Abstract
During Drosophila oogenesis, nurse cells undergo changes in chromosomal morphology, first from the polytenic form to a transient condensed phase known as the five-blob configuration, then into a diffuse polytenic-polyploid state for the remainder of oogenesis. The mechanism by which nurse-cell chromosome dispersal is regulated remains elusive. Mutations in several genes, including the heterogeneous ribonucleoprotein genes squid (sqd) and hrb27C, the alternative splicing factor gene poly U binding factor 68 kDa (pUf68, also known as half-pint), and the germ-line-specific gene ovarian tumor (otu), that produce defects in nurse-cell chromosome dispersal also produce defects in oocyte polarity, suggesting a link between these two processes. here, we characterize a novel gene named poly, which, when mutated in the germ line, disrupts nurse-cell chromosome dispersal, as well as localization of anteroposterior and dorsoventral determinants in the oocyte. We also show that poly interacts genetically with hrb27C and otu. We conclude that poly is required for nurse-cell chromosome dispersal and oocyte polarization in the Drosophila germ-line. In addition, our interaction data suggest that poly is probably a member of the characterized mRNp complex that mediates both processes.
Keywords: germ-line, drosophila, oogenesis, nurse cell, oocyte polarity
Drosophila oogenesis serves as a versatile model for the study of spatiotemporal regulation of transcription, translation and localization of discrete mRNAs and proteins in a developmental context. The processes underlying oogenesis, from stem-cell differentiation to maturation of the oocyte, occur sequentially in long tubes called ovarioles. Each ovary includes approximately 16–20 ovarioles, and each ovariole has a germarium that contains a repository of germ-line and somatic stem cells. Germ-line stem cells self-renew and produce cystoblasts, which subsequently give rise to 15 nurse cells and 1 oocyte per egg chamber, whereas somatic stem cells produce an epithelial layer of follicle cells that covers the germ-line cells. The most prominent roles of nurse cells are the synthesis of enormous amounts of mRNAs and proteins that aid in oocyte differentiation and growth and the accumulation of maternal stockpiles of products to be used during embryogenesis.1 The follicle cells communicate with the germ-line cells throughout oogenesis to support the development and differentiation of the oocyte.2
The anteroposterior (AP) axis is established by localization of oskar (osk) mRNA at the posterior and bicoid mRNA at the anterior of the oocyte during stages 8–10 of oogenesis.3 Microtubule organization plays an important role in many steps in the establishment of oocyte polarity; before stages 6–7 of oogenesis, the plus ends of the microtubules are spread throughout the nurse cells, and the minus ends are focused in the posterior of the oocyte.4 In response to signaling from the posterior follicle cells, the microtubules are disassembled and renucleate, producing a reversion of the polarity of the filaments, such that the plus ends of the microtubules are oriented toward the posterior of the oocyte and the minus ends aggregate at the anterior cortex.5 During stage 9, osk mRNA and Staufen (Stau) (a dsRNA-binding protein that colocalizes with osk mRNA) are both transported by Kinesin, a plus-end microtubule motor, to the posterior of the oocyte.6,7 By stages 7–8, the oocyte nucleus is located at the anterodorsal corner of the oocyte, where Gurken (Grk), an EGFR ligand, is tightly localized, and is involved in dorsoventral axis polarity.8 Mutations that interrupt any of these vital processes, through mutant somatic and/or germ-line cells, can potentially disrupt the AP/DV axis polarities of the oocyte.3,4,9
Recent studies have indicated that heterogeneous ribonucleoproteins (hnRNPs) such as squid (sqd) and hrb27C, the alternative splicing factor poly U binding factor 68 kDa (pUf68, also known as half-pint), and the germ-line-specific gene ovarian tumor (otu) are required for localization of grk mRNA to the dorsoanterior and/or localization of osk mRNA to the posterior of the oocyte.10-12 Intriguingly, these mutants also show chromosomal morphology defects in the nurse cells from stage 6 of oogenesis onward. Nurse cells undergo an asynchronous cell cycle regimen comprising 10–12 endocycles of continuous DNA replication before programmed cell death. After the first five endocycles, the DNA coalesces visibly into the characteristic five-blob structure during stages 4–5 of oogenesis. Each blob represents one of the major polytenic chromosomal arms (X, 2L, 2R, 3L and 3R). Evidence supports the hypothesis that a transient mitosis-like phase follows the fifth endocycle, during which the polytene chromosomes disperse throughout the nurse-cell nucleus, resulting in a loss of the visible five-blob structure in all nurse cells by stage 6 of oogenesis.13 Many of the mutants in which the nurse-cell chromosomes fail to disperse after the five-blob stage also show defects in localization of AP and DV axis determinants, but determination of a functional relationship between this chromosome-dispersal event and formation of the AP/DV axis polarities has been elusive. Nurse-cell chromosome dispersal has been suggested to facilitate rapid ribosomal synthesis of proteins needed for the remainder of oogenesis from stage 6 onward.13
Here we report that, from a flipase-flipase recognition-target (FLP-FRT) mosaic screen, we isolated a new allele of poly (poly2) that caused prominent defects in Stau-GFP localization and nurse-cell chromosome dispersal in germ-line clones. poly is a novel gene that was originally identified for the aberrant nuclear morphology it produces in the brains of third-instar larvae (Heck A, Dros Res Conf 36, Flybase; named poly1). We conclude that the novel gene poly is required for nurse-cell chromosome dispersal and oocyte polarity and suggest that poly can interact with members of the mRNP complex that are involved in similar processes.
Results
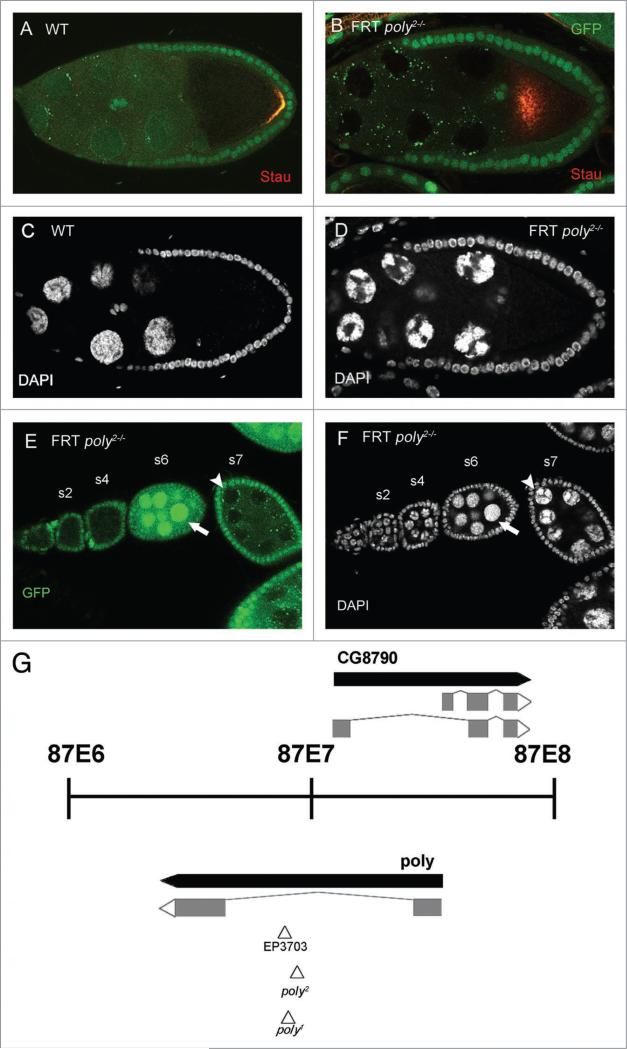
poly is required in the germ-line for efficient Stau localization and nurse-cell chromosomal dispersal. To uncover new genes that are involved in oocyte polarity formation, we performed an FLP-FRT mosaic screen for Stau-GFP localization with approximately 400 FRT mutant lines containing P-element insertions, across the right arm of the third chromosome.14,15 Stau-GFP colocalizes with osk mRNA at the posterior of the oocyte in wild-type stage-9 egg chambers (Fig. 1A and data not shown).16 One of the lines that affected the localization of Stau-GFP in egg chambers with mutant germ-line clones was 8131 (an FRT-containing P-element line from Szeged stock center); out of 66 stage-9 8131 germ-line clones examined, 24% displayed complete mislocalization (center of oocyte; Fig. 1B), 53% displayed partial mislocalization (center and posterior of oocyte), and 23% retained wild-type localization (posterior of oocyte). An additional phenotype observed in 8131 germ-line clones was the alteration of nurse-cell chromosomal morphology (compare Fig. 1C and D). Normally, wild-type egg chambers in early oogenesis possess polytenic chromosomes that progress to a distinct five-blob configuration at stage 4, before dispersing into a diffuse state during stages 4–5. By stage 6, all of the nurse-cell chromosomes exhibit a diffuse state. The polytenic nurse cells of early-stage 8131 germ-line clones are indistinguishable from those of the wild type, but after stage 5 8131 germ-line clones remain in the five-blob configuration (Fig. 1E and F). The nurse-cell chromosomal dispersal defect is completely penetrant in poly2 clones; all of the 66 stage-8 and stage-9 poly2 germ-line clones examined maintained the nurse-cell dispersal defect.
Figure 1.
The identification of the novel gene poly isolated by a flipase-flipase recognition-target mosaic screen for mutations affecting anteroposterior axis polarity. (A) In wild-type stage-9 egg chambers, stau is localized at the posterior of the oocyte, and nurse-cell chromosome morphology is diffuse (c). (B) stau localization is disrupted in the majority of poly2 germ-line clones; complete mislocalization of stau is seen in the center of the oocyte, and nurse-cell chromosomes fail to disperse (D). The wild-type ovariole displays a polytenic configuration in the very early stages before rearrangement to a five-blob configuration that precedes dispersal to a diffuse state in stages 5–6 of oogenesis (early stages at the anterior/left and later stages at the posterior/right). (e and F) An ovariole that contains poly2 germ-line clones, early stages at the anterior/left and later stages at the posterior/right. poly2 germ-line clones, before stage 5, are indistinguishable from wild-type polytenic configurations. The wild-type egg chamber that precedes the posteriormost germ-line clone has dispersed nurse-cell chromosomes (arrow), whereas the germ-line clone maintains a five-blob configuration, indicating a failure to disperse (arrowhead). (G) poly is a partially nested novel gene that encodes a 251-amino-acid protein and has been cytologically mapped to the 87e region. All three p-elements are inserted in close proximity to each other in the sole intron of poly.
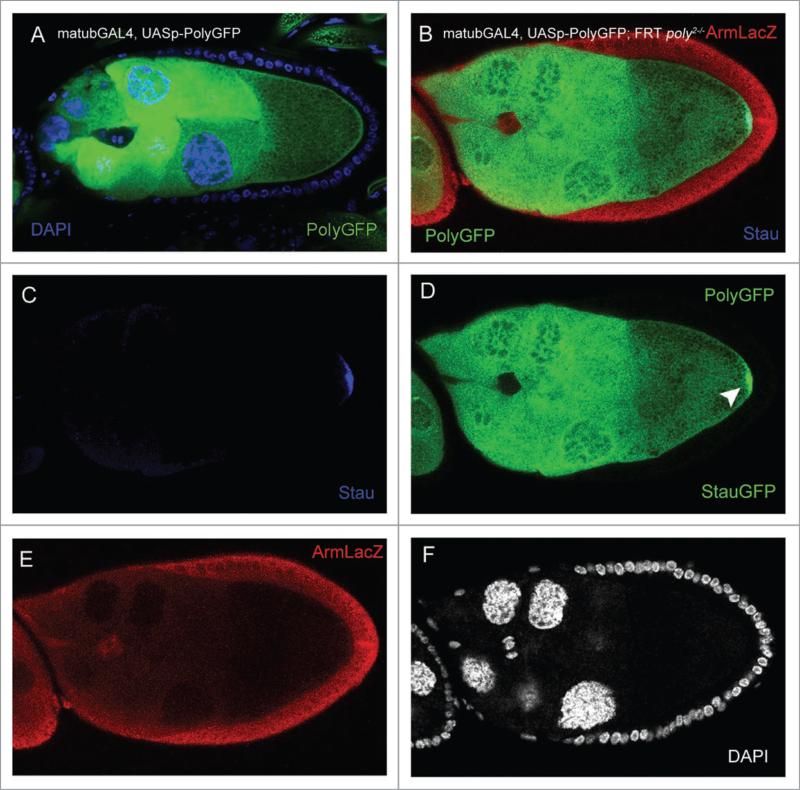
To identify the target gene disrupted by the P-element in the 8131 line, we used inverse PCR to amplify the regions flanking the P-element; sequencing determined the site of insertion in the sole intron of the gene poly (naming based on Flybase nomenclature; Fig. 1G). This finding is supported by complementation analysis of the 8131 (designated poly2) allele with poly1, which also resides in the only intron of poly; poly2 failed to complement poly1, resulting in decreased viability, similar to that of poly2 homozygous mutants (Table 1). In addition, both poly2 homozygous and poly1/poly2 transheterozygous larvae exhibited delayed development, reaching third-instar and pupal stages later than their heterozygous siblings. To ensure that mutations in poly are responsible for the phenotypes seen in poly2 germ-line clones, we performed rescue experiments in which we overexpressed UASp-polyGFP specifically in the germ-line with a maternal-α-tubulin GAL4-VP16 (mat-GAL4) driver (Fig. 2A) in the background of poly2 germ-line clones.17 While the overexpression of PolyGFP may not reflect the endogenous localization of wild-type Poly, we have found PolyGFP to be ubiquitous in the germ-line and localized in nurse-cell nucleoplasm, as well as nurse-cell and oocyte cytoplasm. During midoogenesis, overexpression of PolyGFP was also enriched in the nurse-cell chromatin and oocyte cortex (Fig. 2A). Overexpression of PolyGFP in poly2 germ-line clones fully alleviated Stau localization and nurse-cell chromosomal dispersal defects in all stage-9 and stage-10 egg chambers (Fig. 2B–F; n = 21), indicating that AP axis defects and nurse-cell chromosomal dispersal failure are caused by loss of poly function.
Table 1.
Analysis of the temperature sensitivity of poly2/poly2 and poly1/poly2 mutant flies
| Rearing temperature | poly2/poly2 | poly2/TM3 | Percentage of viability | % SE | Total |
|---|---|---|---|---|---|
| 18°C | 347 | 4751 | 14.6% | 0.61% | 5098 |
| 25°C | 9 | 1085 | 1.66% | 1.31% | 1094 |
| 29°C | 0 | 636 | 0% | 1.98% | 636 |
| Rearing temperature | poly1/poly2 | poly1 or poly2/TM3 | Percentage of viability | % SE | Total |
|---|---|---|---|---|---|
| 18°C | 205 | 1474 | 27.8% | 1.06% | 1679 |
| 25°C | 80 | 469 | 34.1% | 1.85% | 549 |
| 29°C | 0 | 204 | 0% | 3.03% | 204 |
Viability of poly homozygous escapers is low at 18°C, decreases further with increasing temperature, and is zero at 29°C. poly transheterozygous escapers are more viable than their homozygous counterparts at the intermediate temperature (possibly as a result of intragenic complementation) but still do not survive at 29°C. % SE is related to the standard deviation of the sample and is the sampling error, i.e., the probability that data obtained from the sample size is inaccurate.
Figure 2.
In poly germ-line clones, stau localization and nurse-cell chromosome-dispersal are affected and are rescued by germ-line-specific over-expression of UAsp-polyGFp. (A) Overexpression of UAsp-polyGFp transgene with the mat-GAL4 driver in a stage-9 egg chamber reveals ubiquitous expression of polyGFp in the nurse-cell nuclei and cytoplasm, as well as the oocyte, with enrichment around the oocyte cortex. Overexpression of polyGFp frequently causes a modest granulation of the nurse-cell nuclei in ovaries from older females. (B) Overexpression of UAsp-polyGFp in the background of poly2 germ-line clones fully alleviates the five-blob-arrest phenotype and stau mislocalization. (c) stau is localized to the posterior of the oocyte. (D) UAsp-polyGFp overexpression in the egg chamber, with concurrent stau-GFp localization at the posterior of the oocyte (arrowhead). polyGFp protein is not enriched at the posterior of the oocyte. (e) Germ-line clone marked by the absence of arm-lacZ. (F) DApI channel revealing the diffuse nature of the nurse-cell chromosomes.
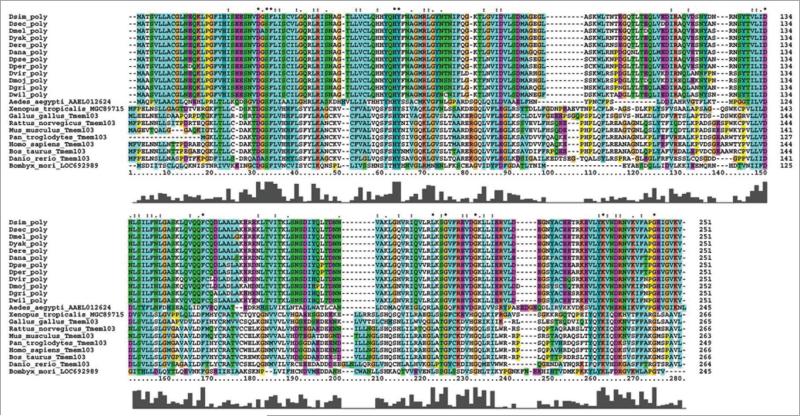
poly encodes a 251-amino-acid protein with no known domains. In an effort to deduce the function of poly, we used ClustalX to find putative homologs of poly in other model organisms (Fig. 3). Virtually identical poly coding sequences were found in all other Drosophila species; homologs in the Aedes aegypti mosquito and Bombyx mori silkworm show 53% and 47% sequence similarity, respectively, to Drosophila melanogaster Poly. The Poly protein sequence displays an extensive divergence from putative homologs from mammals, including Homo sapiens and Mus musculus, which retain 22–25% sequence identity and are known collectively as TMEM103 (transmembrane protein 103) or ATP1 (Angiotonin-transactivated protein 1) proteins and have unknown functions. The lengths of these proteins range from 211 to 256 amino acids, suggesting that, despite significant divergence between Poly and mammalian TMEM103 proteins, Poly still possesses similar functions in Drosophila species.
Figure 3.
Clustalx alignment of poly with putative homologues. Drosophila melanogaster poly is virtually identical to homologues in other drosophilids and displays moderate sequence homology to those in other insect species; it diverges most greatly from mammalian homologues. each protein sequence is labeled by genus and species and the name of the protein; for drosophilids, the genus is denoted as D and the first three letters of the species name are added. In the alignment, the bar graph under each amino acid delineates the conservation of that amino acid in the alignment of all protein sequences.
poly affects the localization of gurken and microtubule integrity
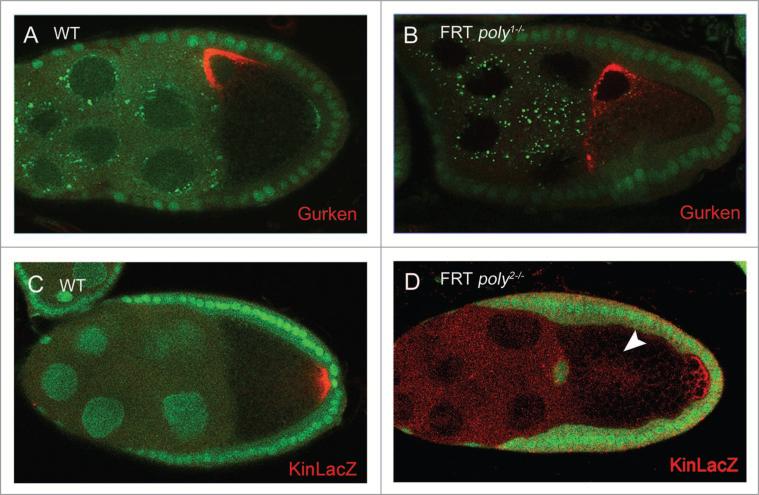
To characterize further the involvement of poly in oocyte polarity, we examined the expression of Grk, a determinant of DV polarity of the egg chamber, in poly1 and poly2 germ-line clones. Wild-type egg chambers displayed a dense localization of Grk at the dorsoanterior corner of the oocyte, tightly associated with the oocyte nucleus during stages 7–10 (Fig. 4A).8poly1 and poly2 stage-8 and stage-9 germ-line clones exhibited a weak Grk mislocalization phenotype (Fig. 4B), in which a small amount of Grk mislocalized to the ventral corner and/or aggregated into foci in the periphery of the oocyte nucleus. Although the Grk mislocalization phenotype was less pronounced than that in hrb27C and sqd mutants,11 it was consistently present. Out of 67 stage-8 and stage-9 poly2 germ-line clones examined, 42.4% showed wild-type Grk localization; 13.6% showed a reduced amount of Grk at the dorsoanterior corner of the oocyte, 27.3% showed some Grk foci, and 16.7% showed a trail of Grk toward the dorsoventral corner. The extent of Grk's effect on eggshell formation is not known, because both poly2 mutant mothers and poly2 germ-line-mutant mothers derived from the dominant female sterile technique both failed to deposit eggs.
Figure 4.
In poly germ-line clones, Grk and kin-lacZ localization are affected during oogenesis. (A) Grk is localized at the dorsoanterior corner of the oocyte in wild-type stage-8 and stage-9 egg chambers. (B) poly1 stage-8 and stage-9 germ-line clones display weak Grk mislocalization that consists of trails of Grk protein to the ventral corner. (c) Kinesin also localizes at the posterior of the oocyte in wild-type stage-9 egg chambers, as evidenced by kin-lacZ staining. (D) In poly2 stage-9 germ-line clones, Kin::β-gal localization is also disrupted, and Kin::β-gal levels are occasionally reduced (arrowhead points to mislocalized Kin::β-gal in the center of the oocyte).
Localization of Stau and Grk depends on an intact microtubule network in the germ-line cells. To determine the microtubule integrity in poly germ-line clones, we examined the localization of Kinesin::β-Galactosidase (Kin::β-Gal), a microtubule plus-end marker in the oocyte.18 Kin::β-Gal in stage-9 wild-type egg chambers localized at the posterior of the oocyte (Fig. 4C). In poly2 germ-line clones, Kin::β-Gal was frequently absent from the posterior or, occasionally, present in only low amounts, implying defective microtubule organization (Fig. 4D). Overall, our data suggest that poly is required in the germ-line for proper microtubule organization and oocyte polarity.
poly interacts with hrb27C and otu
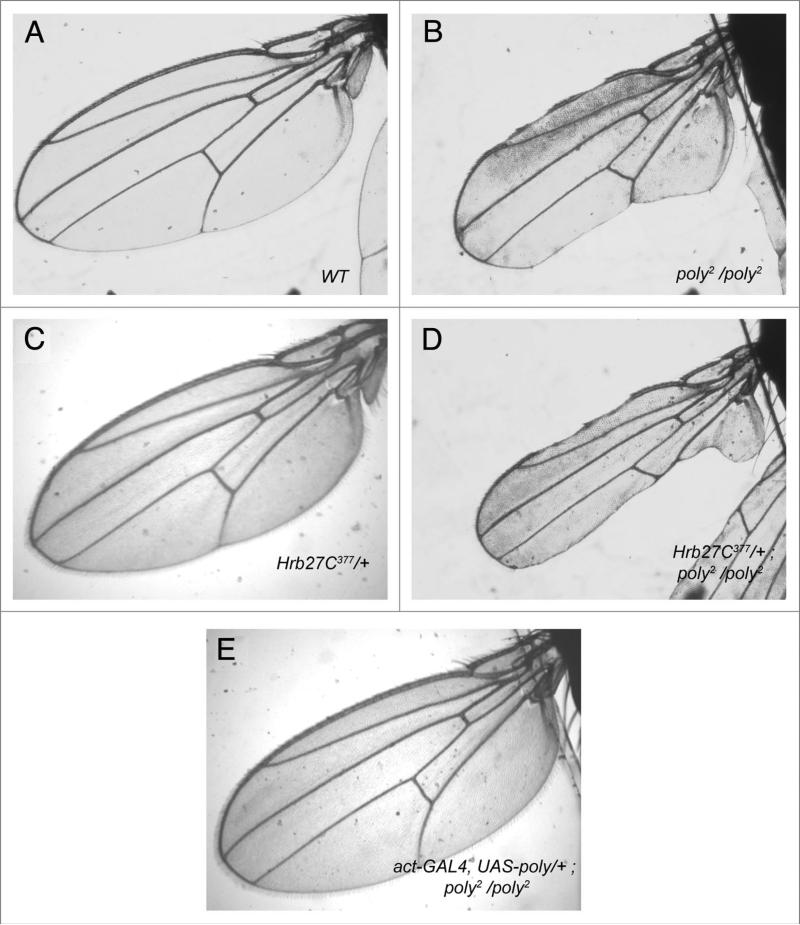
poly germ-line clones display oogenic phenotypes reminiscent of some characterized members of the mRNP complex, including hrb27C. We therefore performed genetic-interaction experiments, using hrb27C alleles with poly2 homozygous flies; poly1 is fully recessive lethal, but poly2 homozygous adults can be recovered and display many of the same oogenic phenotypes described above for poly germ-line clones (Fig. 6A). These studies did not produce conclusive results for ovarian phenotypes, because of the extremely small ovaries found in poly2 homozygous mutants, but evidence of genetic interactions was revealed by modification of additional external phenotypes found in poly mutants. Specifically, poly2 homozygotes and poly1/poly2 transheterozygotes display notching of the wing margin, although the severity varies (Fig. 5B); P-element hop-outs of poly2 yielded precise excision lines that showed intact wing-margins (data not shown). The wings of hrb27C377/+ flies are wild-type in appearance (Fig. 4C), whereas a single copy of the hrb27C377 mutation in the poly2 homozygous mutant background strengthens the wing margin phenotype over that of poly2 homozygosity alone (Fig. 5D). To ensure further that loss of poly caused the wing-margin loss in homozygous mutants, we successfully alleviated the wing-margin loss in poly2 mutants by expressing a UASp-poly transgene with the actP-GAL4 driver (Fig. 5E).
Figure 6.
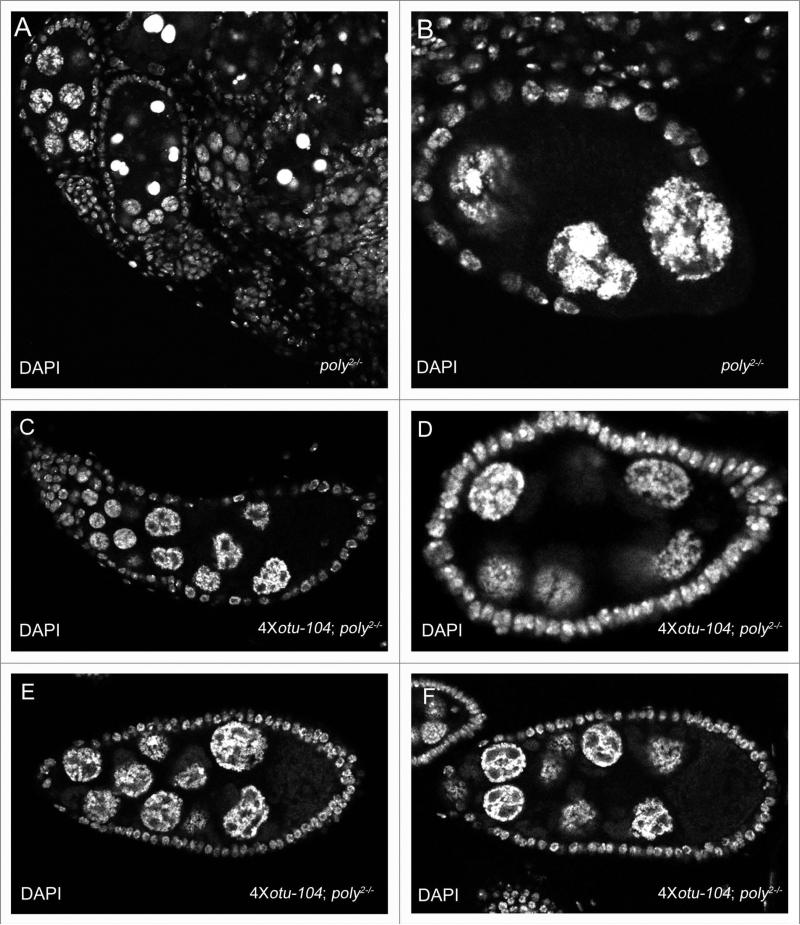
Multiple copies of the otu-104 transgene can partially rescue poly2 mutant ovaries. (A) poly2 mutant ovary displaying apoptosis, egg chamber fusion, and nurse-cell chromosome morphology defects. (B) poly2 egg chamber retaining the five-blob configuration in the nurse-cell nuclei. (c) egg chamber fusion of multiple egg chambers in a poly2 ovariole with overexpression of Otu-104. (D) stage-6 4x otu-104, poly2 mutant egg chamber displaying nurse-cell chromosomal abnormalities distinct from the five-blob configuration. (e and F) Rare stage-8 and stage-9 poly2 egg chambers with overexpression of Otu-104 with granulated nurse-cell morphology defects.
Figure 5.
The wing-margin-loss phenotype of poly2 mutant adults is strengthened by reduction of hrb27C. (A) Light microscopy of a wild-type Drosophila melanogaster wing. (B) Variable wing-margin loss is present in all poly2/poly2 escapers. (c) hrb27C377/+ flies show no defects in the wing margins. (D) A single copy of the hrb27C377 mutation strengthens the wing-margin loss of poly2 homozygotes. (e) Overexpression of UAsp-poly with the act-GAL4 driver in the background of poly2 mutant flies alleviates the wing-margin loss defect.
On the basis of the wing-notching enhancement seen with reduction of hrb27C in the background of poly2 homozygotes, we further studied the genetic interaction between otu and poly. The majority of alleles of identified mRNP components with germ-line functions phenocopy nurse-cell dispersal defects seen in heterozygous otu11 and otu13 mutants. Because mutations in a few of these components have been shown specifically to affect the Otu-104 kDa isoform, we performed genetic interaction studies with otu mutants and poly2 germ-line clones. We detected no prominent changes in nurse-cell chromosomal dispersal in otu13/+; poly2 germ-line clones (data not shown), but multiple copies of the otu-104 transgene facilitated further development of poly2 homozygous ovaries. poly2 mutant ovaries were small and displayed a wide variety of defects, including egg-chamber fusion, apoptosis and nurse-cell chromosome morphology defects (Fig. 6A and B). The enhancement of phenotypes in poly2 mutant egg chambers in comparison to poly2 germline clones is most likely due to loss of Poly function in the somatic tissue in conjunction with loss of Poly in the germ-line, as induction of poly2 follicleclones frequently undergo apoptosis (data not shown). In addition, poly1/poly2 transheterozygotes and poly2/Df6168 hemizygotes also presented similar defects in egg-chamber fusion and nurse-cell nuclei dispersal defects. In poly2 mutant ovaries with four copies of the otu-104 transgene, egg chambers progressed further in development, and the five-blob-configuration phenotype was rescued (Fig. 6C), but in poly2 homozygous ovaries, nurse-cell chromosomal morphology defects persisted that were distinct from the five-blob configuration, and egg chamber fusion was still frequently seen (Fig. 6C–F). We conclude that poly functions downstream of otu and those higher levels of the Otu-104 kDa protein can partially suppress poly phenotypes.
Discussion
In addition to hrb27C, sqd and pUf68, mutations in genes encoding transcription factors E2F1 and DP, the chromo-domain protein Rhino, the RNA helicase P68, and Spoonbill all affect nurse-cell chromosome dispersal and grk mRNA localization.19-23 Similar ovarian phenotypes displayed in mutants for these genes suggest a significant spatiotemporal link between nurse-cell chromosomal organization and oocyte polarity. The great functional diversity represented by these genes highlights the complexity of this essential yet poorly understood process. Here, we report that the novel gene, poly, is required for nurse-cell chromosome dispersal and AP/DV polarity. Our evidence of genetic interactions among poly, hrb27C and otu suggests that Poly is involved in processes similar to those of these previously characterized proteins. The otu-104 transgene (containing a large genomic fragment of otu that contains exon 6a) alleviates the Grk mislocalization and nurse-cell chromosome-dispersal failure defects of pUf68, sqd and hrb27C.11 Overexpression of Otu-104 partially rescues the poly mutant ovarian phenotypes, but the persistent fusion of egg chambers and nurse-cell nuclear defects in this rescue experiment suggest that poly mutations affect the expression of additional genes that mediate nurse-cell chromosome architecture and oocyte polarity formation/maintenance.
The interactions among different members of the mRNP complexes in Drosophila oogenesis cataloged to date are either protein-protein (such as cup-eIF4E)24 or RNA-dependent (hrb27C-sqd).11 The lack of any known protein domains in Poly prevents formation of a clear idea of Poly's possible functions in Drosophila oogenesis. From our results, we conclude that poly mutations affect the expression of genes independently or downstream of otu. Further studies are needed that can reveal the molecular mechanisms of poly and can identify the downstream targets that are affected by loss of poly function.
Methods
Fly stocks
Fly stocks bearing the following mutations were used: poly11648 (a P-element insertion allele from the Bloomington stock center that was recombined to the FRT82B chromosome; designated poly1) [FBal0035465], poly2 (known as P-element insertion 8131; from the Szeged stock center, Hungary) [FBal0190755], P[otu-104] (ovarian tumor 104-kDa isoform on the third chromosome, a gift from Dr. Lillian Searles; on the second chromosome, a gift from Dr. Trudi Schupbach) [FBtp0004944]. The kin-lacZ line (a gift from Dr. Hannele Ruohola-Baker)18 [FBti0002915] was used to determine the integrity of microtubules in poly germ-line clones. Although it is not known if poly1 is a null allele, the complete larval lethality of the poly1 line, as well as the difficulty in creation of stage-9 poly1 germ-line clones, allows classification of poly2 as a hypomorph allele.
To generate poly germ-line clones, we used hsFLP, αTub67C-GFP-Stau;25,26 FRT82B ubi-GFP/TM6B Hu Tb or TM3 Sb, and hsFLP; FRT82BarmlacZ/TM6B Hu Tb stocks (original FRT82B-ubiGFP and FRT82BarmLacZ stocks with FBIDs FBst0005188 and FBst0008218, respectively). For overexpression of poly, a full length cDNA, kindly supplied by Dr. Margarete Heck, was cloned into a pUASp vector by the FSU Molecular Cloning Facility, with or without the eGFP tag at the amino terminus, and injected into embryos according to standard procedures (performed by Duke University Model System Genomics). The P[mat-α-tubulin-GAL4VP16] driver [FBst0007062] on the second chromosome was used for germ-line–specific overexpression of poly. The P[actin-GAL4] driver [FBst0004414] on the second chromosome was used for ubiquitous overexpression of poly.
poly population analysis
Multiple vials of poly2 stocks were expanded, transferred repeatedly, and kept at 18°C, 25°C or 29°C for population analysis of poly2 homozygotes for one generation. Multiple vials of poly1 flies crossed to poly2 flies were given identical experimental treatment and examined for the presence of poly1/poly2 transheterozygotes for one generation as well. The percentage of viability was defined as the actual number of poly mutant flies scored, divided by the expected number of poly mutant flies, which is half of the number of balanced siblings. SE stands for standard (sampling) error and is related to standard deviation, i.e., the probability that data collected from the sample size is inaccurate. For the standard error of poly homozygote/transheterozygote viability, the following formula was used:
where the standard deviation for 1 out of the 4 possible genotypic classes of progeny is the square root of (0.25 × 0.75), approximately 0.433.31 Subsequent calculation of the standard error from standard-deviation values demonstrated that the smaller the SE value was, the more statistically significant the data collected from the sample size was; this result provides additional evidence that the sample size was adequate to demonstrate temperature-sensitive viability in poly homozygotes/transheterozygotes.
Generation of mosaic clones by the FLP-FRT technique
Fly crosses and rearing were performed under standard conditions.27 The FLP-FRT system was used to induce mitotic recombination and produce clones in the ovary through the heat-shock flipase (hsFLP) on the X chromosome. To obtain germ-line clones, we heat shocked second- and third-instar larvae for 2 hours on two consecutive days; the larvae were then reared at 25°C for eight days, sorted and moved to fresh vials with yeast for two additional days before dissection.28 Clones were marked by the absence of ubi-GFP29 or arm-lacZ.
Genetic interaction studies
hrb27C377 (located on the second chromosome) was crossed to a Sp/CyO; poly2/TM6B stock and crossed to poly2/TM6B again to produce poly2 escapers with heterozygous loss of hrb27C in the background. Progeny were reared at 18°C, a temperature that maximized number of escapers for study.
Inverse PCR
Inverse PCR was performed according to the standard BDGP protocol with primers for amplifying flanking sequences of the P{LacW} element. Recovery of flanking sequence data for the 8131 insertion revealed the insertion in the sole intron of the poly gene.
Immunocytochemistry
Immunocytochemistry was performed under standard conditions.30 The antibodies used were mouse anti-Gurken (1:20; Developmental Studies Hybridoma Bank), rabbit anti-Staufen (1:2,000; a gift from D. St. Johnston), and mouse anti-β-galactosidase G4644 at 1:1,000 dilution (Sigma-Aldrich).
Multiple alignment analysis
poly coding sequences from other species of Drosophila and putative homologues from nondipteran species were aligned with ClustalX 2.0.10 and modified in Adobe Photoshop CS2.
Acknowledgements
We thank Anne B. Thistle, Jamila Horabin and Yoichiro Tamori for critical reading and helpful input into the manuscript; Margarete Heck for sharing reagents before publication and for critical reading of the manuscript; Trudi Schupbach for hrb27C mutants; Lillian Searles for the otu-104 transgene; Daniel St. Johnston for staufen antibodies; the Molecular Cloning Lab (Michael Asher, Rani Dhanarajan, Andre Irsigler) at Florida State University for cloning of the poly gene into the UASp vector; and Kimberly Riddle and the FSU Biological Science Imaging Resource facility for assistance with electron microscopy and confocal imaging. We also thank David Houle and his lab for assistance with wing imaging; Chris Green, Amanda Novak and Yi-Chun Huang for stock maintenance and food preparation; Andrea Bixler for assistance in experiments; and Jianjun Sun, Laila Smith and John Poulton for discussion and interpretation of results and for critical reading of the manuscript. W.-M. Deng is supported by a National Institutes of Health grant R01 GM072562.
Abbreviations
- armLacZ
armadillo-lacZ
- eGFP
enhanced GFP
- GFP
green fluorescent protein
- Hu
humeral (marker)
- kDa
kilodalton
- PCR
polymerase chain reaction
- Sb
stubble (marker)
- TM3
third multiple three (balancer)
- TM6B
third multiple six, B structure (balancer)
- Tb
tubby (marker)
- UASp
upstream activation sequence P-element construct
- ubi-GFP
ubiquitin-GFP
Footnotes
Authors’ contributions: S.K. performed all experiments and drafted the manuscript. W.D. coordinated and supervised the experiments and helped to draft the manuscripts. Both authors read and approved the final manuscript.
References
- 1.Cooley L, Theurkauf WE. Cytoskeletal functions during Drosophila oogenesis. Science. 1994;266:590–6. doi: 10.1126/science.7939713. [DOI] [PubMed] [Google Scholar]
- 2.Spradling AC, editor. Developmental genetics of oogenesis. Cold Spring Harbor Laboratory Press; New York: 1993. [Google Scholar]
- 3.Riechmann V, Ephrussi A. Axis formation during Drosophila oogenesis. Curr Opin Genet Dev. 2001;11:374–83. doi: 10.1016/s0959-437x(00)00207-0. [DOI] [PubMed] [Google Scholar]
- 4.Steinhauer J, Kalderon D. Microtubule polarity and axis formation in the Drosophila oocyte. Dev Dyn. 2006;235:1455–68. doi: 10.1002/dvdy.20770. [DOI] [PubMed] [Google Scholar]
- 5.Theurkauf WE, Smiley S, Wong ML, Alberts BM. Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and inter-cellular transport. Development. 1992;115:923–36. doi: 10.1242/dev.115.4.923. [DOI] [PubMed] [Google Scholar]
- 6.St. Johnston D, Beuchle D, Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- 7.Brendza RP, Serbus LR, Duffy JB, Saxton WM. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science. 2000;289:2120–2. doi: 10.1126/science.289.5487.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neuman-Silberberg FS, Schupbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGFalpha-like protein. Cell. 1993;75:165–74. [PubMed] [Google Scholar]
- 9.Poulton JS, Deng WM. Cell-cell communication and axis specification in the Drosophila oocyte. Dev Biol. 2007;311:1–10. doi: 10.1016/j.ydbio.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Buskirk C, Schupbach T. Half pint regulates alternative splice site selection in Drosophila. Dev Cell. 2002;2:343–53. doi: 10.1016/s1534-5807(02)00128-4. [DOI] [PubMed] [Google Scholar]
- 11.Goodrich JS, Clouse KN, Schupbach T. Hrb27C, Sqd and Otu cooperatively regulate gurken RNA localization and mediate nurse cell chromosome dispersion in Drosophila oogenesis. Development. 2004;131:1949–58. doi: 10.1242/dev.01078. [DOI] [PubMed] [Google Scholar]
- 12.Huynh JR, Munro TP, Smith-Litière K, Lepesant JA, St. Johnston D. The Drosophila hnRNPA/B homolog, Hrp48, is specifically required for a distinct step in osk mRNA localization. Dev Cell. 2004;6:625–35. doi: 10.1016/s1534-5807(04)00130-3. [DOI] [PubMed] [Google Scholar]
- 13.Dej KJ, Spradling AC. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development. 1999;126:293–303. doi: 10.1242/dev.126.2.293. [DOI] [PubMed] [Google Scholar]
- 14.Deak P, Omar MM, Saunders RD, Pal M, Komonyi O, Szidonya J, et al. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster: correlation of physical and cytogenetic maps in chromosomal region 86E-87F. Genetics. 1997;147:1697–722. doi: 10.1093/genetics/147.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellotto M, Bopp D, Senti KA, Burke R, Deak P, Maroy P, et al. Maternal-effect loci involved in Drosophila oogenesis and embryogenesis: P element-induced mutations on the third chromosome. Int J Dev Biol. 2002;46:149–57. [PubMed] [Google Scholar]
- 16.Lopez-Schier H, St. Johnston D. Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev. 2001;15:1393–405. doi: 10.1101/gad.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rorth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–8. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 18.Clark IE, Jan LY, Jan YN. Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development. 1997;124:461–70. doi: 10.1242/dev.124.2.461. [DOI] [PubMed] [Google Scholar]
- 19.Volpe AM, Horowitz H, Grafer CM, Jackson SM, Berg CA. Drosophila rhino encodes a female-specific chromo-domain protein that affects chromosome structure and egg polarity. Genetics. 2001;159:1117–34. doi: 10.1093/genetics/159.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Royzman I, Hayashi-Hagihara A, Dej KJ, Bosco G, Lee JY, Orr-Weaver TL. The E2F cell cycle regulator is required for Drosophila nurse cell DNA replication and apoptosis. Mech Dev. 2002;119:225–37. doi: 10.1016/s0925-4773(02)00388-x. [DOI] [PubMed] [Google Scholar]
- 21.Motola S, Neuman-Silberberg FS. Spoonbill, a new Drosophila female-sterile mutation, interferes with chromosome organization and dorsal-ventral patterning of the egg. Dev Dyn. 2004;230:535–45. doi: 10.1002/dvdy.20066. [DOI] [PubMed] [Google Scholar]
- 22.Buszczak M, Spradling AC. The Drosophila P68 RNA helicase regulates transcriptional deactivation by promoting RNA release from chromatin. Genes Dev. 2006;20:977–89. doi: 10.1101/gad.1396306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuman-Silberberg FS. Drosophila female sterile mutation spoonbill interferes with multiple pathways in oogenesis. Genesis. 2007;45:369–81. doi: 10.1002/dvg.20303. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 25.Schuldt AJ, Adams JH, Davidson CM, Micklem DR, Haseloff J, St. Johnston D, Brand AH. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 1998;12:1847–57. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin SG, Leclerc V, Smith-Litière K, St. Johnston D. The identification of novel genes required for Drosophila anteroposterior axis formation in a germline clone screen using GFP-Staufen. Development. 2003;130:4201–15. doi: 10.1242/dev.00630. [DOI] [PubMed] [Google Scholar]
- 27.Poulton JS, Deng WM. Dystroglycan downregulation links EGF receptor signaling and anterior-posterior polarity formation in the Drosophila oocyte. Proc Natl Acad Sci USA. 2006;103:12775–80. doi: 10.1073/pnas.0603817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng WM, Ruohola-Baker H. Laminin A is required for follicle cell-oocyte signaling that leads to establishment of the anterior-posterior axis in Drosophila. Curr Biol. 2000;10:683–6. doi: 10.1016/s0960-9822(00)00514-5. [DOI] [PubMed] [Google Scholar]
- 29.Maines JZ, Stevens LM, Tong X, Stein D. Drosophila dMyc is required for ovary cell growth and endoreplication. Development. 2004;131:775–86. doi: 10.1242/dev.00932. [DOI] [PubMed] [Google Scholar]
- 30.Deng WM, Althauser C, Ruohola-Baker H. Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development. 2001;128:4737–46. doi: 10.1242/dev.128.23.4737. [DOI] [PubMed] [Google Scholar]
- 31.Freedman D, Pisani R, Purves R. Statistics. 1st edn. Norton; New York: 1978. [Google Scholar]