Abstract
Understanding the mechanisms responsible for tube formation by endothelial cells (ECs) is of major interest and importance in medicine and tissue engineering. Endothelial cells of the human cell line EA.hy926 behave ambivalently when cultured on a random positioning machine (RPM) simulating microgravity. Some cells form tube-like three-dimensional (3D) aggregates, while other cells (AD) continue to grow adherently. Between the fifth and seventh day of culturing, the two types of cell growth achieve the greatest balance. We harvested ECs that grew either adherently or as 3D aggregates separately after five and seven days of incubation on the RPM, and applied gene array analysis and PCR techniques to investigate their gene expression profiles in comparison to ECs growing adherently under normal static 1 g laboratory conditions for equal periods of time. Using gene arrays, 1,625 differentially expressed genes were identified. A strong overrepresentation of transient expression differences was found in the five-day, RPM-treated samples, where the number of genes being differentially expressed in comparison to 1 g cells was highest as well as the degree of alteration regarding distinct genes. We found 27 genes whose levels of expression were changed at least 4-fold in RPM-treated cells as compared to 1 g controls. These genes code for signal transduction and angiogenic factors, cell adhesion, membrane transport proteins or enzymes involved in serine biosynthesis. Fifteen of them, with IL8 (interleukin 8) and VWF (von Willebrand factor) the most prominently affected, showed linkages to genes of another 20 proteins that are important in cell structure maintenance and angiogenesis and extended their network of interaction. Thus, the study reveals numerous genes, which mutually influence each other during initiation of 3D growth of endothelial cells.
Introduction
The inner surface of healthy blood vessels is lined with endothelial cells (ECs) that play an active role in phenomena such as transporting molecules, guiding cell migration, regulating blood pressure and coagulation [1]. In addition, ECs are very important in neoangiogenesis, which occurs in vivo during wound healing, placenta formation or tumor neovascularization [2], [3], [4]. In these cases, some endothelial cells of existing vessels start growing. A tip cell is selected and pushed forward by proliferating stalk cells to form a vessel wall [5]. Most of our knowledge about ECs comes from in vitro experiments with human umbilical vein endothelial cells (HUVEC) [6]. In addition, permanent cell lines are often used in angiogenesis research [7]. One of the most frequently used and best characterized permanent human vascular EC lines is EA.hy926, which was generated by fusion of HUVEC with the human lung carcinoma cell line A549 [8]. EA.hy926 cells have proven especially useful for studying the formation of new vessels [9].
When we cultured EA.hy926 cells on a random positioning machine (RPM), a device created to simulate microgravity on Earth, adherently growing cells as well as three-dimensional (3D) aggregates were observed [10], [11]. The adherently growing cells maintained a shape comparable to cells that were cultured under normal 1 g conditions, but possessed altered molecular features [12]. ECs forming 3D aggregates had detached from the bottom of the culture flasks [10]. The 3D aggregates were columnar and had a central lumen surrounded by one (single layered) or more layers of ECs [11]. They had never been detected under static 1 g conditions. Therefore, we concluded that annulling gravitational forces can trigger ECs to form tubes.
Microgravity affects several molecular features of ECs [13]. Even short-term cancellation of gravity (22s) generated by parabolic flights significantly influences signaling pathways [14]. After four and 12 hours of cultivation on the RPM, a number of proteins were up- or downregulated in comparison to control cells and apoptosis was enhanced [10]. During further incubation, apoptosis remained below 30% while the mRNA- and protein levels of several extracellular matrix components and growth-regulating factors changed. After two weeks, a very interesting subtype of 3D-aggregates was observed in the culture supernatants. Its central lumen was surrounded by one layer of ECs. These single-layered tubular structures (TS) resembled the intimas of blood vessels. Characterization of these TS revealed that they might originate from double-row cell assemblies formed between the fifth and seventh days of culture under simulated microgravity, while the percentage of apoptotic cells was about twice as high as in control cell populations at this time [12].
The formation of a blood vessel is accompanied by changes in transcriptional regulation in ECs [15], [16]. Under simulated microgravity, EC exhibit alterations in the expression of various genes and proteins including protein kinase A catalytic subunit, protein kinase C-alpha, and extracellular signal-regulated kinases 1 and 2 [12], [17], [18]. In order to extend our knowledge about changes in gene expression levels that occur when EA.hy926 cells are grown on the RPM simulating microgravity for either five or seven days as compared to control cells incubated under normal laboratory conditions, we applied microarray analysis and quantitative PCR techniques to search for differentially expressed transcripts in cells cultured on the RPM and those incubated in a normal laboratory incubator (1 g). Genes found to be changed more than 4-fold on the RPM were compared with genes known to be involved in angiogenesis but altered less than 4-fold in these experiments. The results obtained suggested that IL-8 is driving the 3D growth of ECs in a microgravity-dependent system via a network of genes and proteins involved in cell structure maintenance, while serine biosynthesis is enhanced.
Materials and Methods
Random positioning machine
Microgravity was simulated by a desktop RPM manufactured by Dutch Space, an EADS-Astrium company (Leiden, The Netherlands). Under the control of dedicated software, the RPM enables a random 3D positional change in the biological specimen [19]. The samples were mounted close to the center of the platform on the RPM, which was then placed in an incubator under standard cell culture conditions at 37°C and 5% CO2. The method was previously published in detail [10], [11], [12].
Cell culture procedure
Human endothelial EA.hy926 cells [8] were grown in Dulbecco's modified Eagle's medium (Invitrogen, Eggenstein, Germany) supplemented with 10% fetal bovine serum (Biochrom, Berlin, Germany), 100 units penicillin/ml, and 100 µg streptomycin/ml. The cells were grown in 25-cm2 culture flasks (Sarstedt, Nümbrecht, Germany) until subconfluent monolayers were formed. After reaching a subconfluency of 70%, the monolayers (106 cells/flask) were randomized into the following study groups: 120 static control cultures (1 g static ground controls are cultures kept in the same incubator as the RPM at 37°C); and 120 samples for the simulated microgravity experiments. All flasks were completely filled without air bubbles. RPM cultures were then incubated immediately on the desktop RPM and under static control conditions (1 g) according to the experimental design. Every 48 hours, the cell populations were fed by removing 50% of the cell medium with a pipette, carefully avoiding sucking off the floating 3D aggregates or damaging adherent cells, and adding new culture medium. On days five and seven the cells were harvested as previously described in detail [12].
Morphology of the three-dimensional structures
Immunohistochemical investigation was performed using monoclonal antibodies against laminin (Sigma-Aldrich Chemie, Taufkirchen, Germany). Antigen-antibody complexes were visualized by the streptavidin–biotin method [12], [20].
RNA isolation
Twenty cell culture flasks from each group were used for RNA extraction. The cells were scraped off using cell scrapers (Sarstedt, Nümbrecht, Germany), transferred to tubes and pelleted by centrifugation (2500×g, 10 min, 4°C). The RNeasy Mini Kit (Qiagen, Hilden, Germany) was used according to the manufacturer's instructions to isolate total RNA [12]. RNA concentrations and quality were determined spectrophotometrically at 260 nm using a NanoDrop instrument (Thermo Scientific, Wilmington, DE, USA). The isolated RNA had an A260/280 ratio of >1.5. cDNA designated for quantitative real-time PCR was then obtained with the First Strand cDNA Synthesis Kit (Fermentas, St. Leon-Rot, Germany) using 1 µg of total RNA in a 20-µl reverse transcription reaction mixture. The method was published previously [14], [21].
Gene-array technique
The endothelial cell line EA.hy926 was cultivated for five and seven days on the RPM and in parallel five and seven days under normal conditions (1 g static controls). During culture on the RPM, some adherent (AD) cells detached to form freely floating 3D aggregates (tubular structures, TS) in the culture medium. Both AD and TS were collected separately for RNA extraction. Each time, four independent RNA preparations from the resulting three different conditions were processed and hybridized using the Illumina HumanWG-6_V2_0_R3_11223189_A array. The resulting profiles had quantile normalization without background correction using the BeadStudio Gene Expression Module v3.3.7. After exclusion of low or unexpressed genes (maximum detection p-value >0.05), the arrays were checked for outliers in samples using principal component analysis (PCA), a correlation dispersion matrix and normalized Eigenvector scaling. No outliers were detected. Probes and samples were analyzed for significant expression differences using a two-way ANOVA approach differentiating between the conditions 1 g, AD and 3D on the one hand and the time points five days and seven days on the other. Multiple testing correction of the test statistic was performed using the Benjamini Hochberg FDR procedure [22]. Probes that underwent a 5% FDR between the three conditions, the time-course or its interaction and that exceeded a two-fold difference between any of the six conditions were selected as differentially expressed. Individual profile data were separated by k-means clustering using an average Euclidean distance function and k = 9 according to a local minimum in the Davies Bouldin k estimation [23]. Clustering was performed after normalization of probes to a mean of zero and a standard deviation of one. The nine resulting k-mean clusters were further investigated by functional enrichment using g:Profiler [24] with a simulation-based analytical threshold for significance estimation. Test statistics and clustering were performed using the Partek Genomic Suite version 6.4.
STRING analysis
Physical and functional interactions between proteins were determined using the STRING platform [25], [26] using a low confidence score of 0.15 for all but two exceptions. With respect to the high number of genes in the STRING analyses over combined clusters, proteins were preselected using a high confidence score (0.9). Resulting singletons were rejected before network visualization using a low confidence score of 0.15.
Quantitative real-time PCR
Quantitative real-time PCR [12], [14], [21] was used to determine the expression levels of the genes of interest. The Primer Express® software was utilized to design appropriate primers with a Tm of about 60°C (Table S1). The primers were synthesized by TIB Molbiol (Berlin, Germany). All assays were run on a 7500 Fast Real-Time PCR System using the Fast SYBR®Green PCR Master Mix (both Applied Biosystems, Darmstadt, Germany). The reaction volume was 25 µl, including 1 µl of template cDNA and a final primer concentration of 300 nM. PCR conditions were as follows: 20 s at 95°C, and then 40 cycles of 3 s at 95°C and 30 s at 60°C, followed by a melting curve analysis step (temperature gradient from 60°C to 95°C with +0.3°C per cycle). If all amplicons showed a single Tm similar to the one predicted by the Primer Express software, the PCR reactions were considered specific. Every sample was measured in triplicates and we utilized the comparative CT (DDCT) method for the relative quantification of transcription levels. 18S rRNA was used as a housekeeping gene to normalize our expression data.
Statistical analysis
Statistical analysis was performed using SPSS 16.0 (SPSS, Inc., Chicago, IL, USA). All data are expressed as means ± standard deviation (SD). Differences were considered significant at the level of p<0.05, employing the Mann-Whitney U-test.
Results
EA.hy926 cells were grown under normal laboratory conditions (60 culture flasks) and on the RPM (60 culture flasks) for five and seven days (n = 30/time point), respectively. After five days on the RPM, a considerable number of cells had detached from the bottoms of the culture flasks. Some of the detached cells had formed small, oblong three-dimensional aggregates that were laminin-positive (Fig. 1A). By this time point, a nearly confluent monolayer had developed in a normal laboratory incubator under 1 g gravity (Fig. 1B). Harvesting normal monolayer cultures (Fig. 1B), the cells were scraped off and collected. During the harvest of RPM cultures (Fig. 1A), aggregates and detached cells were collected together with the culture medium first. Then the remaining adherent cells were scraped off and collected. In this way, we obtained six populations of EA.hy926 cells (Table 1). Each cell population was divided into aliquots, of which one was used for microarray analysis and another one for PCR.
Figure 1. Immunocytochemistry.
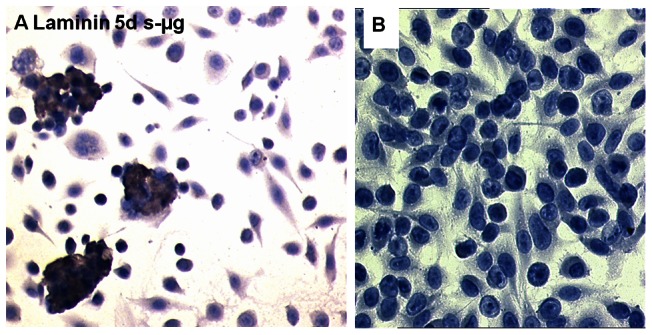
(A) Laminin-immunocytochemistry of EA.hy926 cells cultured for five days on the RPM. (B) Laminin-immunocytochemistry of EA.hy926 cells cultured for five days in a normal laboratory incubator.
Table 1. Time periods and conditions of culturing the various EA.hy926 cell populations.
| Time period | Condition | Kind of growth at harvest | Referred to as in text |
| 5 days | 1g | adherent | 5d 1g |
| 5 days | RPM | adherent | 5d AD |
| 5 days | RPM | 3D aggregates | 5d 3D |
| 7 days | 1g | adherent | 7d 1g |
| 7 days | RPM | adherent | 7d AD |
| 7 days | RPM | 3D aggregates | 7d 3D |
After harvest, each population was divided and aliquots were used for gene array technique or quantitative real-time PCR; 1 g: incubation in a normal laboratory incubator; RPM: incubation under simulated microgravity on the Random positioning machine; d: days.
Microarray analysis
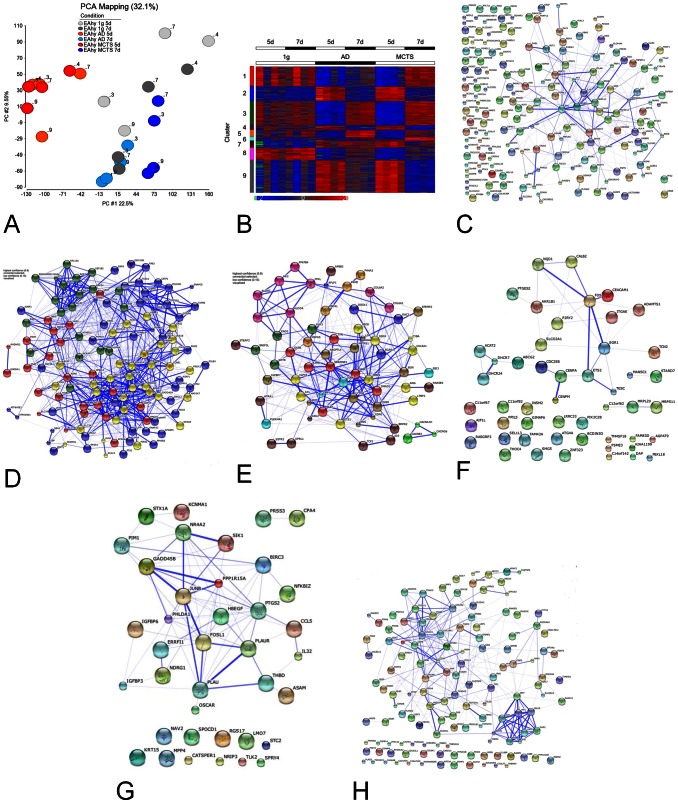
Using the Illumina HumanWG-6_V2_0_R3_11223189_A array was a first approach to finding genes that are expressed differently depending on the different gravitational conditions under which the cells had grown. Of 31,991 possible transcripts, 1,625 met the criteria of a False Discovery Rate (FDR) below 5% and an expression difference of at least two-fold between any of the tested conditions. These genes were defined as differentially expressed. Biological functions retrievable from the gene ontology database (GO) could be assigned to 1,174 products of the 1,625 differentially expressed transcripts. Several of these functions are related to vessel formation, others to apoptosis, cell structure maintenance, cell migration, and cell adhesion (Table S2). Obvious transient transcriptome differences in the five-day, RPM-treated samples were detected by principal component analysis (PCA) (Fig. 2A). Separating the 1,625 significantly differentially expressed individual profile data by k-means clustering resulted in 9 clusters (Fig. 2B).
Figure 2. Microarray Analysis.
(A) The first two PCs of the PCA of 31,991 expressed transcripts describe 32.1% of the dataset variance. The five-day RPM samples showed a separation from 1 g controls and seven-day RPM samples in PC #1 (22.5% of the variance). (B) K-mean clustering of the 1625 significantly regulated probes. The color scale represents upregulation (red) or downregulation (blue) of probes. Outliers are marked in orange (upregulated) or green (downregulated). (C–H) STRING visualization of known and predicted physical and functional interactions between the proteins organized in cluster 1 (C), clusters 2, 7 and 9 (D), clusters 3 and 5 (E), cluster 4 (F), cluster 6 (G) and cluster 8 (H).
Cluster 1
A widely unspecific set of 163 genes recognized by g:Profiler and 182 proteins recognized by STRING analysis are organized in cluster 1 and characterized by upregulation in seven-day 3D cells, a heterogeneous expression profile in the five-day and seven-day controls and downregulation in AD cells (Fig. 2B). Only a few GO biological processes were detected as significantly enriched: signal transduction (53 genes, GO:0007165, p = 6.04×10−6) and regulation of phosphorus metabolic processes (19 genes, GO:0051174, p = 3.98×10−6). In addition, we found single genes that are involved in angiogenesis (CALD1, PNPLA6, MAPK11, THY1), apoptosis (IL10, MAP3K11, NOX4, BIRC3), extracellular matrix organization (NFKB2) or actin cytoskeleton organization (INF2) (Fig. 2C).
Clusters 2, 7 and 9
These clusters represent genes that are upregulated in both AD and 3D cells by day five on the RPM (Fig. 2B). In all three clusters, general metabolic processes were significantly enriched; for example, GO:0044281 (small molecule metabolic processes) with p = 2.42×10−6 and 38 genes in cluster 2, and GO:0008152 (metabolic processes) with p = 4.03×10−6 and 35 genes with p = 2.90×10−8 and 177 genes in clusters 7 and 9, respectively (Fig. 2D). An increase in general expression activity is indicated by increases in expression of DNA-dependent RNA polymerase (POLR2L), of exportin (XPOT) (which mediates the nuclear export of aminoacylated tRNAs) of ribosomal proteins (RPL6, RPL9, RPL10a, RPL29, and RPLP0) and the tRNA synthetases for serine (SARS), methionine (MARS), tyrosine (YARS), isoleucine (IARS), and glycine (GARS). Moreover, especially in clusters 2 and 7, we found more genes involved in angiogenesis (HIF1A, IL8), apoptosis (SFN, BECL2L1, GLRX2, IL1A, IL6), extracellular matrix organization (MMP10), and cytoskeleton organization (TUBB6, ABLIM1).
Clusters 3 and 5
Clusters 3 and 5 are characterized by day-five downregulation in AD and 3D samples (Fig. 2B). The enrichment analysis indicated that these clusters are characterized by genes involved in anatomical structure development (GO:0048856, 79 genes, p = 2.00×10−9 in cluster 3 and 29 genes, p = 6.98×10−7 in cluster 5) as well as in cell-cell and cell-environment interactions (GO:0050896: response to stimulus, 121 genes, p = 1.61×10−8; GO:0007155: cell adhesion, 31 genes, p = 3.23×10−8; GO:0007154: cell communication 91 genes, p = 5.51×10−9, all cluster 3). Cluster 3 includes one half of the regulated collagens (COL13A1, COL4A5, COL5A1, COL6A1, and COL6A2), and both clusters include a multitude of angiogenic genes (for example, ENG in cluster 3 and CAV1 and CAV2, SERPINE1 and MMP2 in cluster 5) (Fig. 2E).
Cluster 4
Cluster 4 is characterized by an upregulation of gene expression by day five and a downregulation by day seven in AD and 3D samples cultured on the RPM (Fig. 2B). No significant enrichment of a GO biological process was found, but single genes involved in angiogenesis (CEACAM1), apoptosis (DAP, DHCR24, FOS, PSME3), and adhesion (ITGAE) belong to this cluster (Fig. 2F).
Cluster 6
Cluster 6 includes genes upregulated in 3D cells by day seven on the RPM (Fig. 2B). The most significantly enriched biological process is programmed cell death (GO:0016256, 14 genes, p = 7.78×10−7). Some of the other processes comprising a relevant number of genes include cell communication (GO:0007154, 21 genes, p = 7.55×10−6) and locomotion (GO:0040011, 12 genes, p = 3.81×10−7) (Fig. 2G).
Cluster 8
The genes in cluster 8 are generally downregulated (with an emphasis on day five) in AD and 3D samples and have some similarities in their expression patterns to genes from clusters 3 and 5 (Fig. 2B). Again, anatomical structure development (GO:0048856, 44 genes, p = 1.74×10−6) and angiogenic processes (GO:0001525, 11 genes, p = 2.79×10−6) are significantly enriched, including such genes as COL4A1, COL4A2, COL5A2, COL18A1, EGFL7, EPHB4, NOTCH1, PTPRB, ROBO4, SOX18, VASH1, and ADAM15 (Fig. 2G). In addition, genes regulating cell adhesion (GO:0007155, 22 genes, p = 1.81×10−8) and the extracellular matrix (GO:0031012, 17 genes, p = 2.98×10−10) are also prominently represented (Fig. 2H).
Quantification of differences
Looking at each individual cluster (Fig. 2B), no major differences between five- and seven-day samples of the 1 g control groups were observable. In contrast and corresponding to the PCA results (Fig. 2A), we could identify a strong overrepresentation of transient expression differences in the five-day, RPM-treated samples, where both AD and 3D samples showed a similar pattern (Fig. 2B). The similarity of the gene profiles of AD and 3D cells in five-day RPM treated samples is also notable because the expression of only two genes (IFIT1, CTGF) differs by a factor of two, while regarding the seven-day RPM treated samples, 3 genes (NFKBIZ, NR4A2, FOS) in 3D cells are more than three-fold upregulated as compared to the corresponding AD cells, and the expression of another 30 genes including MMP2, MMP9, and MMP10 as well as FOSL, PRDX3 and TNRSF25 in the 3D and AD cells differs by a factor of two (Table S2).
The overrepresentation of transient expression differences in the five-day, RPM-treated samples (Fig. 2B) relates not only to the number of genes being differentially expressed in comparison to 1 g cells, but also to the degree of change regarding distinct genes. Most noticeable is the IL8 gene. After 5 days on the RPM, the expression of this gene is 6-fold higher in 3D cells and 4-fold higher in AD cells as compared to 1 g cells. Further genes strongly affected by incubating the cells on the RPM are summarized in Table 2. It clearly shows that more genes exhibited greater than two-fold changes in expression levels in five-day RPM-treated cells than in seven-day RPM treated cells as compared to 1 g cells, respectively. Most of the genes showing large differences in expression code for signal transduction factors (ASAP3, IFIT1, CBS, GNG10, STC1). But major changes in genes for angiogenic factors (ANGPTL4, IL-8, HMOX1, LOX), cell adhesion (VWF, SPP1, ITGB4) and membrane transport (SLCO4A1, SLC2A1, SLC3A2) proteins also became obvious (Table 2). Interestingly, two genes were observed which code for enzymes involved in transforming pyruvate to serine (PHGDH, PSAT1).
Table 2. Number of genes, whose expression was changed more than twofold during incubation on the RPM.
| Fold change | 5d AD vs. 1g | 5d 3D vs. 1g | 7d AD vs. 1g | 7d 3D vs. 1g |
| 6 fold | – | 11 | – | 12 |
| 5 fold | 43 | 74 | – | 25 |
| 4 fold | 136 | 117 | 18 | – |
| 3 fold | 28 | 42 | 4 | 8 |
| 2 fold | 264 | 346 | 33 | 51 |
IL-8 (+); 2 VWF (−); 3SERPINE2 (+), SLCO4A1 (−), PHGDH (+), SEMA4B (−); 4 ANGPTL4 (−), SPP1 (+), SERPINE2 (+), SLCO4A1 (−), TSC22D1 (+), MMP10(+), PHGDH (+); 5 IL-8 (+); MMP10(+) 6 ANGPTL4 (−), HMOX1 (+), IL-8 (+), MX1 (−), TXNIP (−), ITGB4 (−), ASAP3 (−), SPP1 (+), IFIT1(−), ZNF467 (−), IFI44L (−), PSAT1 (+), PTPRR (+); 7HMOX1 (+), MX1 (−), TXNIP(−), ITGB4 (−), CBS (+), GNG10 (+), STC1 (+), SLC2A1 (−), SLC3A2 (+), SEMA4B (−), PSAT1 (+); 8 LOX (+); + = up-regulation; – = down-regulation.
Quantitative real-time PCR of selected transcripts involved in the process of angiogenesis
In addition to genes showing outstanding degrees of alteration in expression, genes coding for proteins that had been recognized earlier to be involved in cell aggregation on the RPM [27], [28] were detected in the microarray analysis (Table S2). The list of these genes included TUBB6, TGM2, SPATAN1, GSN, SERPINE1, ANXA2, CALD1, CAV1, SPAG9, PECAM1, ENG, IL-6 and IL8. We added the ICAM1, ITGB1, MSN, RDX, TLN1, VIL2, and VIM genes, although they did not emerge in the microarray, because their products had already attracted our attention earlier [26]. Then we tested the expression of all the genes mentioned above using quantitative real-time PCR (qPCR) (Table 3). We could not detect a significant transcriptional regulation for ANXA2, CAV1, ENG, and SPTAN1, although in most cases (with ENG as the most prominent example) their general expression patterns matched those observed in the microarray analysis (Table S2). The changes in expression determined by qPCR did not correlate to those from the microarray experiments for TGM2 and CALD1; in these cases, qPCR indicated an increase in both AD and 3D samples after five days, whereas a decrease was observed in the microarray analysis (Table 3). A correlation between the microarray and qPCR experiments, however, was seen for TUBB6, SPAG9, IL6, and IL8 after five days and for SERPINE1, PECAM1, IL6, and IL8 after 7 days in AD and 3D cells. In addition, qPCR revealed significant up-regulation for ITGB1, MSN, RDX, and VIL2 genes after 5 days.
Table 3. Comparison of gene array results and the relative quantities of mRNA determined by qPCR.
| 5d change | 7d change | ||||
| Gene symbol | Protein name | 3D vs. 1g FC Microarray/qPCR | AD vs. 1g FC Microarray/qPCR | 3D vs. 1g FC Microarray/qPCR | AD vs. 1g FC Microarray/qPCR |
| TUBB6 | Tubulin beta-6 chain | 2.02/162:100* | 2.07/182:100* | 1.12/135:100* | 1.24/105:100 |
| TGM2 | Protein-glutamine-γ-glutamyltransferase | −1.50/210:100* | −1.56/147:100 | 1.31/92:100 | 1.23/104:100 |
| SPTAN1 | Spectrin alpha chain, brain | −2.21/95:100 | −2.03/87:100 | −1.43/72:100 | −1.27/74:100 |
| GSN | Gelsolin | −1.40/123:100 | −1.45/111:100 | −1.89/91:100 | −2.36/54:100* |
| SERPINE1 | Plasminogen activatorinhibitor 1 | −1.47/63:100 | 1.08/96:100 | 1.52/245:100* | 1.46/187:100* |
| ANXA2 | Annexin A2 | 2.84/75:100 | 2.55/106:100 | −1.09/117:100 | 1.48/116:100 |
| CALD1 | Caldesmon | −1.51/193:100* | −1.23/207:100* | 1.46/124:100 | −1.28/109:100 |
| CAV1 | Caveolin-1 | −1.46/75:100 | −1.19/93:100 | −1.54/75:100 | 1.11/98:100 |
| SPAG9 | C-Jun-amino-terminalkinase interacting protein 4 | 1.97/237:100* | 1.93/169:100 | 1.04/134:100 | 1.23/100:100 |
| PECAM1 | Platelet endothelial cell adhesion mol. | −1.8/63:100 | −1.7/65:100 | −1.86/48:100* | −1.34/45:100* |
| ENG | Endoglin | −2.6/69:100 | −2.2/64:100 | −1.16/95:100 | −1.06/107:100 |
| IL6 | Interleukin-6 | 2.37/346:100* | 1.91/354:100* | 2.32/728:100* | 1.34/197:100 |
| IL8 | Interleukin-8 | 6.18/1734:100* | 4.66/550:100* | 5.4/1123:100* | 3.36/564:100* |
| ICAM1 | Intercellular adhesionmolecule 1 | n.d./72:100 | n.d./79:100 | n.d./131:100 | n.d./54:100* |
| ITGB1 | Integrin beta-1 | n.d./250:100* | n.d./207:100* | n.d./136:100 | n.d./150:100 |
| MSN | Moesin | n.d./175:100* | n.d./149:100 | n.d./104:100 | n.d./79:100 |
| RDX | Radixin | n.d./197:100* | n.d./171:100 | n.d./87:100 | n.d./90:100 |
| TLN1 | Talin-1 | n.d./157:100 | n.d./193:100* | n.d./143:100 | n.d./97:100 |
| VIL2 | Ezrin | n.d./180:100* | n.d./231:100* | n.d./117:100 | n.d./127:100 |
| VIM | Vimentin | n.d./150:100 | n.d./183:100 | n.d./129:100 | n.d./128:100 |
significant changes; n.d. not detected.
Interactions of observed genes
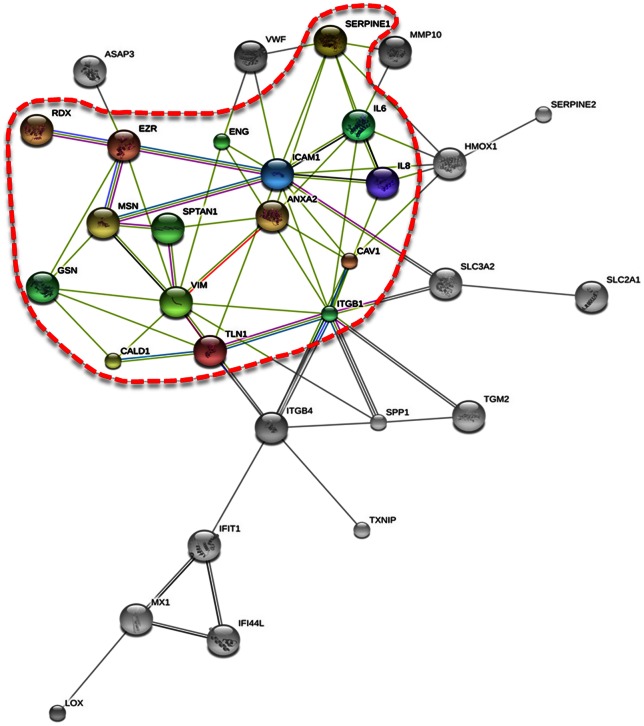
After a large number of genes were determined to be up- or downregulated when ECs were incubated on the RPM, it was of interest to see whether there are interactions between them. Application of STRING analysis revealed that the corresponding proteins of genes playing a role in 3D aggregation and shown in Table 3 form an interacting network, with the exception of TUBB6 and SPAG9 (Fig. 3, surrounded by a red line). Proteins of genes whose level of interaction changed at least 4-fold (Table 2) did not show significant interaction by themselves. Combining both groups, however, revealed that 15 of the 27 genes indicated in Table 2 clearly fitted into and extended the network formed by the earlier group indicated in Table 3 (Fig. 3, whole graph). These results suggest that an entire system of genes whose products are involved in maintaining cell structure and contact is changed when ECs start to form tubes on the RPM.
Figure 3. Interaction Analysis.
STRING visualization of interactions between the proteins of genes playing a role in 3D aggregation and shown in Table 3 (surrounded by a red line) and genes whose level of interaction was changed at least 4-fold (Table 2). IL-8 belongs to both groups.
Discussion
Looking for genes involved in the formation of 3D aggregates formed by EA.hy926 endothelial cells (Fig. 1), the genomic approach described in this paper was applied to search for differentially expressed transcripts in cells cultured on the RPM as compared to those incubated in a normal laboratory incubator (1 g). In accordance with the microscopic observation that oblong 3D aggregates, which we consider as precursors of the single-layered tubular structures observed after two weeks of incubation on the RPM [12], appear in cell cultures incubated on the RPM for 5 days (Fig. 1), we found a strong overrepresentation of transient expression differences in the five-day, RPM-treated samples. At that time, when the early stages of tubular structures appear, some clusters of genes were down-regulated, while others were up-regulated (Fig. 2B). However, amongst the 1,625 genes affected by annulling gravity, there were only 27 genes whose level of expression was up- or down-regulated at least 4-fold (Table 2). Fifteen of them, including IL8 and VWF as the most prominently affected, showed linkages to genes known to be involved in angiogenesis (Fig. 3), while two other genes are important for serine biosynthesis (Table 2).
Evaluating the results of our experiments, we focused on two groups of genes: those whose expression was dramatically changed (>4-fold, Table 2) and those that are known to be involved in angiogenesis but were up- or down-regulated less than 4-fold (Table 3). In comparison to EC growing under normal 1 g conditions, the VWF gene was 6-fold down-regulated in 3D aggregates after 7 days of culturing on the RPM. Von Willebrand factor is a protein important in hemostasis and angiogenesis [29]. Its down-regulation during formation of vascular-like networks by HUVEC cells encapsulated in hydrogel was already reported [30]. A 6-fold up-regulation was observed for the IL8 gene, while the IL6 gene was 2–3 fold up-regulated (Tables 2, 3). IL-8 enhances endothelial cell proliferation, induces capillary tube organization and up-regulates the expression of anti-apoptotic genes [31]. IL-6 also induces proliferation, migration and tube formation in various types of endothelial cells [32], [33]. In our experiments, up-regulation of the IL6 and IL8 genes was most prominent in 3D aggregates after 5 days of culturing on the RPM (Table 3). In human ovarian cancer cells, IL8 is up-regulated together with IL6 by the nuclear factor-kappa B (NFκB) pathway [34]. Thus, our findings further support the idea that NFκB may play a special role when human cells start to form 3D aggregates under simulated microgravity [35].
According to our STRING analysis, IL-6 and IL-8 interact together with the HMOX1, ICAM1 and ITGB1 proteins, which are nodal points in a network of interaction of the detected genes (Fig. 3). The HMOX1 gene was up-regulated more than 4-fold after 5 days of incubation on the RPM and less than 2-fold after 7 days (Table S2). It encodes heme-oxygenase-1, which facilitates angiogenesis [36]. The intercellular adhesion molecule-1 (ICAM-1) is an endothelial-associated transmembrane protein. It is important in stabilizing cell-cell interactions and facilitating leukocyte endothelial transmigration [37]. In accordance with a previous study [12], we did not see significant regulation of the ICAM-1 gene. ICAM-1 has been shown to interact with ezrin and moesin [38], whose genes (VIL2, MSN) were significantly upregulated in AD cells and 3D aggregates after 5 days. Ezrin and moesin belong to the ERM protein family, which also includes radixin. These proteins have the ability to interact with both the plasma membrane and filamentous actin [39]. Moesin is required in ECs for in vivo tubulogenesis [40]. Its regulation resembles TUBB6 gene regulation (Table 3). MSN extends the network of interaction via GSN, VIM and SPTAN1 (Fig. 3). GSN codes for the actin-binding protein gelsolin, which has multiple cellular functions including apoptosis and is reported to be down-regulated in various cancers and premalignant lesions [41]. The SPTAN1 gene encodes another cytoskeletal protein known as spectrin, the non-erythroid alpha chain [42]. Both genes showed a tendency of being down-regulated when the ECs were incubated on the RPM, while VIM gene expression was not significantly changed on the RPM (Table 3).
When we investigated the ITGB1 gene by qPCR, significant upregulation was observed, although it was not detected in the gene array analysis (Table 3). The increase in mRNA levels was substantial after five days, but only slightly elevated after seven days of culturing on the RPM (Table 3). In vivo, the cytoplasmic tail of the β-integrin subunit is coupled to F-actin by talin. This link is required to transmit force from the actin cytoskeleton to the extracellular matrix [43]. ECs lacking talin 1 in vivo are unable to undergo the cell spreading and flattening required for vessel formation [44]. We found that TLN1 gene expression was significantly elevated on day five in AD cells of RPM cultures, but not on day seven (Table 3). At the same time, SPP1, another interaction partner of ITGB1, was 4-fold up-regulated, while ITGB4 was 4-fold down-regulated as measured in the gene array analysis. Integrin beta 4 chains are involved in neoangiogenesis just like beta 1 chains. SPP1 codes for osteopontin, a ligand of integrins [45]. Enhancement of cellular osteopontin has been observed in ECs in vitro when gravity was reduced during incubation [10] and in vivo during vascular remodelling [46]. These findings suggest that IL-8 and IL-6 may trigger a cascade of reactions of cytoskeleton related genes and proteins in the early phases of tube formation.
According to our STRING analysis, the cascade of possible mutual interactions that leads to tube formation includes further nodes (Fig. 3). Endoglin (ENG) is found on the surface of cells and is important for angiogenesis, as it regulates endothelial signaling and function during blood vessel development [47], [48]. Caveolin-1 (CAV1) supports the assembly of caveolae, which are the sites in the cell membrane responsible for concentrating an array of signaling molecules critical for endothelial cell function [49], [50]. Serpine peptidase inhibitor-1 (SERPINE1) interacts with extracellular matrix proteins as well as with transmembrane receptors and other links to the intracellular signaling machinery. Thus, it modulates cell migration, cell-matrix interactions, signaling pathways and angiogenesis [51]. We detected a non-significant down-regulation of the ENG gene on day five, whereas on day seven there was no change. Similarly, CAV1 remained unchanged on days five and seven in AD and exhibited a moderate down-regulation in 3D aggregates. As caveolin-1 is thought to play a role as a negative regulator of signal transduction [52], downregulation of CAV1 may indicate a contribution to tube formation. SERPINE1 (PAI-1) gene expression was significantly up-regulated on day seven in AD and 3D cultures, while on day five it was significantly down-regulated in 3D cultures and unaltered in AD. Earlier measurements of this factor's presence in culture supernatants indicated that less PAI-1 is secreted in the supernatants of RPM cultures than in control 1 g cultures [18]. The difference may be explained by a strong binding of PAI-1 to the cells on the RPM. PAI-1 promotes tumour angiogenesis by preventing excessive proteolysis [53]. Thus, its association with cells may better protect them against plasmin activity, which is promoted by Annexin A2 (ANXA2), whose gene expression remained unchanged in all groups. Considering that proteolytic digestion of spectrin [54] favours angiogenesis, while plasmin activity is reduced [53], the present results support the idea of a complicated regulation of proteolysis during the initial phase of tube formation, as already suspected in a previous publication [11].
A prominent role for balanced proteolysis in EC tube formation also became obvious when gene profiles of AD and 3D cells harvested after 7 days of incubation on the RPM were compared. At that time, when the initial phase of cell aggregation is over, 10% of the genes (i.e. MMP2, MMP9, MMP10), whose expression differed at least 2-fold between AD and 3D cells encode metalloproteinases, which are involved in remodeling the extracellular matrix of blood vessels [55]. In addition, the NFKBZI and NR4A2 genes were up-regulated 3-fold in 3D as compared to AD cells after 7 days of incubation on the RPM. The expression of these genes is important for maintaining IL-6 and IL-8 production, respectively [56], [57]. Hence, it appears reasonable that these interleukins are also important in the elongation of formed tubes.
Taken together, the genomic approach applied in this study proved that a network of genes responsible for maintaining cellular structures and contacts is modulated when endothelial cells transit from a two- to a three-dimensional type of growth. Similar results were obtained when we studied spheroid formation of thyroid cancer cells [26], [28]. But investigating the thyroid cells, we noticed several enzymes of glycolysis that were modulated when gravity was reduced during cell culturing [58]. In this study, the PHGDH and PSATI genes were noted to be upregulated 5- and 4-fold, respectively. These genes encode for phosphoglycerate dehydrogenase and phosphoserine aminotransferase, which are involved in metabolizing pyruvate to L-serine. To our best knowledge, a role of these enzymes in in vitro tube formation has not been described so far. But they are critical for neurogenesis [59] and enhance the aggressiveness of various types of cancer cells [60], [61].
Supporting Information
Primers used for quantitative real-time PCR.
(DOCX)
Biological functions retrievable in the gene ontology database (GO) could be assigned to 1174 of the 1625 differentially expressed transcripts.
(DOCX)
Acknowledgments
The authors would like to thank Mrs. Heidi Schou Knudsen for her excellent technical support. We would also like to thank the team of PRS & EJE Hertfordshire, UK, for academic proofreading of the manuscript.
Funding Statement
This study was supported by the European Space Agency (ESA-CORA-GBF-2010–201, ESTEC contract number 4000102118, Acronym TUBES), and the German Space Agency DLR (DLR/BMWi grant 50WB1124). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cines DB, Pollak ES, Buck CA, Loscalzo J, Zimmerman GA, et al. (1998) Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood 91: 3527–3561. [PubMed] [Google Scholar]
- 2. Arnold F, West DC (1991) Angiogenesis in wound healing. Pharmacol Ther 52: 407–422. [DOI] [PubMed] [Google Scholar]
- 3. Reynolds LP, Redmer DA (2001) Angiogenesis in the placenta. Biol Reprod 64: 1033–1040. [DOI] [PubMed] [Google Scholar]
- 4. Folkman J (1995) Angiogenesis in cancer, vascular, rheumatoid and other disease. Nature Med 1: 27–31. [DOI] [PubMed] [Google Scholar]
- 5. Herbert SP, Stainier DYR (2011) Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nature Rev Mol Cell Biol 12: 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jaffe EA, Nachman RL, Becker CG, Mimick CR (1973) Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J Clin Invest 52: 2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bouis D, Hospers GAP, Meijer C, Molema G, Mulder NH (2001) Endothelium in vitro: A review of human vascular endothelial cell lines for blood-vessel related research. Angiogenesis 4: 91–102. [DOI] [PubMed] [Google Scholar]
- 8. Edgell CJS, McDonald CC, Graham JB (1983) Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc Natl Acad Sci USA 80: 3734–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benndorf R, Boger RH, Ergun S, Steenpass A, Wieland T (2003) Angiotensin II type 2 receptor inhibits vascular endothelial growth factor-induced migration and in vitro tube formation of human endothelial cells. Circ Res 93: 438–447. [DOI] [PubMed] [Google Scholar]
- 10. Infanger M, Kossmehl P, Shakibaei M, Baatout S, Witzing A, et al. (2006) Induction of three-dimensional assembly and increase in apoptosis of human endothelial cells by simulated microgravity: Impact of vascular endothelial growth factor. Apoptosis 11: 749–764. [DOI] [PubMed] [Google Scholar]
- 11. Grimm D, Infanger M, Westphal K, Ulbrich C, Pietsch J, et al. (2009) A delayed type of three-dimensional growth of human endothelial cells under simulated weightlessness. Tissue Eng Part A 15: 2267–2275. [DOI] [PubMed] [Google Scholar]
- 12. Grimm D, Bauer J, Ulbrich C, Westphal K, Wehland M, et al. (2010) Different responsiveness of endothelial cells to vascular endothelial growth factor and basic fibroblast growth factor added to culture media under gravity and simulated microgravity. Tissue Eng Part A 16: 1559–1573. [DOI] [PubMed] [Google Scholar]
- 13. Pietsch J, Bauer J, Egli M, Infanger M, Wise P, et al. (2011) The effects of weightlessness on the human organism and mammalian cells. Curr Mol Med 11: 350–364. [DOI] [PubMed] [Google Scholar]
- 14. Grosse J, Wehland M, Pietsch J, Ma X, Ulbrich C, et al. (2012) Short-term weightlessness produced by parabolic flight maneuvers altered gene expression patterns in human endothelial cells. FASEB J 26: 639–655. [DOI] [PubMed] [Google Scholar]
- 15. Minami T, Aird WC (2005) Endothelial cell gene regulation. Trends Cardiovasc Med 15: 174–184. [DOI] [PubMed] [Google Scholar]
- 16. Bell SE, Mavila A, Salazar R, Bayless KJ, Kanagala S, et al. (2001) Differential gene expression during capillary morphogenesis in 3D collagen matrices: regulated expression of genes involved in basement membrane matrix assembly, cell cycle progression, cellular differentiation and G-protein signaling. J Cell Sci 114: 2755–2773. [DOI] [PubMed] [Google Scholar]
- 17. Kang CY, Zou L, Yuan M, Wang Y, Li TZ, et al. (2011) Impact of simulated microgravity on microvascular endothelial cell apoptosis. Eur J Appl Physiol 111: 2131–2138. [DOI] [PubMed] [Google Scholar]
- 18. Infanger M, Ulbrich C, Baatout S, Wehland M, Kreutz R, et al. (2007) Modeled gravitational unloading induced downregulation of endothelin-1 in human endothelial cells. J Cell Biochem 101: 1439–1455. [DOI] [PubMed] [Google Scholar]
- 19. Ulbrich C, Westphal K, Pietsch J, Winkler HD, Leder A, et al. (2010) Characterization of human chondrocytes exposed to simulated microgravity. Cell Physiol Biochem 25: 551–560. [DOI] [PubMed] [Google Scholar]
- 20. Grimm D, Jabusch HC, Kossmehl P, Huber M, Fredersdorf S, et al. (2002) Experimental diabetes and left ventricular hypertrophy: effects of beta-receptor blockade. Cardiovasc Pathol 11: 229–237. [DOI] [PubMed] [Google Scholar]
- 21. Ulbrich C, Pietsch J, Grosse J, Wehland M, Schulz H, et al. (2011) Differential gene regulation under altered gravity conditions in follicular thyroid cancer cells: Relationship between the extracellular matrix and the cytoskeleton. Cell Physiol Biochem 28: 185–198. [DOI] [PubMed] [Google Scholar]
- 22. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300. [Google Scholar]
- 23. Davies DL, Bouldin DW (1979) A clustering separation measure. IEEE Transactions on PAMI 1: 224–227. [PubMed] [Google Scholar]
- 24.Reimand J, Kull M, Peterson H, Hansen J, Vilo J (2007) g:Profiler – a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res 35(Web Server issue): W193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Snel B, Lehmann G, Bork P, Huynen MA (2000) STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res 28: 3442–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pietsch J, Riwaldt S, Bauer J, Sickmann A, Weber G, et al. (2013) Interaction of proteins identified in human thyroid cells. Int J Mol Sci 14: 1164–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ulbrich C, Westphal K, Baatout S, Wehland M, Bauer J, et al. (2008) Effects of basic fibroblast growth factor on endothelial cells under conditions of simulated microgravity. J Cell Biochem 104: 1324–1341. [DOI] [PubMed] [Google Scholar]
- 28. Pietsch J, Sickmann A, Weber G, Bauer J, Egli M, et al. (2011) A Proteomic approach to analyzing spheroid formation of two human thyroid cell lines cultured on a random positioning machine. Proteomics 11: 2095–2104. [DOI] [PubMed] [Google Scholar]
- 29. Lenting PJ, Casari C, Christophe OD, Denis CV (2012) von Willebrand factor: the old, the new and the unknown. J Thrombos Haemostas 10: 2428–2437. [DOI] [PubMed] [Google Scholar]
- 30. Kraehenbuehl TP, Ferreira LS, Zammaretti P, Hubbell JA, Langer R (2009) Cell-responsive hydrogel for encapsulation of vascular cells. Biomaterials 30: 4318–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li A, Dubey S, Varney ML, Dave BJ, Singh RK (2003) IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J Immunol 170: 3369–3376. [DOI] [PubMed] [Google Scholar]
- 32. Yao JS, Zhai W, Young WL, Yang GY (2006) Interleukin-6 triggers human cerebral endothelial cells proliferation and migration: the role for KDR and MMP-9. Biochem Biophys Res Com 342: 1396–1404. [DOI] [PubMed] [Google Scholar]
- 33. Fan Y, Ye J, Shen F, Zhu Y, Yeghiazarians Y, et al. (2008) Interleukin-6 stimulates circulating blood-derived endothelial progenitor cell angiogenesis in vitro . J Cereb Blood Flow Metab 28: 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu JS, Yang G, Thompson-Lanza JA, Glassman A, Hayes K, et al. (2004) Genetically defined model of human ovarian cancer. Cancer Res 64: 1655–1663. [DOI] [PubMed] [Google Scholar]
- 35. Grosse J, Wehland M, Pietsch J, Schulz H, Saar K, et al. (2012) Gravity-sensitive signaling drives 3-dimensional formation of multicellular thyroid cancer spheroids. FASEB J 26: 5124–5140. [DOI] [PubMed] [Google Scholar]
- 36. Dulak J, Deshane J, Jozkowicz A, Agarwal A (2008) Heme oxygenase-1 and carbon monoxide in vascular pathology. Circulation 117: 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lawson C, Wolf S (2009) ICAM-1 signaling in endothelial cells. Pharmacol Rep 61: 22–32. [DOI] [PubMed] [Google Scholar]
- 38. Barreiro O, Yanez-Mo M, Serrador JM, Montoya MC, Vicente-Manzanares M, et al. (2002) Dynamic interaction of VCAM-1 and ICAM-1 with moesin and ezrin in a novel endothelial docking structure for adherent leukocytes. J Cell Biol 157: 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neisch AL, Fehon RG (2011) Ezrin, Radixin and Moesin: key regulators of membrane-cortex interactions and signaling. Curr Opin Cell Biol 23: 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y, Kaiser MS, Larson JD, Nasevicius A, Clark KJ, et al. (2010) Moesin1 and Ve-cadherin are required in endothelial cells during in vivo tubulogenesis. Development 137: 3119–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cho JE, Il Park W, Kim DC, Kim HJ, Kim SW, et al. (2012) Down-regulation of gelsolin may play a role in the progression of inverted papilloma through an antiapoptotic mechanism. Am J Rhinol Allergy 26: 177–182. [DOI] [PubMed] [Google Scholar]
- 42. Mehboob S, Song YL, Witek M, Long F, Santarsuero BD, et al. (2010) Structure of the nonerythroid alpha-spectrin tetramerization site reveals differences between erythroid and nonerythroid spectrin tetramer formation. J Biol Chem 285: 14572–14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Critchley DR (2009) Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Ann Rev Biophys 38: 235–254. [DOI] [PubMed] [Google Scholar]
- 44. Monkley SJ, Kostourou V, Spence L, Petrich B, Coleman S, et al. (2011) Endothelial cell talin1 is essential for embryonic angiogenesis. Dev Biol 349: 494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schneider JG, Amend SR, Weilbaecher KN (2011) Integrins and bone metastasis: Integrating tumor cell and stromal interactions. Bone 48: 54–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Infanger M, Grosse J, Westphal K, Leder A, Ulbrich C, et al. (2008) Vascular endothelial growth factor induces extracellular matrix proteins and osteopontin in umbilical artery. Ann Vasc Surg 22: 273–284. [DOI] [PubMed] [Google Scholar]
- 47. Duff SE, Li CG, Garland JM, Kumar S (2003) CD105 is important for angiogenesis: evidence and potential applications. FASEB J 17: 984–992. [DOI] [PubMed] [Google Scholar]
- 48. Lee NY, Golzio C, Gatza CE, Sharma A, Katsanis N, et al. (2012) Endoglin regulates PI3-kinase/Akt trafficking and signaling to alter endothelial capillary stability during angiogenesis. Mol Biol Cell 23: 2412–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sowa G, Xie L, Xu L, Sessa WC (2008) Serine 23 and 36 phosphorylation of caveolin-2 is differentially regulated by targeting to lipid raft/caveolae and in mitotic endothelial cells. Biochemistry 47: 101–111. [DOI] [PubMed] [Google Scholar]
- 50. Minshall RD, Sessa WC, Stan RV, Anderson RGW, Malik AB (2003) Caveolin regulation of endothelial function. Am J Physiol 285: L1179–L1183. [DOI] [PubMed] [Google Scholar]
- 51. Balsara RD, Ploplis VA (2008) Plasminogen activator inhibitor-1: the double-edged sword in apoptosis. Thromb Haemost 100: 1029–1036. [PMC free article] [PubMed] [Google Scholar]
- 52. Labrecque L, Royal I, Surprenant DS, Patterson C, Gingras D, et al. (2003) Regulation of vascular endothelial growth factor receptor-2 activity by caveolin-1 and plasma membrane cholesterol. Mol Biol Cell 14: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bajou K, Masson V, Gerard RD, Schmitt PM, Albert V, et al. (2001) The plasminogen activator inhibitor PAI-1 controls in vivo tumour vascularization by interaction with proteases, not vitronectin: Implications for antiangiogenic strategies. J Cell Biol 152: 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ma H, Tochigi A, Shearer TR, Azuma M (2009) Calpain inhibitor SNJ-1945 attenuates events prior to angiogenesis in cultured human retinal endothelial cells. J Ocul Pharmacol Ther 25: 409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Turner NA, Warburton P, O'Regan DJ, Ball SG, Porter KE (2010) Modulatory effect of interleukin-1 alpha on expression of structural matrix proteins, MMPs and TIMPs in human cardiac myofibroblasts: Role of p38 MAP kinase. Matrix Biol 29: 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Matsuo S, Yamazaki S, Takeshige K, Muta S (2007) Crucial roles of bionding sites for NF-kappa B and C/EBPs in I kappa B-zeta-mediated transcriptional activation. Biochem J 405: 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Aherne CM, McMorrow J, Kane D, FitzGerald O, Mix KS, Murphy EP (2009) Identification of NR4A2 as a transcriptional activator of IL-8 expression in human inflammatory arthritis. Mol Immunol 46: 3345–3357. [DOI] [PubMed] [Google Scholar]
- 58. Pietsch J, Sickmann A, Weber G, Bauer J, Egli M, et al. (2012) Metabolic enzyme diversity in different human thyroid cell lines and their sensitivity to gravitational forces. Proteomics 12: 2539–2546. [DOI] [PubMed] [Google Scholar]
- 59. Kinoshita MO, Shinoda Y, Sakai K, Hashikawa T, Watanabe M, et al. (2009) Selective upregulation of 3-phosphoglycerate dehydrogenase (Phgdh) expression in adult subventricular zone neurogenic niche. Neurosci Letters 453: 21–26. [DOI] [PubMed] [Google Scholar]
- 60. Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, et al. (2011) Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature Genetics 43: 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vie N, Copois V, Bascoul-Mollevi C, Denis V, Bec N, et al.. (2008) Overexpression of phosphoserine aminotransferase PSATI stimulates cell growth and increases chemoresistance of colon cancer cells. Molecular Cancer 7: article number 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for quantitative real-time PCR.
(DOCX)
Biological functions retrievable in the gene ontology database (GO) could be assigned to 1174 of the 1625 differentially expressed transcripts.
(DOCX)