Abstract
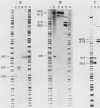
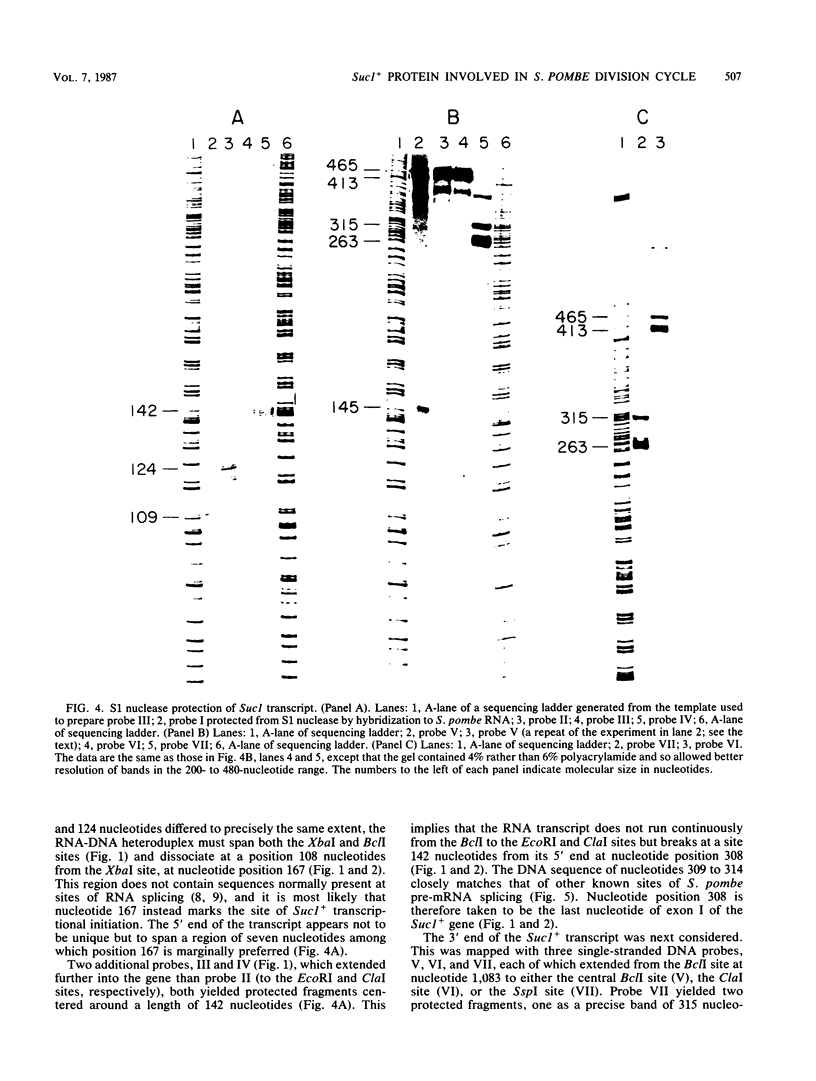
Sucl+ was originally identified as a DNA sequence that, at high copy number, rescued Schizosaccharomyces pombe strains carrying certain temperature-sensitive alleles of the cdc2 cell cycle control gene. We determined the nucleotide sequence of a 1,083-base-pair Sucl+ DNA fragment and S1 mapped its 866-nucleotide RNA transcript. The protein-coding sequence of the gene is interrupted by two intervening sequences of 115 and 51 base pairs. The predicted translational product of the gene is a protein of 13 kilodaltons. A chromosomal gene disruption of Sucl+ was constructed in a diploid S. pombe strain. Germinating spores carrying a null allele of the gene were capable of very limited cell division, following which many cells became highly elongated. The Sucl+ gene was also strongly overexpressed under the control of a heterologous S. pombe promoter. Overexpression of Sucl+ is not lethal but causes a division delay such that cells are approximately twice the normal length at division. These data suggest that Sucl+ encodes a protein which plays a direct role in the cell division cycle of S. pombe.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beach D., Durkacz B., Nurse P. Functionally homologous cell cycle control genes in budding and fission yeast. Nature. 1982 Dec 23;300(5894):706–709. doi: 10.1038/300706a0. [DOI] [PubMed] [Google Scholar]
- Beach D., Piper M., Nurse P. Construction of a Schizosaccharomyces pombe gene bank in a yeast bacterial shuttle vector and its use to isolate genes by complementation. Mol Gen Genet. 1982;187(2):326–329. doi: 10.1007/BF00331138. [DOI] [PubMed] [Google Scholar]
- Beach D., Rodgers L., Gould J. ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr Genet. 1985;10(4):297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Booher R., Beach D. Site-specific mutagenesis of cdc2+, a cell cycle control gene of the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 1986 Oct;6(10):3523–3530. doi: 10.1128/mcb.6.10.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Benoist C., O'Hare K., Gannon F., Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Burke J. F. High-sensitivity S1 mapping with single-stranded [32P]DNA probes synthesized from bacteriophage M13mp templates. Gene. 1984 Oct;30(1-3):63–68. doi: 10.1016/0378-1119(84)90105-7. [DOI] [PubMed] [Google Scholar]
- Durkacz B., Carr A., Nurse P. Transcription of the cdc2 cell cycle control gene of the fission yeast Schizosaccharomyces pombe. EMBO J. 1986 Feb;5(2):369–373. doi: 10.1002/j.1460-2075.1986.tb04221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y., Kaziro Y. Molecular cloning and sequence analysis of a ras gene from Schizosaccharomyces pombe. EMBO J. 1985 Mar;4(3):687–691. doi: 10.1002/j.1460-2075.1985.tb03684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J., Beach D., Durkacz B., Nurse P. The fission yeast cell cycle control gene cdc2: isolation of a sequence suc1 that suppresses cdc2 mutant function. Mol Gen Genet. 1986 Feb;202(2):291–293. doi: 10.1007/BF00331653. [DOI] [PubMed] [Google Scholar]
- Hindley J., Phear G. A. Sequence of the cell division gene CDC2 from Schizosaccharomyces pombe; patterns of splicing and homology to protein kinases. Gene. 1984 Nov;31(1-3):129–134. doi: 10.1016/0378-1119(84)90203-8. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y., Toda T., Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984 Dec;39(2 Pt 1):349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Plasmids carrying the yeast OMP decarboxylase structural and regulatory genes: transcription regulation in a foreign environment. Cell. 1983 Feb;32(2):371–377. doi: 10.1016/0092-8674(83)90456-7. [DOI] [PubMed] [Google Scholar]
- Lörincz A. T., Reed S. I. Primary structure homology between the product of yeast cell division control gene CDC28 and vertebrate oncogenes. Nature. 1984 Jan 12;307(5947):183–185. doi: 10.1038/307183a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Nurse P., Bissett Y. Gene required in G1 for commitment to cell cycle and in G2 for control of mitosis in fission yeast. Nature. 1981 Aug 6;292(5823):558–560. doi: 10.1038/292558a0. [DOI] [PubMed] [Google Scholar]
- Nurse P., Thuriaux P. Regulatory genes controlling mitosis in the fission yeast Schizosaccharomyces pombe. Genetics. 1980 Nov;96(3):627–637. doi: 10.1093/genetics/96.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. I., Hadwiger J. A., Lörincz A. T. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4055–4059. doi: 10.1073/pnas.82.12.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T. M., Swanberg S. L., Poteete A., Riedel G., Backman K. A plasmid cloning vehicle allowing a positive selection for inserted fragments. Gene. 1980 Dec;12(1-2):123–127. doi: 10.1016/0378-1119(80)90022-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Simanis V., Nurse P. The cell cycle control gene cdc2+ of fission yeast encodes a protein kinase potentially regulated by phosphorylation. Cell. 1986 Apr 25;45(2):261–268. doi: 10.1016/0092-8674(86)90390-9. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Adachi Y., Hiraoka Y., Yanagida M. Identification of the pleiotropic cell division cycle gene NDA2 as one of two different alpha-tubulin genes in Schizosaccharomyces pombe. Cell. 1984 May;37(1):233–242. doi: 10.1016/0092-8674(84)90319-2. [DOI] [PubMed] [Google Scholar]