Abstract
In bacteria, both promoters and 5′-untranslated regions (5′-UTRs) of mRNAs play vital regulatory roles in gene expression. In this study, we identified 1203 active promoter candidates in Bacillus thuringiensis through analysis of the genome-wide TSSs based on the transcriptome data. There were 11 types of σ-factor and 34 types of transcription factor binding sites found in 723 and 1097 active promoter candidates, respectively. Moreover, within the 1203 transcriptional units (TUs), most (52%) of the 5′-UTRs were 10–50 nucleotides in length, 12.8% of the TUs had a long 5′-UTR greater than 100 nucleotides in length, and 16.3% of the TUs were leaderless. We then selected 20 active promoter candidates combined with the corresponding 5′-UTR DNA regions to screen the highly active promoter-5′-UTR DNA region complexes with different characteristics. Our results demonstrate that among the 20 selected complexes, six were able to exert their functions throughout the life cycle, six were specifically induced during the early-stationary phase, and four were specifically activated during the mid-stationary phase. We found a direct corresponding relationship between σ-factor-recognized consensus sequences and complex activity features: the great majority of complexes acting throughout the life cycle possess σA-like consensus sequences; the maximum activities of the σF-, σE-, σG-, and σK-dependent complexes appeared at 10, 14, 16, and 22 h under our experimental conditions, respectively. In particular, complex Phj3 exhibited the strongest activity. Several lines of evidence showed that complex Phj3 possessed three independent promoter regions located at −251∼−98, −113∼−31, and −54∼+14, and that the 5′-UTR +1∼+118 DNA region might be particularly beneficial to both the stability and translation of its downstream mRNA. Moreover, Phj3 successfully overexpressed the active β-galactosidase and turbo-RFP, indicating that Phj3 could be a proper regulatory element for overexpression of proteins in B. thuringiensis. Therefore, our efforts contribute to molecular biology research and the biotechnological application of B. thuringiensis.
Introduction
Unlike archaea and eukaryotes, bacteria contain only one form of RNA polymerase (RNAP) core enzyme comprised of five subunits (α2ββ′ω). However, bacteria possess multiple forms of a specific σ subunit (σ-factor) and thus multiple forms of RNAP holoenzymes, which, in turn, bind to their cognate promoters to initiate transcription of specific genes (or operons) [1]–[4]. In bacteria, a promoter is a specific DNA sequence that provides secure initial binding sites for RNAP to initiate transcription of a particular gene (or operon) [1], [2]. The core promoter includes a transcription start site (TSS) and two hexameric elements centered at or near –10 and –35 positions relative to the TSS. Some promoters contain one or more upstream promoter (UP) elements and the TGn extended –10 element, among others [1]–[4].
A TSS is an important marker of an active promoter, and mapping the TSSs is therefore a novel and effective strategy for the identification of active promoters. McGrath et al. mapped 769 TSSs and subsequently identified 27 promoter motifs in Caulobacter crescentus using a high-density array that was specifically designed to detect the TSS positions [5]. Mendoza-Vargas et al. mapped more than 1700 TSSs and identified a large number of promoters that control the expression of approximately 800 genes in Escherichia coli by combining a modified 5′ RACE protocol and an unbiased high-throughput pyrosequencing strategy [6]. However, the active promoter candidates acquired by them were not verified through further experimentation. Recently, the high-throughput and unbiased sequencing of the cDNA (RNA-seq) technique has been used for whole-genome transcriptomics analyses of diverse bacteria [7]. Sharma et al. reported that the genome-wide TSSs could be directly detected from RNA-seq data using a novel differential approach selective for the 5′ triphosphate (5′-PPP) ends of the primary transcripts [8]. Although Sharma et al. did not report the data of active promoter identification, the knowledge of TSSs could provide us with a promising opportunity for the high-throughput identification of active promoters from RNA-seq data.
Besides the promoters, the 5′-untranslated regions (5′-UTRs) of bacterial mRNA are also known to play important regulatory roles in gene expression, which possibly occur at the transcriptional, post-transcriptional, or translational levels [9]. Extremely diverse mechanisms are employed by the cis-acting RNA regulatory elements in 5′-UTRs to strictly adjust the cellular levels of their downstream genes, including: (i) the ability of many 5′-UTRs to recognize a specific regulatory signal, such as T-boxes, riboswitches and RNA thermometers [10]–[12]; (ii) the capability of some 5′-UTRs to provide binding sites for small regulatory RNAs [9], [13]; and (iii) more 5′-UTRs being able to regulate the expression of the downstream gene, presumably by RNase III-mediated cleavage modification [14], preventing degradation of the mRNA [15], or other unknown mechanisms. Therefore, besides promoters, some 5′-UTR DNA regions have a significant applied potential in molecular biology research and improvement of recombinant protein expression [9], [12], [16], [17].
Bacillus thuringiensis is characterized by the formation of parasporal crystals consisting of insecticidal crystal proteins (ICPs) during sporulation. Moreover, the accumulation of ICPs can account for 20–30% of the cell’s dry weight [18]. This unique advantage enables B. thuringiensis to be not only the most widely used environmentally compatible biopesticide [19], [20] but also a promising gene expression system. In the Bacillus species, the sporulation-specific σ-factors SigH, SigF, SigE, SigG, and SigK are spatially and temporally activated to control the process of sporulation [21]. SigF and SigE regulate early compartmentalized gene expression, whereas SigG and SigK activate transcription of the genes that build the structural components of the spore [21]–[23]. SigE and SigK also promote transcription of the ICP genes for the formation of parasporal crystals in B. thuringiensis [24]. Consequently, to thoroughly investigate the regulation of gene expression and/or construct a novel gene expression system in B. thuringiensis, high-throughput identification and screening of promoter-5′-UTR DNA region complexes (to avoid redundancy, “complex” refers to the promoter region and the 5′-UTR DNA region) with specific characteristics (intrinsic strength and temporal activation) are of great practical significance.
B. thuringiensis subsp. chinensis CT-43 is the first sequenced strain harboring ICP genes [25]. Moreover, the whole-genome transcriptomics analysis of CT-43 at four different growth phases in GYS medium [26] was performed by the RNA-seq technique. In the RNA-seq data, the average length of the clean-reads was 110 nucleotides, and the number of the clean-reads in the four different libraries was 577,810 to 1,493,721. Thus, the sequencing coverage of the four growth phases was 10- to 27-fold. Moreover, the percentages of the clean-reads that were mapped to the CT-43 genome were approximately 90 to 96% [27]. In this study, 1203 active promoter candidates were identified from the RNA-seq data, and 20 highly active promoter candidates combined with the corresponding 5′-UTRs were selected to perform further analyses to screen the highly active promoter-5′-UTR DNA region complexes with different characteristics.
Materials and Methods
Bacterial Strain and Plasmids
The bacterial strains and plasmids used in this study are listed in Table S1.
Genome-wide TSS Mapping and Identification of Active Promoter Candidates
Using RNA-seq method, we previously acquired transcriptome data of B. thuringiensis strain CT-43 at four growth phases when grown in GYS medium [26] at 28°C and 200 rpm: 7 h (the mid-exponential growth phase), 9 h (the early-stationary growth phase), 13 h (the mid-stationary growth phase, sporulation), and 22 h (the spore maturation and mother cell lysis phase) [27]. To map genome-wide TSSs, the clean-reads of each sample were mapped to the CT-43 genome using BlastN with a threshold e value of 0.00001 and the “−F F” parameter [28], and then the number of unambiguously mapped reads per nucleotide was calculated and visualized by R and Origin version 8.0. According to the mapping data, all 5′-ends that showed obvious cDNA coverage enrichment were annotated to predict the TSSs.
The regions located ≤500 nucleotides upstream of the mapped TSS were taken as the active promoter candidates. Then, these 500-nucleotide sequences were submitted to DBTBS [29] (http://dbtbs.hgc.jp/) to identify the recognition sites for σ-factors and transcription factors (TFs) through “Weight Matrix Search (by sequence)”. During the “advanced search”, the threshold of the p-value was set as 0.05.
Construction of Plasmids
All promoter-5′-UTR DNA region complexes were designated as Phj with the corresponding serial numbers.
Construction of translational fusion plasmids
All primers used in this study are listed in Table S2. The translational fusion plasmid pHT1K-Phj1-lacZ was constructed through the experimental procedure shown in Figure S1. Briefly, the promoter-5′-UTR DNA region complex of Phj1 was amplified from the genomic DNA of CT-43 using the primer pair Phj1-F/Phj1-R that carried additional recognition sites of the restriction endonucleases NcoI, XbaI and NotI at the 5′-end and BamHI and SmaI at the 3′-end. The PCR products were digested and ligated with the shuttle plasmid pHT1K [30] at the 5′ BglII and 3′ PstI restriction sites and then transformed into E. coli strain DH5α to construct the plasmid pHT1K-Phj1. The lacZ gene without the 5′-UTR DNA region was amplified from the plasmid pHT304-18Z [31]. The amplified products were digested with BamHI and KpnI, inserted into the plasmid pHT1K-Phj1 and then transformed into E. coli DH5α to acquire the plasmid pHT1K-Phj1-lacZ. All other translational fusion plasmids were obtained by replacing Phj1 with amplified promoter-5′-UTR DNA region complexes at 5′ NcoI and 3′ BamHI sites (Figure S1).
Construction of transcriptional fusion plasmids using fragments from Phj3
To analyze the characteristics of complex Phj3 in detail, the lacZ gene with its 5′-UTR DNA region was digested with BamHI and KpnI from the plasmid pHT304-18Z and inserted into the plasmid pHT1K to obtain the plasmid pHT1K-lacZ(UTR). Seven fragments of complex Phj3, including −251∼−98, −251∼−31, −251∼+14, −113∼−31, −54∼+14, −54∼+118, and −6∼+118 were amplified with the cognate primer pairs (Table S1). Subsequently, the PCR products of the seven fragments were separately digested with NcoI and BamHI and inserted into the plasmid pHT1K-lacZ(UTR) to construct the corresponding transcriptional fusion plasmids.
Construction of chimeric complexes
The 5′-UTR DNA fragment +1∼ +118 of complex Phj3 was separately fused at the 3′-ends of the promoter regions of complexes Phj12 and Phj17 to construct the chimeric complexes named as cPhj12 and cPhj17 by overlapping PCR. Next, the PCR products were used to replace Phj1 in the plasmid pHT1K-Phj1-lacZ to acquire the translational fusion plasmids pHT1K-cPhj12-lacZ and pHT1K-cPhj17-lacZ (see Figure S1).
Construction of plasmids for protein overexpression
The turbo-rfp gene was amplified by PCR using rfp-F/rfp-R as the primers and the plasmid pRP1028 (a gift from Scott Stibitz, Center for Biologics Evaluation and Research, Food and Drug Administration, Bethesda, Maryland, USA) as the template. The amplified products were digested with BamHI and KpnI and inserted into the plasmid pHT1K-Phj3 to construct the plasmid pHT1K-Phj3-turbo-rfp (Figure S2).
Transformation of the Plasmids to B. thuringiensis BMB171
After confirmation by sequencing, the plasmids were extracted from E. coli DH5α and transformed (electroporation) into B. thuringiensis BMB171 [32]. Various transformants were harvested by screening the clones in LB plates with 25 µg/mL erythromycin. Here, each transformant was not designated as a new strain, but rather expressed as BMB171 containing a specific plasmid.
Determination of β-Galactosidase Activity
The B. thuringiensis strain BMB171 containing each translational fusion plasmid or transcriptional fusion plasmid with the lacZ reporter gene was grown at 28°C in an orbital shaker at 200 rpm in GYS medium with 25 µg/mL erythromycin. Samples were taken at 2 h intervals for the determination of β-galactosidase activities. The growth curve was obtained by determining the optical density (OD) at 600 nm (OD600) combined with observation under a phase contrast microscope (Nikon ECLIPSE E6000, Nikon Corp., Tokyo, Japan). The β-galactosidase specific activities were determined and converted to Miller units as previously described [33]. The values shown represent the average of three independent experiments.
SDS-PAGE Analysis of Overexpressed Proteins
Each recombinant BMB171 strain containing pHT1K-Phj3-lacZ or pHT1K-Phj3-turbo-rfp plasmid was grown at 28°C for 22 h in LB medium with 25 µg/mL erythromycin. The culture was harvested by centrifugation and the crude proteins were extracted by boiling. SDS-PAGE was performed with 5% (w/v) stacking gels and 12% (w/v) separating gels, and proteins were visualized by Coomassie Blue R-250 staining.
Accession Number
The RNA-seq data from this article are available as raw short read data in the NCBI’s GEO database under accession number GSE39479.
Results
Identification of Active Promoter Candidates from RNA-seq Data
Genome-wide TSS mapping
After calculating the number of unambiguously mapped reads per nucleotide, we observed the cDNA coverage enrichment at all 5′-ends of the highly expressed genes that showed high redundancy in RNA-seq data. Generally, a TSS is manually determined once (i) a substantially sharp cDNA coverage enrichment is observed at the 5′-end, or (ii) a sharp cDNA coverage enrichment at the 5′-end appears in at least two libraries of the four growth phases [8], [34]; the TSSs of the remaining genes with low expression levels were unable to be unambiguously determined due to the relatively low signal-to-noise ratio. Following this principle, 1203 TSSs were mapped in the CT-43 genome, of which 1125 and 78 TSSs were shared by chromosome and plasmids, respectively (Table S3). Interestingly, 76 genes located within specific operons were found to have their own TSSs, such as the gene CT-43_CH1330 (indicated as “operon (intra)” in Table S3). Figure S3 shows the substantially sharp cDNA coverage enrichment at TSS positions of the 20 complex candidates Phj1-Phj20, which were selected for further analyses in this study.
Prediction of σ-factor and TF binding sites
The mapped 1203 TSSs represented 1203 active promoter candidates. To analyze the putative binding sites for σ-factors and TFs, 500-nucleotide sequences located upstream of the mapped TSSs were submitted one by one to DBTBS [29] (http://dbtbs.hgc.jp/). Using the “Weight Matrix Search (by sequence)” with the threshold set at a p-value 0.05, we identified the putative binding sites for SigA (209, 17.4%), SigB (78, 6.5%), SigD (26, 2.2%), SigE (129, 10.7%), SigF (105, 8.7%), SigG (112, 9.3%), SigH (190, 15.8%), SigK (72, 6.0%), SigL (22, 1.8%), SigW (49, 4.1%), and SigX (25, 2.1%). However, the putative σ-factor binding sites of 480 (about 40%) active promoter candidates could not be predicted (Table S3). Among the 723 active promoter candidates that could be predicted to possess the putative σ-factor binding sites, 495 (68.5%) were possibly controlled by a single σ-factor, while 228 (31.5%) were possibly controlled by multiple σ-factors. It is worth mentioning that 491 (68.0%) promoters were found to possess the putative binding sites for the sporulation-specific σ-factors SigH, SigF, SigE, SigG, and SigK (Table S3), reflecting that transcription of the corresponding genes was temporally activated during sporulation.
There were 34 different TF binding sites found in 1097 active promoter candidates (Table S3). The most frequently found TF binding sites were those for DegU (437), ComK (267), PerR (217), CodY (196), Fur (150), AbrB (125), AhrC (125), Zur (119), PurR (106), and ResD (101) (Table S3). These results indicated that a complicated TF regulatory network was involved in gene expression in B. thuringiensis, and that the TFs DegU, ComK, PerR, CodY, Fur, AbrB, AhrC, Zur, PurR, and ResD played more important roles than the others under our experimental conditions.
Length of the 5′-UTRs
In terms of the 5′-UTR length (ranging from the TSS to the first annotated start codon ATG of the corresponding DNA rigion) for the 1203 transcriptional units (TUs), we found that: i) most (52.0%) of the 5′-UTRs were 10–50 nucleotides in length; ii) the length of 18.9% 5′-UTRs varied between 50 and 100 nucleotides; iii) 12.8% of TUs had a long 5′-UTR (between 100 and 350 nucleotides in our data); and iv) 16.3% of TUs were leaderless (typically, a mRNA is considered as “leaderless” if the length of 5′-UTRs is less than ten nucleotides [8]) (Figure S4 and Table S3). In addition, the TSS of the gene pCT127.010 is located two nucleotides downstream of the first annotated ATG codon, perhaps owing to an error annotation. For the 5′-UTRs that were longer than 50 nucleotides, we searched them in the Rfam database [35] to identify known regulatory RNA elements. We found that five TUs most likely have an RNA regulatory element, including the CH1169 gene (T-box), rplS operon (L19_leader), rplU operon (L21_leader), infC operon (L20_leader), and CH5446 (SAM-riboswitch).
Using lacZ as a reporter gene, 20 active promoter candidates together with their corresponding 5′-UTR DNA rigions (promoter-5′-UTR DNA region complexes) were selected to further investigate their activity features, including intrinsic strength, temporal activation, and the consensus sequences recognized by σ-factor (Tables S4 and S5). According to the RNA-seq data, nine complex candidates could be able to exert their functions throughout the life cycle, seven could be specifically induced in the early-stationary phase and four could be specifically activated in the mid-stationary phase.
The Life Cycle of Strain BMB171 in GYS Medium
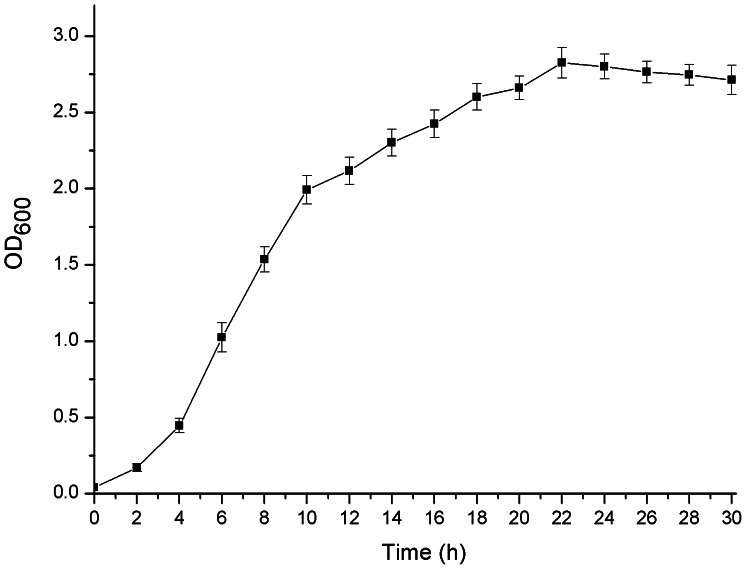
The life cycle of B. thuringiensis can be differentiated into two distinctively different stages: vegetative growth and sporulation. Because various σ-factors are temporally and/or spatially activated at different growth phages to control the process of vegetative growth and sporulation [21]–[23], the determination of the life cycle is necessary to analyze the features of the complexes with specific characteristics. By measuring the OD600, a growth curve of strain BMB171 containing the control plasmid pHT1K in GYS medium with 25 µg/mL erythromycin was obtained (Figure 1). These results combined with the obervation under a phase contrast microscope indicated that: 1) the growth of strain BMB171 containing pHT1K entered the early-stationary phase after appproximately 10 h of growth and the cells began to aggregate; 2) the 16 h time point represented the mid-stationary phase and the percentage of sporulating cells reached approximately 30%; 3) from approximately 22 h, BMB171 containing pHT1K entered the spore maturation and mother cell lysis phase, and approximately 30% mother cells were lysed with some spore release.
Figure 1. Growth curve of strain BMB171 containing the control plasmid pHT1K in GYS medium.
The strain BMB171 containing the control plasmid pHT1K was grown in GYS medium with 25 µg/mL erythromycin. The y-axis presents the average optical densities of triplicate bacterial cultures at 600 nm at each time point. Data are averages of three independent experiments (error bars are SEM from mean values).
Screening of the Highly Active Promoter-5′-UTR DNA Region Complexes with Different Characteristics
The complexes acting throughout the life cycle
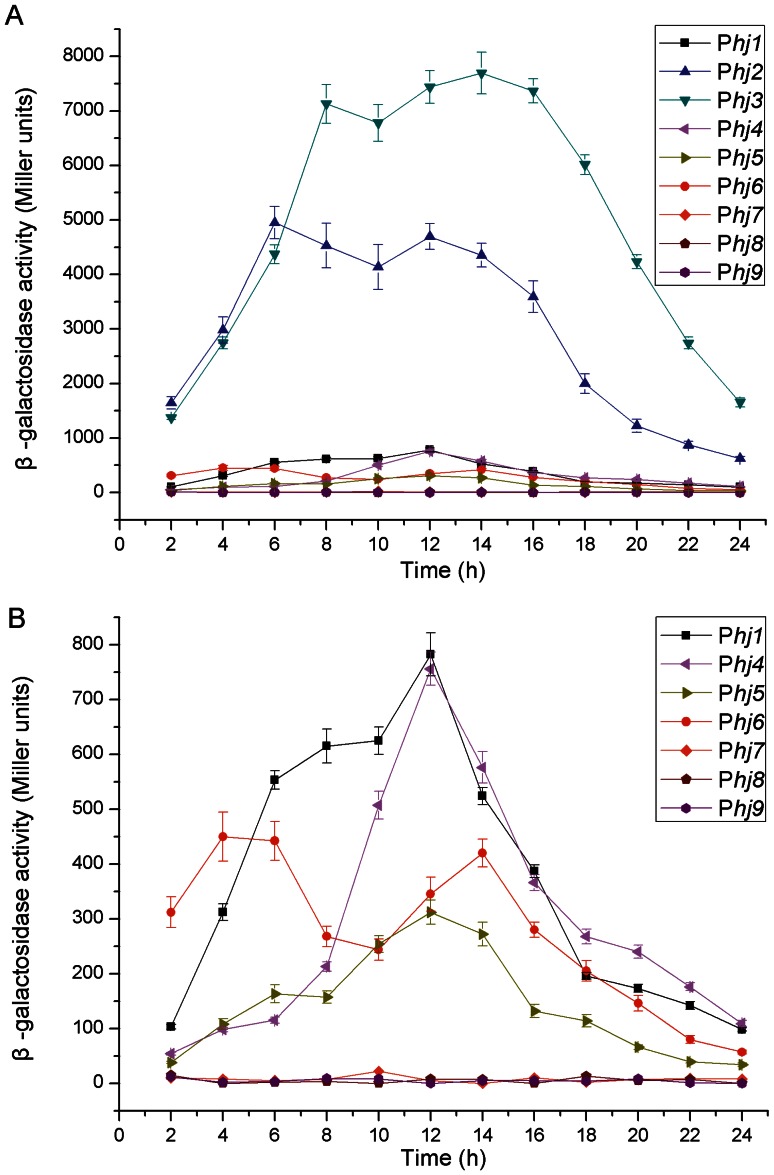
Candidates from Phj1 to Phj9 were selected to screen the highly active promoter-5′-UTR DNA region complexes, which can exploit their activity throughout the life cycle (Table S4). Our results showed that complex Phj3 displayed the strongest activity, followed by Phj2, Phj1, Phj4, and Phj6 (Figure 2A and Figure 2B). The maximum β-galactosidase specific activities directed by complexes Phj3 and Phj2 were approximately 7,600 and 5,000 Miller units in GYS medium, respectively; they reached 11,000 and 8,400 Miller units in LB medium (data not shown), respectively. Moreover, the Phj3-directed β-galactosidase activity could be detected at the onset of growth (2 h). It reached the first and second peaks at 8 and 14 h of growth, respectively, and then remained at a high level throughout the life cycle (Figure 2A). Being similar to the promoter of complex Phj3, the promoters of Phj2 and Phj6 also appeared to exhibit a second induction phenomenon, possibly owing to the fact that these promoters all possess more than one kind of consensus sequences that might be controlled by at least two different σ-factors (Table S5). Unfortunately, the activities of complex candidates Phj7, Phj8, and Phj9 from the plasmids of strain CT-43 could not be detected in strain BMB171 (Figure 2B). It is unclear why complex Phj6 also came from a plasmid of strain CT-43, but it was confirmed to work normally in strain BMB171 (Figure 2B). Thus, the reason for why complex candidates Phj7, Phj8, and Phj9 could not exert their functions remains to be elucidated.
Figure 2. Activity analyses of the complex candidates acting throughout the life cycle.
(A) β-galactosidase specific activities directed by complexes Phj1-Phj9. (B) β-galactosidase specific activities directed by complexes Phj1, and Phj4-Phj9. Data are averages of three independent experiments (error bars are SEM from mean values).
The complexes specifically induced during the early-stationary phase
Further analyses were performed on the seven complex candidates Phj10-Phj16 that could specifically exert their functions in the early-stationary phase (Table S4). Our results showed that complex Phj10 possessed the strongest activity among the seven analyzed complex candidates, followed by complex Phj12 (Figure 3). Interestingly, β-galactosidase activities directed by complexes Phj10, Phj11, Phj12, and Phj14, which have the σE-like consensus sequences (Table S5), all reached the peak values at approximately 14 h (early-stationary phase), whereas the highest activity of complex Phj15 containing the σG-like consensus sequence appeared 2 h later (at 16 h) compared to the σE-dependent complex (Figure 3). These results truly reflect the temporal regulation of SigE and SigG in B. thuringiensis. In addition, the activity of complex Phj13 was very weak, and that of complex Phj16 could not be detected.
Figure 3. Activity analyses of the complex candidates specifically induced in the early-stationary growth phase.
Complexes from Phj10 to Phj16 were separately fused with the gene lacZ and their activities were monitored by detecting the β-galactosidase specific activities. Data are averages of three independent experiments (error bars are SEM from mean values).
The complexes specifically activated during the mid-stationary phase
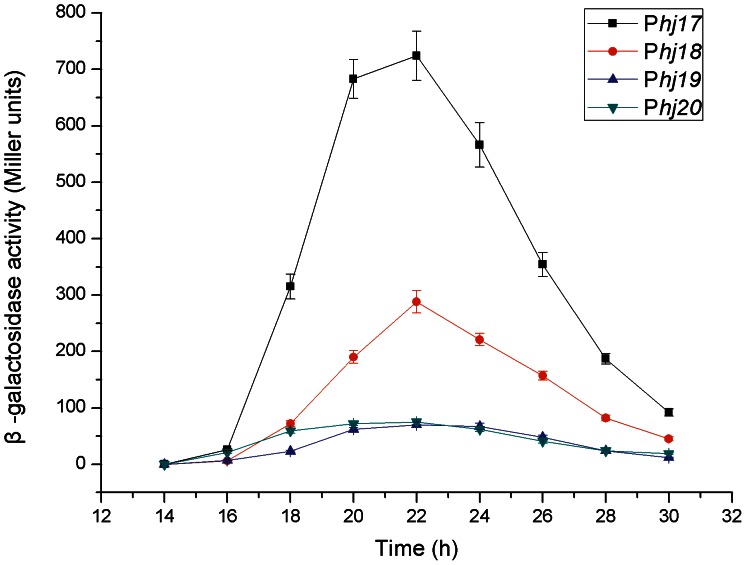
Complex candidates Phj17-Phj20, which are specifically activated in the mid-stationary phase, were selected to be further confirmed by translational fusion analysis. The results indicated that the analyzed complexes all began induction at approximately 16 h and reached the maximum inductions at 22 h of growth (Figure 4). These results were in excellent agreement with the fact that these complexes all contain the σK-like consensus sequences (Table S4). Among them, complex Phj17 shared the strongest activity, whereas complexes Phj19 and Phj20 had weak activities (Figure 4).
Figure 4. Activity analyses of the complex candidates specifically induced in the mid-stationary growth phase.
Complexes Phj17, Phj18, Phj19, and Phj20 were separately fused with the gene lacZ and their activities were monitored by detecting the β-galactosidase specific activities. Data are averages of three independent experiments (error bars are SEM from mean values).
Characteristics of Complex Phj3
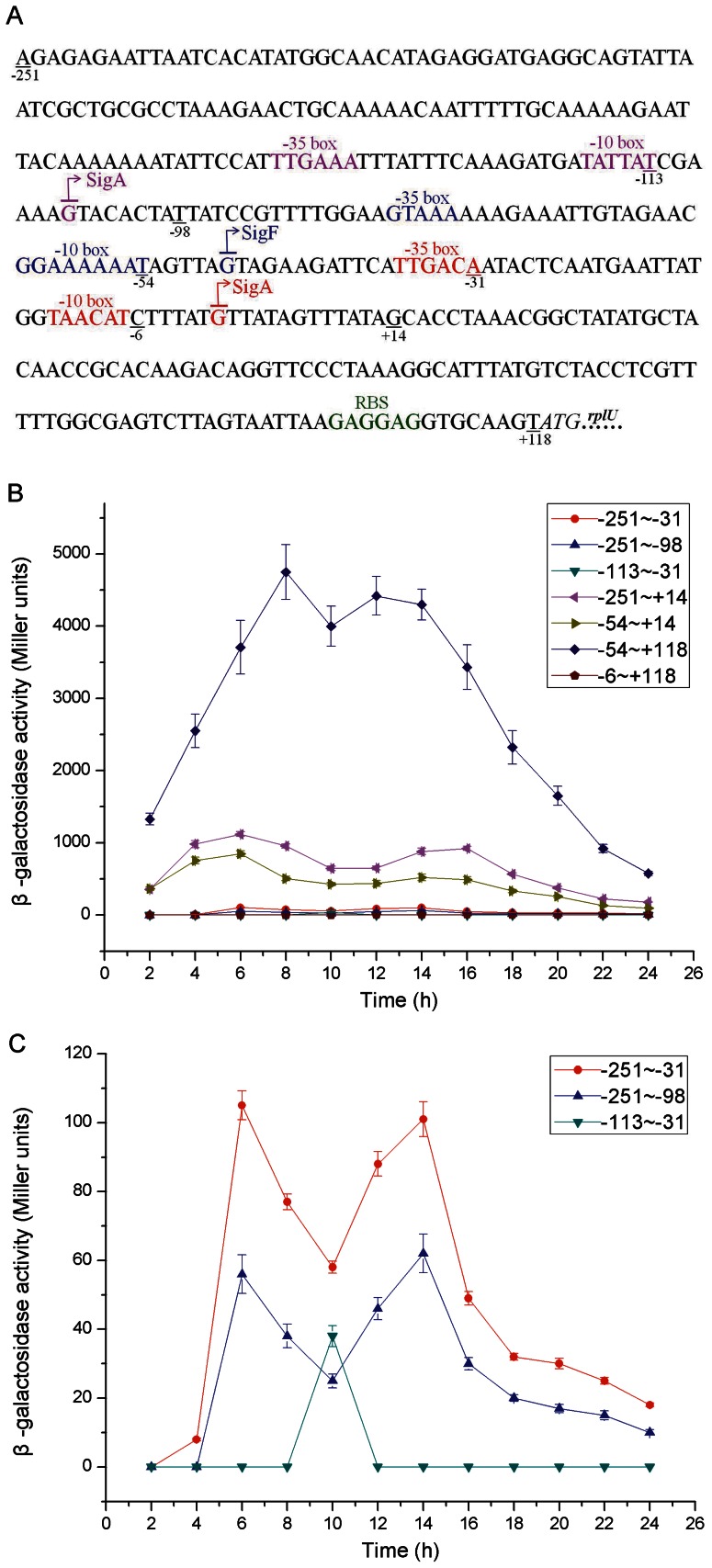
Complex Phj3 was found to share the strongest activity in B. thuringiensis in this study, and therefore we examined its characteristics in more detail. To perform transcriptional fusion analysis, we divided complex Phj3 into 7 different fragments: −251∼−98, −251∼−31, −251∼+14, −113∼−31, −54∼+14, −54∼+118 and −6∼+118 (Figure 5A).
Figure 5. Characteristics of the structure and activity of complex P hj3.
(A) Nucleotide sequence and structure characteristics of complex Phj3. The consensus sequences recognized by σ-factor (σA and σF) and the corresponding TSSs, as well as the RBS are highlighted in different colors. (B) β-galactosidase specific activities directed by the fragments −251∼−98, −251∼−31, −251∼+14, −113∼−31, −54∼+14, −54∼+118 and −6∼+118 of complex Phj3. (C) β-galactosidase specific activities directed by the fragments −251∼−98, −251∼−31, and −113∼−31 of complex Phj3. Data are averages of three independent experiments (error bars are SEM from mean values).
The fragments −251∼−31 and −54∼+14 contain the σA-like consensus sequences TTGAAA and TATTAT in the −35 elements, and TTGACA and TAACAT in the −10 elements (Figure 5A and Table S4). The fragment −113∼−31 has the σF-like consensus sequence (Figure 5A and Table S4). The results demonstrated that each of the three fragments (−251∼−31, −113∼−31, and −54∼+14) could act as an independent promoter (Figure 5B and Figure 5C). Among them, the activity of the promoter −113∼−31 was the weakest, and the activity of the promoter −54∼+14 was 14-fold higher than the promoter −251∼−98. Accordingly, the promoter −54∼+14 would be a major contributor to the promoter of complex Phj3 activity.
The two truncated promoters −251∼−31 and −54∼+14 appeared to have a second induction and exerted their activities throughout the life cycle similar to the full-length promoter. In addition, although the activity of the truncated promoter −113∼−31 was relatively low, it reached the maximum value after 10 h of growth, which was in agreement with the fact that the fragment −113∼−31 contains the σF-like consensus sequences (Figure 5A and 5C).
It is important to note that the β-galactosidase activity directed by fragment −54∼+118 was approximately nine times higher than fragment −54∼+14, but the fragment −6∼+118 did not share the promoter activity (Figure 5B). Accordingly, we hypothesized that the fragment −6∼+118 could play a certain additional regulatory role contributing to the production of β-galactosidase. To investigate this possibility, we examined the RNA secondary structure of the RNA transcript from +1∼+118 through Mfold [36]. Exhilaratingly, the RNA fragment +1∼+118 preferred to fold into a perfect stem-loop structure, and more importantly, the ribosome binding site (RBS) became accessible due to its localization on the loop (Figure S5A). Consequently, the secondary structure of this RNA fragment could be beneficial to both the stability and translation of its downstream mRNA. Similarly, the activity of the fragment −251∼−31 was higher than that of the fragment −251∼−98 (Figure 5C), and the fragment −98∼−31 did not share promoter activity (data not shown). A perfect stem-loop structure was also predicted in the secondary structure of the RNA transcript from −98∼−31 (Figure S5B). Accordingly, this stem-loop structure held by the fragment −98∼−31 could also be beneficial to mRNA stability.
Application of Complex Phj3
Application of the 5′-UTR DNA rigion from complex Phj3
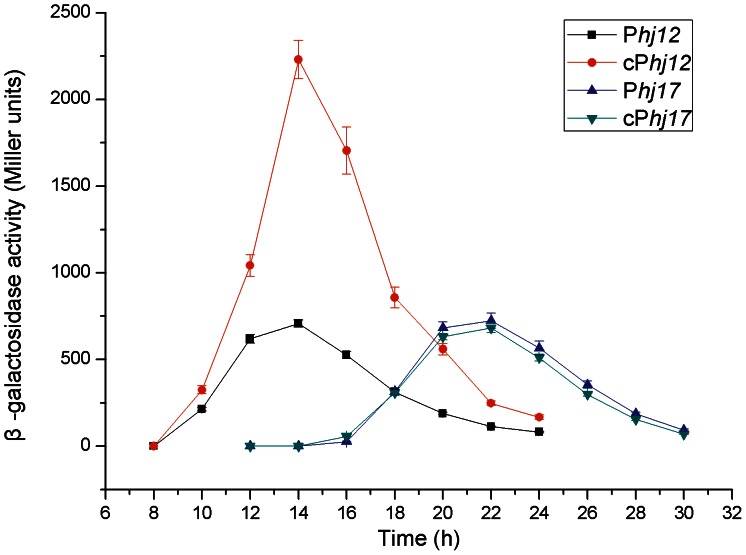
Because the 5′-UTR +1∼+118 transcripted from complex Phj3 would have some important roles in both the stability and translational facilitation of its downstream mRNA, we wondered whether or not this 5′-UTR could improve the gene expression levels directed by other weak promoters. Therefore, the DNA fragment +1∼+118 of complex Phj3 was fused to the 3′-ends of the promoters of Phj12 and Phj17 complexes (deleting their own 5′-UTR DNA rigions) to construct the chimeric complexes cPhj12 and cPhj17, respectively. As expected, the activity of the chimeric complex cPhj13 increased two to three times compared to the original Phj12 (Figure 6). Furthermore, the chimeric complex cPhj12 exhibited the same transcriptional feature of the original complex: initial detection starting at 10 h and reaching the maximum induction at 14 h of growth (Figure 6). Unexpectedly, the activity of the chimeric complex cPhj17 remained almost unchanged (Figure 6). These results imply that there exists some degree of context dependency between the 5′-UTR DNA region and its upstream promoter sequences.
Figure 6. Activity analyses of the chimera complexes cPhj12 and cPhj17.
The 5′-UTR DNA fragment +1∼+118 of complex Phj3 was fused at the 3′-end of the promoters of complexes Phj12 and Phj17 (deleting their own 5′-UTR DNA region) to construct the chimeric complexes cPhj12 and cPhj17, respectively. The chimeric complexes were separately fused with the gene lacZ and their activities were monitored by detecting the β-galactosidase specific activities. Data are averages of three independent experiments (error bars are SEM from mean values).
Overexpression of heterologous proteins directed by complex Phj3
To evaluate whether complex Phj3 could perform overexpression of heterologous proteins, different expression plasmids were constructed and transformed into the strain BMB171. Our results showed that the genes lacZ and turbo-rfp were successfully overexpressed with the active β-galactosidase (Figure 2A and Figure 7) and turbo-RFP (Figure 7 and Figure S6). In addition, complex Phj3 was successfully used to overexpress some endogenous genes from B. thuringiensis, including the genes that encode the response regulators of the two-component system as well as the diguanylate cyclases and phosphodiesterase of the c-di-GMP-mediated signal transduction system (unpublished data).
Figure 7. SDS-PAGE analyses of the β-galactosidase and turbo-RFP.
M: Marker; 1, the strain BMB171 containing pHT1K; 2, the strain BMB171 containing pHT1K-Phj3-lacZ; 3, the strain BMB171 containing pHT1K-Phj3-turbo-rfp. The recombinant BMB171 strains were grown in LB medium with 25 µg/mL erythromycin at 28°C for 22 h. The cultures were harvested by centrifugation and the crude proteins were extracted by boiling. The protein bands of β-galactosidase and turbo-RFP are marked by the arrows in the lanes 2 and 3, respectively.
Discussion
High-throughput Identification of Active Promoter Candidates
According to in silico prediction of the genome-wide operons (http://csbl1.bmb.uga.edu/OperonDB/), there are 4063 transcriptional units (TUs) in the genome of B. thurigiensis CT-43. In fact, only a part of TUs were transcribed under our experimental condition, simultaneously some transcribed mRNA were removed during the experimental process of RNA-seq, so the transcriptional percentages of the TUs encoded by the CT-43 chromosome were only 40.9%, 43.1%, 53.2%, and 17.7% for the four growth phases, respectively [27]. More importantly, TSSs were unable to be unambiguously determined owing to the relatively low signal-to-noise ratio for many genes with low transcriptional level. Based on the transcriptome data of B. thuringiensis CT-43 at four different growth phases, we manually determined the genome-wide TSSs and successfully identified 1203 active promoter candidates. Furthermore, we revealed their different temporal characteristics through the analyses of transcription strength at various phases coupled with secure binding sites for specific σ-factors. Therefore, from a methodological point of view, the strategy has obvious superiority on high-throughput identification of the temporally-activated promoters.
The putative binding sites for 11 different σ-factors were found in 723 active promoter candidates. The most frequently found σ-factor binding sites were those for the housekeeping σ-factor, SigA (17.4%) as well as the sporulation-specific σ-factors, SigH (15.8%), SigE (10.7%), SigG (9.3%), SigF (8.7%), and SigK (6.0%) (Table S3). These results reflect that a large number of genes are controlled by the spatially and temporally activated sporulation-specific σ-factors during sporulation [21]. In addition, these characteristics could have specific applications for gene expression research.
The 5′-UTRs of bacterial mRNAs are also known to play important regulatory roles in gene expression through extremely diverse mechanisms [9]–[17]. Among the 1203 TUs that the TSSs were mapped in this study, the length of most (52%) 5′-UTRs varied between 10 and 50 nucleotides (Table S3). In Helicobacter pylori, approximately 50% of the 5′-UTRs are 20–40 nucleotides in length [8], and the most frequent 5′-UTR length is also between 20 to 40 nucleotides in E. coli [6], whereas only 26.6% of the 5′-UTRs were 20–40 nucleotides in length in our data. In addition, very few 5′-UTRs are shorter than 20 nucleotides in E. coli [6], but 16.3% of the 5′-UTRs were shorter than 10 nucleotides in this study. These results might reflect the significant difference of 5′-UTR length in different species.
The Superiority of BMB171 as a Host Strain
The wild-type strain CT-43 holds ten plasmids with different sizes and its efficiency of transformation by electroporation is very low (103) [25], [37], therefore making genetic operation difficult. Fortunately, the acrystalliferous mutant BMB171 of B. thuringiensis YBT-1463 [32] possesses very high efficiency of electroporation transformation (1010) [37] and has been used as a host strain of genetic studies for a long time. Furthermore, the complete genomes of CT-43 and BMB171 have been sequenced by our laboratory [25], [32], and excellent collinearity exists in the two genomes (Figure S7). Consequently, all recombinant plasmids for the analyses of promoter-5′-UTR DNA region complex activities were transformed into strain BMB171.
Temporal Activation of the Promoter-5′-UTR DNA Region Complex
Our results explicitly reveal the directly corresponding relationship between the σ-factor-recognized consensus sequence and the complex activity feature. The great majority of the complexes acting throughout the life cycle possess the σA-like consensus sequences; some complexes that specifically exert their functions in early-stationary phase and mid-stationary phase have the σE-like and σK-like consensus sequences (Table S5), respectively. Our results indicate that 1) the fragment −113∼−31 of complex Phj3 containing the σF-like consensus sequence reached the maximum induction at 10 h (Figure 5C); 2) the promoters of complexes Phj10, Phj11, Phj12, and Phj14 share the σE-like consensus sequences, and therefore they all reached the maximum activities at approximately 14 h of growth (Figure 3); 3) the maximum activity of the σG-dependent complex Phj15 appeared at 16 h of growth (Figure 3); and 4) the promoters of complexes Phj17, Phj18, Phj19 and Phj20 have the σK-like consensus sequence, and thus they all began induction after approximately 16 h of growth and reached maximum activity at 22 h (Figure 4). These results are consistent with the temporally-activated processes of the sporulation-specific σ-factors SigF, SigE, SigG, and SigK in B. thuringiensis [21]–[23].
Regarding the complexes acting throughout the life cycle, Phj3 was confirmed to have the strongest activity, followed by Phj2 (Figure 2). The genes directed by complexes Phj3 and Phj2 in CT-43 encode the 50S ribosomal protein L21 RplU and the cold shock protein CspB2, respectively. It has been shown that bacterial cold shock proteins can function as mRNA chaperones and transcription antiterminators in response to the temperature downshift and other various stresses [38], [39]. Moreover, both RplU and CspB2 have been confirmed to be highly abundant proteins by our proteomics analysis using isobaric tags for relative and absolute quantitation (iTRAQ) technique (data not shown). Consequently, complexes Phj3 and Phj2 as well as their cognate genes rplU and cspB2 could play important regulatory roles in the process of translation and transcription.
The Application Prospect of the Promoter-5′-UTR DNA Region Complexes
In this study, we identified some important promoter-5′-UTR DNA region complexes that could exert their functions at specific growth phases with different activity levels. Therefore, these complexes would have different applications. For example, they could be used to investigate the gene functions in B. thuringiensis and other species of the B. cereus group. In this respect, the complexes specifically activated at certain growth phases have great significance, because the accuracy of temporal auto-induction could be superior to artificial induction. Thus, these types of complexes could be used to analyze the functions of a gene at different growth phases more precisely. In addition, the complexes with different activity levels could be used to reveal the effects of a gene on bacterial physiologic processes under its different expression levels.
More importantly, some bacilli (such as B. brevis, B. megaterium and B. subtilis) have been the most popular organism for heterologous protein production [40]. Bacilli have some general advantages, such as the lack of the endotoxin lipopolysaccharide, which is a pyrogenic factor in humans or other mammals, and the strong secretion capacity for the production of secreted enzymes [40], [41]. However, these strains also have some disadvantages leading to the poor stability of protein production, mainly because of two reasons: the very high protease activity and poor plasmid stability [42]. In contrast, some B. thuringiensis strains exhibit excellent plasmid compatibility and stability. For example, the strain CT-43 and YBT-1520 hold 10 and 11 plasmids with different size, respectively [25], [43]. Furthermore, the ICP proteins can be assembled into parasporal crystals, protecting the proteins from the proteolytic degradation. Meanwhile, the acrystalliferous mutant BMB171 of B. thuringiensis possesses some unique features, including high efficiency of electroporation transformation (1010) [37], excellent plasmid compatibility and stability, and clear genetic background [32]. Consequently, the strain BMB171 could be reformed to be a novel host strain for the expression of heterologous proteins.
An appropriate promoter-5′-UTR DNA region complex within a plasmid is very important regular element for the optimal overexpression of proteins. Our results confirmed that complex Phj3 could successfully promote expression of the active β-galactosidase and turbo-RFP with sufficiently high levels (Figure 2A, Figure 7 and Figure S6). Moreover, the high expression level of heterologous proteins did not significantly affect the growth features of the recombinant BMB171 strains (data not shown). Thus, Phj3 would be a proper promoter-5′-UTR DNA region complex for the overexpression of proteins in the strain BMB171.
In conclusion, the results of this study provide a substantial contribution to molecular biology research and biotechnological applications of B. thuringiensis, and our work has made the first step in developing a novel protein expression system in this regard.
Supporting Information
Flow chart of the construction of the translational fusion plasmid pHT1K-P hj1 - lacZ .
(TIF)
Flow chart of the construction of the expression plasmid pHT1K-P hj3 - turbo-rfp .
(TIF)
Visualization of TSS mapping for P hj1 -P hj20 . The number of unambiguously mapped reads per nucleotide was calculated and visualized by R and Origin version 8.0. The black, red, blue and dark cyan columns represent the mapped reads per nucleotide at 7 h, 9 h, 13 h and 22 h, respectively. The green and purple arrows represent the coordinates of a complex and the first downstream ORF, respectively.
(PDF)
Distribution of individual 5′-UTR length based on 1203 TSSs of mRNAs.
(TIF)
The predicted RNA secondary structures of the fragments +1∼ +118 (A) and −106∼ −31 (B) transcripted from Phj3. RNA secondary structures were predicted by Mfold (version 2.3) at 28°C based on global minimum free energy principle with no constrains [36], and visualized by VARNA.
(TIF)
Activity analysis of turbo-RFP. The strain BMB171 containing pHT1K (left) and pHT1K-Phj3-turbo-rfp (right) were inoculated on the same LB plate analysis detected in plate with 25 µg/mL erythromycin. The magenta bacterial colonies demonstrated that these bacterial cells produced the active turbo-RFP.
(TIF)
Nucleotide alignment obtained by the program MUMmer. Whole-genome sequence comparison was performed at the nucleotide level using the program MUMmer (http://mummer.sourceforge.net/) with default values, which relies on exact matches of at least 20 base pairs. Each dot in the figure is one such match. The red lines on the two main diagonals result from the high density of points with sequence identity along chromosome of the two Bacillus thuringiensis strains CT-43 and BMB171. The scattered points outside the main diagonals represent other short regions of sequence identity.
(TIF)
Bacterial strains and plasmids used in this study.
(DOC)
Primers used in this study.
(DOC)
Genome-wide TSS mapping and promoter identification.
(XLS)
Highly active complex candidates selected in this study.
(DOC)
Consensus sequences recognized by the σ-factor in the complex candidates.
(DOC)
Acknowledgments
We thank Scott Stibitz (Center for Biologics Evaluation and Research, Food and Drug Administration, Bethesda, Maryland, USA) for kindly providing the plasmid pRP1028 and Chinese National Human Genome Center at Shanghai (Shanghai, China) for the technical supports for the RNA-seq.
Funding Statement
This work was supported by the Chinese National Natural Science Funds (grants 31270105 and 30930004), the National Basic Research Program of China (973 Program, grant 2010CB126105), the National High-tech R&D Program of China (No. 2011AA10A205) and the Fundamental Research Funds for Central Universities of China (grant 2011PY092). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Haugen SP, Ross W, Gourse RL (2008) Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol 6: 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Browning DF, Busby SJ (2004) The regulation of bacterial transcription initiation. Nat Rev Microbiol 2: 57–65. [DOI] [PubMed] [Google Scholar]
- 3. Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, et al. (2006) rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell 125: 1069–1082. [DOI] [PubMed] [Google Scholar]
- 4. Mathew R, Chatterji D (2006) The evolving story of the omega subunit of bacterial RNA polymerase. Trends Microbiol 14: 450–455. [DOI] [PubMed] [Google Scholar]
- 5. McGrath PT, Lee H, Zhang L, Iniesta AA, Hottes AK, et al. (2007) High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat Biotechnol 25: 584–592. [DOI] [PubMed] [Google Scholar]
- 6. Mendoza-Vargas A, Olvera L, Olvera M, Grande R, Vega-Alvarado L, et al. (2009) Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli . PLoS One 4: e7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sorek R, Cossart P (2010) Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nature Rev Genet 11: 9–16. [DOI] [PubMed] [Google Scholar]
- 8. Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, et al. (2010) The primary transcriptome of the major human pathogen Helicobacter pylori . Nature 464: 250–255. [DOI] [PubMed] [Google Scholar]
- 9. Waters LS, Storz G (2009) Regulatory RNAs in bacteria. Cell 136: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gutiérrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E (2009) Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev 73: 36–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garst AD, Batey RT (2009) A switch in time: detailing the life of a riboswitch. Biochim Biophys Acta 1789: 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loh E, Memarpour F, Vaitkevicius K, Kallipolitis BH, Johansson J, et al. (2012) An unstructured 5′-coding region of the prfA mRNA is required for efficient translation. Nucleic Acids Res 40: 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Lay N, Gottesman S (2012) A complex network of small non-coding RNAs regulate motility in Escherichia coli . Mol Microbiol 86: 524–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lioliou E, Sharma CM, Caldelari I, Helfer AC, Fechter P, et al. (2012) Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet 8: e1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bongrand C, Sansonetti PJ, Parsot C (2012) Characterization of the promoter, MxiE box and 5′ UTR of genes controlled by the activity of the type III secretion apparatus in Shigella flexneri . PLoS One 7: e32862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berg L, Kucharova V, Bakke I, Valla S, Brautaset T (2012) Exploring the 5′-UTR DNA region as a target for optimizing recombinant gene expression from the strong and inducible Pm promoter in Escherichia coli . J Biotechnol 158: 224–230. [DOI] [PubMed] [Google Scholar]
- 17. Lale R, Berg L, Stüttgen F, Netzer R, Stafsnes M, et al. (2011) Continuous control of the flow in biochemical pathways through 5′ untranslated region sequence modifications in mRNA expressed from the broad-host-range promoter Pm. Appl Environ Microbiol 77: 2648–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aronson A (2002) Sporulation and delta-endotoxin synthesis by Bacillus thuringiensis . Cell Mol Life Sci 59: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanahuja G, Banakar R, Twyman RM, Capell T, Christou P (2011) Bacillus thuringiensis: a century of research, development and commercial applications. Plant Biotechnol J 9: 283–300. [DOI] [PubMed] [Google Scholar]
- 20. van Frankenhuyzen K (2009) Insecticidal activity of Bacillus thuringiensis crystal proteins. J Invertebr Pathol 101: 1–16. [DOI] [PubMed] [Google Scholar]
- 21. Higgins D, Dworkin J (2012) Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36: 131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abee T, Groot MN, Tempelaars M, Zwietering M, Moezelaar R, et al. (2011) Germination and outgrowth of spores of Bacillus cereus group members: diversity and role of germinant receptors. Food Microbiol 28: 199–208. [DOI] [PubMed] [Google Scholar]
- 23. Paredes-Sabja D, Setlow P, Sarker MR (2011) Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol 19: 85–94. [DOI] [PubMed] [Google Scholar]
- 24. Ibrahim MA, Griko N, Junker M, Lee A (2010) Bacillus thuringiensis: A genomics and proteomics perspective. Bioengineered Bugs 1: 31–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He J, Wang J, Yin W, Shao X, Zheng H, et al. (2011) Complete genome sequence of Bacillus thuringiensis subsp. chinensis strain CT-43. J Bacteriol 193: 3407–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nickerson KW, St Julian G, Bulla Jr LA (1974) Physiology of sporeforming bacteria associated with insects: radiorespirometric survey of carbohydrate metabolism in the 12 serotypes of Bacillus thuringiensis . Appl Microbiol 28: 129–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang JP, Mei H, Zheng C, Qian HL, Cui C, et al. (2013) The metabolic regulation of sporulation and parasporal crystal formation in Bacillus thuringiensis revealed by transcriptomics and proteomics. Mol Cell Proteomics doi: 10.1074/mcp.M112.023986. [DOI] [PMC free article] [PubMed]
- 28. Yoder-Himes DR, Chain PS, Zhu Y, Wurtzel O, Rubin EM, et al. (2009) Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc Natl Acad Sci USA 106: 3976–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sierro N, Makita Y, de Hoon MJL, Nakai K (2008) DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res 36: D93–D96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kang JN, Kim YS, Wang Y, Choi H, Li MS, et al. (2005) Construction of a high-efficiency shuttle vector containing the minimal replication origin of Bacillus thuringiensis . Int J Indust Entomol 11: 125–127. [Google Scholar]
- 31. Agaisse H, Lereclus D (1994) Expression in Bacillus subtilis of the Bacillus thuringiensis cryIIIA toxin gene is not dependent on a sporulation-specific sigma factor and is increased in a spo0A mutant. J Bacteriol 176: 4734–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He J, Shao X, Zheng H, Li M, Wang J, et al. (2010) Complete genome sequence of Bacillus thuringiensis mutant strain BMB171. J Bacteriol 192: 4074–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller JH (1972) Experiments in molecular genetics. NY: Cold Spring Harbor Laboratory. 352–355.
- 34. Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, et al. (2011) An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA 108: 2124–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, et al. (2009) Rfam: updates to the RNA families database. Nucleic acids res 37: D136–D140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peng DH, Luo Y, Guo S, Zeng H, Ju S, et al. (2009) Elaboration of an electroporation protocol for large plasmids and wild-type strains of Bacillus thuringiensis . J Appl Microbiol 106: 1849–1858. [DOI] [PubMed] [Google Scholar]
- 38. Phadtare S (2004) Recent developments in bacterial cold-shock response. Curr Issues Mol Biol 6: 125–136. [PubMed] [Google Scholar]
- 39. Sachs R, Max KE, Heinemann U, Balbach J (2012) RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution. RNA 18: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Terpe K (2006) Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol 72: 211–222. [DOI] [PubMed] [Google Scholar]
- 41. Bron S, Bolhuis A, Tjalsma H, Holsappel S, Venema G, et al. (1998) Protein secretion and possible roles for multiple signal peptidases for precursor processing in bacilli. J Biotechnol 64: 3–13. [DOI] [PubMed] [Google Scholar]
- 42. Wong SL (1995) Advances in the use of Bacillus subtilis for the expression and secretion of heterologous proteins. Curr Opin Biotechnol 6: 517–522. [DOI] [PubMed] [Google Scholar]
- 43. Zhong C, Peng D, Ye W, Chai L, Qi J, et al. (2011) Determination of plasmid copy number reveals the total plasmid DNA amount is greater than the chromosomal DNA amount in Bacillus thuringiensis YBT-1520. PLoS One 6: e16025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow chart of the construction of the translational fusion plasmid pHT1K-P hj1 - lacZ .
(TIF)
Flow chart of the construction of the expression plasmid pHT1K-P hj3 - turbo-rfp .
(TIF)
Visualization of TSS mapping for P hj1 -P hj20 . The number of unambiguously mapped reads per nucleotide was calculated and visualized by R and Origin version 8.0. The black, red, blue and dark cyan columns represent the mapped reads per nucleotide at 7 h, 9 h, 13 h and 22 h, respectively. The green and purple arrows represent the coordinates of a complex and the first downstream ORF, respectively.
(PDF)
Distribution of individual 5′-UTR length based on 1203 TSSs of mRNAs.
(TIF)
The predicted RNA secondary structures of the fragments +1∼ +118 (A) and −106∼ −31 (B) transcripted from Phj3. RNA secondary structures were predicted by Mfold (version 2.3) at 28°C based on global minimum free energy principle with no constrains [36], and visualized by VARNA.
(TIF)
Activity analysis of turbo-RFP. The strain BMB171 containing pHT1K (left) and pHT1K-Phj3-turbo-rfp (right) were inoculated on the same LB plate analysis detected in plate with 25 µg/mL erythromycin. The magenta bacterial colonies demonstrated that these bacterial cells produced the active turbo-RFP.
(TIF)
Nucleotide alignment obtained by the program MUMmer. Whole-genome sequence comparison was performed at the nucleotide level using the program MUMmer (http://mummer.sourceforge.net/) with default values, which relies on exact matches of at least 20 base pairs. Each dot in the figure is one such match. The red lines on the two main diagonals result from the high density of points with sequence identity along chromosome of the two Bacillus thuringiensis strains CT-43 and BMB171. The scattered points outside the main diagonals represent other short regions of sequence identity.
(TIF)
Bacterial strains and plasmids used in this study.
(DOC)
Primers used in this study.
(DOC)
Genome-wide TSS mapping and promoter identification.
(XLS)
Highly active complex candidates selected in this study.
(DOC)
Consensus sequences recognized by the σ-factor in the complex candidates.
(DOC)