] [Google Scholar]
- Ehrich TH, Kenney JP, Vaughn TT, Pletscher LS, Cheverud JM. Diet, obesity, and hyperglycemia in LG/J and SM/J mice. Obes Res. 2003;11:1400–1410. doi: 10.1038/oby.2003.189. [DOI] [PubMed] [Google Scholar]
- Falconer DS, Mackay TF. Introduction to Quantitative Genetics. Essex: Pearson; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, Eckman CB, Tanzi RE, Selkoe DJ, Guenette S. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci U S A. 2003;100:4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett GL, Jarvis JP, Roseman CC, Wang B, Wolf JB, Cheverud JM. Fine-Mapping of Obesity-Related Quantitative Trait Loci in and F9/10 Advanced Intercross Line. Obesity (Silver Spring) 2009 doi: 10.1038/oby.2009.411. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett GL, Roseman CC, Jarvis JP, Wang B, Wolf JB, Cheverud JM. Genetic architecture of adiposity and organ weight using combined generation QTL analysis. Obesity (Silver Spring) 2008;16:1861–1868. doi: 10.1038/oby.2008.300. [DOI] [PubMed] [Google Scholar]
- Goodale HD. Progress Report on Possibilities in Progeny-Test Breeding. Science. 1941;94:442–443. doi: 10.1126/science.94.2445.442. [DOI] [PubMed] [Google Scholar]
- Hager R, Cheverud JM, Wolf JB. Relative contribution of additive, dominance, and imprinting effects to phenotypic variation in body size and growth between divergent selection lines of mice. Evolution. 2009;63:1118–1128. doi: 10.1111/j.1558-5646.2009.00638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Hill-Baskin AE, Markiewski MM, Buchner DA, Shao H, DeSantis D, Hsiao G, Subramaniam S, Berger NA, Croniger C, Lambris JD, Nadeau JH. Diet-induced hepatocellular carcinoma in genetically predisposed mice. Hum Mol Genet. 2009;18:2975–2988. doi: 10.1093/hmg/ddp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huopio H, Otonkoski T, Vauhkonen I, Reimann F, Ashcroft FM, Laakso M. A new subtype of autosomal dominant diabetes attributable to a mutation in the gene for sulfonylurea receptor 1. Lancet. 2003;361:301–307. doi: 10.1016/S0140-6736(03)12325-2. [DOI] [PubMed] [Google Scholar]
- Junyent M, Tucker KL, Smith CE, Garcia-Rios A, Mattei J, Lai CQ, Parnell LD, Ordovas JM. The effects of ABCG5/G8 polymorphisms on plasma HDL cholesterol concentrations depend on smoking habit in the Boston Puerto Rican Health Study. J Lipid Res. 2009;50:565–573. doi: 10.1194/jlr.P800041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabagambe EK, Glasser SP, Ordovas JM, Warodomwichit D, Tsai MY, Hopkins PN, Borecki IB, Wojczynski MK, Arnett DK. TCF7L2 polymorphisms and inflammatory markers before and after treatment with fenofibrate. Diabetol Metab Syndr. 2009;1:16. doi: 10.1186/1758-5996-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney-Hunt JP, Vaughn TT, Pletscher LS, Peripato A, Routman E, Cothran K, Durand D, Norgard E, Perel C, Cheverud JM. Quantitative trait loci for body size components in mice. Mamm Genome. 2006;17:526–537. doi: 10.1007/s00335-005-0160-6. [DOI] [PubMed] [Google Scholar]
- Kitami T, Rubio R, O’Brien W, Quackenbush J, Nadeau JH. Gene-environment interactions reveal a homeostatic role for cholesterol metabolism during dietary folate perturbation in mice. Physiol Genomics. 2008;35:182–190. doi: 10.1152/physiolgenomics.00294.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MG, Vaugn TT, Pletscher S, King-Ellison K, Adams E, Erikson C, Cheverud JM. Genetic variation in body weight gain and composition in the intercross of Large (LG/J) and Small (SM/J) inbred strains of mice. Genetics and Molecular Biology. 1998;21:211–218. [Google Scholar]
- Lawson HA, Zelle KM, Fawcett G, Wang B, Pletscher LS, Maxwell TJ, Ehrich TH, Kenney-Hunt J, Wolf J, Semenkovich CF, Cheverud JM. Genetic, epigenetic, and gene-by-diet interaction effects underlie variation in serum lipids in a LG/J x SM/J murine model. Journal of Lipids Research. 2010a doi: 10.1194/jlr.M006957. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson HA, Zelle KM, Fawcett GL, Wang B, Pletscher LS, Maxwell TJ, Ehrich TH, Kenney-Hunt JP, Wolf JB, Semenkovich CF, Cheverud JM. Genetic, epigenetic, and gene-by-diet interaction effects underlie variation in serum lipids in a LG/J x SM/J murine model. J Lipid Res. 2010b doi: 10.1194/jlr.M006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Li J, Jiang T. Computing the minimum recombinant haplotype configuration from incomplete genotype data on a pedigree by integer linear programming. J Comput Biol. 2005;12:719–739. doi: 10.1089/cmb.2005.12.719. [DOI] [PubMed] [Google Scholar]
- Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J. Genetics of body size and related characters. American Naturalist. 1944;78:142–157. [Google Scholar]
- Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantey C, Brockmann GA, Kalm E, Reinsch N. Mapping and exclusion mapping of genomic imprinting effects in mouse F2 families. J Hered. 2005;96:329–338. doi: 10.1093/jhered/esi044. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Zeggini E. Genome-wide association studies in type 2 diabetes. Curr Diab Rep. 2009;9:164–171. doi: 10.1007/s11892-009-0027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National_Diabetes_Information_Clearinghouse; Department_of_Health_and_Human_Services, editor. National Diabetes Statistics. Bethesda: 2005. [Google Scholar]
- Neel JV. Diabetes mellitus: a ‘thrifty’ genotype rendered detrimental by ‘progress’? American Journal of Human Genetics. 1962:353–362. [PMC free article] [PubMed] [Google Scholar]
- Ordovas JM, Shen J. Gene-environment interactions and susceptibility to metabolic syndrome and other chronic diseases. J Periodontol. 2008;79:1508–1513. doi: 10.1902/jop.2008.080232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permutt MA, Wasson J, Cox N. Genetic epidemiology of diabetes. J Clin Invest. 2005;115:1431–1439. doi: 10.1172/JCI24758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampersaud E, Mitchell BD, Naj AC, Pollin TI. Investigating parent of origin effects in studies of type 2 diabetes and obesity. Curr Diabetes Rev. 2008;4:329–339. doi: 10.2174/157339908786241179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanti G, Southam L, Altshuler D, Ardlie K, Barroso I, Boehnke M, Cornelis MC, Frayling TM, Grallert H, Grarup N, Groop L, Hansen T, Hattersley AT, Hu FB, Hveem K, Illig T, Kuusisto J, Laakso M, Langenberg C, Lyssenko V, McCarthy MI, Morris A, Morris AD, Palmer CN, Payne F, Platou CG, Scott LJ, Voight BF, Wareham NJ, Zeggini E, Ioannidis JP. Underlying genetic models of inheritance in established type 2 diabetes associations. Am J Epidemiol. 2009;170:537–545. doi: 10.1093/aje/kwp145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Burrage LC, Sinasac DS, Hill AE, Ernest SR, O’Brien W, Courtland HW, Jepsen KJ, Kirby A, Kulbokas EJ, Daly MJ, Broman KW, Lander ES, Nadeau JH. Genetic architecture of complex traits: large phenotypic effects and pervasive epistasis. Proc Natl Acad Sci U S A. 2008;105:19910–19914. doi: 10.1073/pnas.0810388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson KL, Von Smith R, Magnani PA, Suetin HR, Paigen B, Naggert JK, Li R, Churchill GA, Peters LL. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J Appl Physiol. 2007;102:2369–2378. doi: 10.1152/japplphysiol.01077.2006. [DOI] [PubMed] [Google Scholar]
- Taylor BA, Tarantino LM, Phillips SJ. Gender-influenced obesity QTLs identified in a cross involving the KK type II diabetes-prone mouse strain. Mamm Genome. 1999;10:963–968. doi: 10.1007/s003359901141. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Population Genetics and Microevolutionary Theory. Hoboken: Wiley-Liss; 2006. [Google Scholar]
- Weinstein LS, Xie T, Qasem A, Wang J, Chen M. The role of GNAS and other imprinted genes in the development of obesity. Int J Obes (Lond) 2009 doi: 10.1038/ijo.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson CR, Blake A, Thomas S, Beechey CV, Hancock J, Cattanach BM, Peters J. World Wide Web Site - Mouse Imprinting Data and References. Oxfordshire; 2009. [Google Scholar]
- Wolf JB, Hager R, Cheverud JM. Genomic imprinting effects on complex traits: a phenotype-based perspective. Epigenetics. 2008;3:295–299. doi: 10.4161/epi.3.6.7257. [DOI] [PubMed] [Google Scholar]
- Wolfs MG, Hofker MH, Wijmenga C, van Haeften TW. Type 2 Diabetes Mellitus: New Genetic Insights will Lead to New Therapeutics. Curr Genomics. 2009;10:110–118. doi: 10.2174/138920209787847023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Chen M, Gavrilova O, Lai EW, Liu J, Weinstein LS. Severe obesity and insulin resistance due to deletion of the maternal Gsalpha allele is reversed by paternal deletion of the Gsalpha imprint control region. Endocrinology. 2008;149:2443–2450. doi: 10.1210/en.2007-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York B, Lei K, West DB. Sensitivity to dietary obesity linked to a locus on chromosome 15 in a CAST/Ei x C57BL/6J F2 intercross. Mamm Genome. 1996;7:677–681. doi: 10.1007/s003359900204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.