Abstract
We report how cell rheology measurements can be performed by monitoring the deformation of a cell in a microfluidic constriction, provided that friction and fluid leaks effects between the cell and the walls of the microchannels are correctly taken into account. Indeed, the mismatch between the rounded shapes of cells and the angular cross-section of standard microfluidic channels hampers efficient obstruction of the channel by an incoming cell. Moreover, friction forces between a cell and channels walls have never been characterized. Both effects impede a quantitative determination of forces experienced by cells in a constriction. Our study is based on a new microfluidic device composed of two successive constrictions, combined with optical interference microscopy measurements to characterize the contact zone between the cell and the walls of the channel. A cell squeezed in a first constriction obstructs most of the channel cross-section, which strongly limits leaks around cells. The rheological properties of the cell are subsequently probed during its entry in a second narrower constriction. The pressure force is determined from the pressure drop across the device, the cell velocity, and the width of the gutters formed between the cell and the corners of the channel. The additional friction force, which has never been analyzed for moving and constrained cells before, is found to involve both hydrodynamic lubrication and surface forces. This friction results in the existence of a threshold for moving the cells and leads to a non-linear behavior at low velocity. The friction force can nevertheless be assessed in the linear regime. Finally, an apparent viscosity of single cells can be estimated from a numerical prediction of the viscous dissipation induced by a small step in the channel. A preliminary application of our method yields an apparent loss modulus on the order of 100 Pa s for leukocytes THP-1 cells, in agreement with the literature data.
INTRODUCTION
Cell mechanical properties play an important role in the transit of circulating cells in blood microvasculature and are involved in various cardiovascular and immunological pathological disorders. However, in vivo investigation of microscopic events occurring in the microvasculature is a difficult task, especially for deep organs. Non-invasive imagery techniques (e.g., magnetic resonance imaging, tomography, nuclear and positron imagery, fibroscopy imagery) lack the spatial resolution necessary to analyze microscopic events, whereas optical microscopy requires surgical windows to reach organs of interest,1 restricting applications to humans. In this context, microfluidics has emerged in the last decade as a powerful tool to mimic blood microcirculation and probe the behavior of circulating cells in microenvironments.2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24 Indeed, microfluidic systems circumvent the lack of appropriate living models and provide simplified testing procedures of pharmacological therapeutic treatments.8, 9, 10, 21, 25, 26, 27, 28 However, if devices for qualitative probing of cells passage in a microfluidic constriction have fast developed, these approaches remain to be adapted into quantitative tests for circulating cell rheological parameters.
Physicists have developed various tools to measure the rheological properties of living cells. Micropipette experiments yielded the first quantitative data on cell rheology with erythrocytes29, 30, 31 and leukocytes.32, 33, 34, 35, 36, 37, 38 The principle is to suck a single cell into a narrow glass capillary at a controlled pressure and to monitor simultaneously the rate of deformation. More sophisticated techniques allowing direct measurement of forces during cell deformation have later been developed. These techniques are based on the mechanical micromanipulation of cells and involve different equipments, such as atomic force microscopes, microplates, or optical and magnetic traps.39 They correspond to microscopic versions of standard rheometers and allow stress relaxation, creep recovery or frequency sweep experiments. Different groups have used these setups to establish the existence of a universal viscoelastic behavior of living cells, which is characterized by dynamic moduli that follow a power law dependence with the excitation frequency. The main asset of these recent techniques is their accuracy. However, they have a low throughput and are difficult to implement into a microfluidic device to analyze non-adherent circulating cells. Conversely, the principle of micropipette experiments is well adapted to microfluidics technologies. The kinetics of cell shape deformation during entry into a microfluidic constriction has, for instance, already been used to infer qualitative information on cell compliance properties of leukocytes,5, 6, 9, 22, 23 erythrocytes,10, 27, 40, 41 or cancer cells.3, 42 However, performing a quantitative measurement of cell rheology with a microfluidic constriction is still challenging.
Microfluidic research is largely dominated by devices fabricated via soft lithography in polydimethylsiloxane (PDMS) due to fabrication simplicity, cost-efficiency, low cytotoxicity, and permeability to oxygen and carbon dioxide. Nevertheless, performing cell deformation measurements similar to the micropipette technique with standard PDMS microchannels of rectangular cross section presents some difficulties. Indeed, the rounded shape of the cells cannot accommodate the sharp corners of the channels, contrary to the case of pipettes of circular cross-section. The driving liquid may then leak along the edges of the channel. Whereas in micropipette experiments, the force acting on the cell is simply given by the pressure drop across the cell times the tube cross-section, the shear force acting on the cell is more difficult to estimate with a microfluidic constriction due to this additive flow. Although PDMS circular microchannels have been fabricated with the aim of imitating actual blood vessels,43, 44 the fabrication process is not adapted for controlling precisely the shape of complex circuitries. Moreover, developing systems involving rectangular channels presents specific advantages. Indeed, flat channel walls are well adapted to observe cell-surface interactions by optical microscopy during cell transit in a capillary. Efforts in modeling have been undertaken for the entry of a cell in a simple constriction,45, 46 but they omit leakage and friction effects. A microfluidic micropipette method was recently described to measure the cell cortical tension, but not the viscous properties, or loss modulus, of cells.47 Spatz et al.48 have also proposed a system with a cantilever-based PDMS force sensor incorporated into a flow cell chip. This device has the advantage to provide a direct measurement of the compression force that the cantilever exerts on the cell. Herein, we present a new solution to perform micropipette-like experiments with rectangular PDMS micro-channels. To circumvent the difficulty related to the leakage flow, we consider a pair of successive constrictions of increasing confinement. Single cells are navigated in the microfluidic circuitry without deformation and introduced in a first constriction channel designed to be geometrically clogged by the deformed cell. The following constriction of narrower gauge is subsequently used to perform the quantitative force vs. deformation experiment. Measurements of the steady-state velocity of cells flowing in a straight channel allowed us to show, for the first time to the best of our knowledge, that cell friction with walls is nonlinear with velocity and can have a significant effect in the entry of cells in constrictions. Interferometry imaging of the cell/wall interface can be used to calculate the local pressure drop pushing the cell in the presence of corner leaks. We finally show how our method permits in-line determination of the apparent loss modulus of circulating cells once friction and leakage flow effects have been taken into account. The method is suitable for cells with a wide range of deformability, which makes it a versatile tool to investigate cell stiffness in the context of pathologies.6
MATERIAL AND METHODS
Microfabrication
Microfluidic devices were fabricated using standard soft lithography routines.49 A positive mould was created with SU-8 2010 negative resin (Microchem, Newton, MA, USA) spin-coated on silicon wafers (Siltronix, Archamps, France) and insulated/aligned with a photolithography device (Süss Microtec, Munich, Germany). Replicas of the moulds were prepared in PDMS (Sylgard 184 Silicone Elastomer Kit, Dow Corning, Midland, USA). Ports to plug inlet and outlet reservoirs were punched with a 1 mm outer diameter needle and the devices were finalized by sealing the PDMS piece on a 170 μm thick glass coverslip via O2-plasma activation (Harricks plasma) of both surfaces. All channels were incubated with a 1% Pluronic F108 solution (BASF, Mount Olive, NJ, USA) for 2 h to deter cell adhesion.
Cell preparation
We used the human monocytic THP-1 cell line (TIB-202™, ATCC)50 maintained as previously described.51
Microscopy setup
Force vs. deformation measurements were carried at 37 °C with a Zeiss axiovert 200 inverted microscope (Carl Zeiss, Jena, Germany) equipped with a Zeiss Plan-Neofluar 100×/1.30 Oil Ph3 objective, and a camera BURLE (model TC65) for fluorescence and phase contrast video-microscopy at 24 frames/s. For reflection interference contrast microscopy (RICM), we used adjustable field and aperture stops, a crossed-polarisers Cube, an Antiflex EC Plan-Neofluar 63×/1.25 Oil Ph3 objective, a high-speed camera Fastcam SA4 (Photron, CA, USA), and an X-cite 120Q lamp (Exfo, Mississauga, Canada) coupled to a narrow bandpass filter (λ = 546 nm ± 12 nm). RICM measurements were performed with an illumination numerical aperture 1.25. We used μmanager52 to pilot the set-up and matlab to process images.
Reflection interference contrast microscopy modeling
Details on the model can be found in previous publication.53 We consider two reflecting surfaces, the glass/solution, and the solution/cell interfaces and calculate the reflected intensity as a function of the thickness of the film of water separating the cell from the glass substrate using refractive indices of n0 = 1.525 for the glass substrate, n1 = 1.334 for water, n = 1.45 for PDMS, and an effective value of 1.36 for the cell. Theoretical and experimental reflected intensities are normalized by the intensities and reflected at the glass/water and glass/PDMS interfaces
| (1) |
where the subscript index N stands for normalized, and the superscript index i = t,e stands, respectively, for theoretical or experimental. The theoretical curve allows us to convert the experimental intensities into a local thickness h of the interstitial film between the cell and the glass substrate.
EXPERIMENTAL DEVICE
Device rationale of the double constriction device
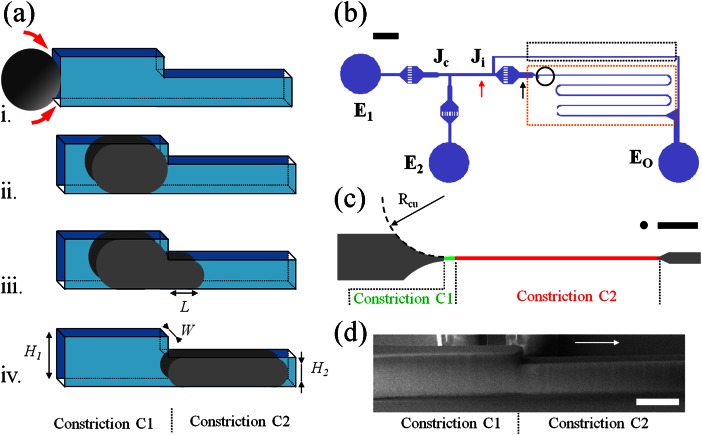
Since a cell inserted in a narrow rectangular channel cannot perfectly match the angular shape of the corners, the surrounding fluid may leak along the edges upon a pressure gradient (Figure 1a-i). The force acting on the cell thus results from a combination of pressure gradient and shear stress, which is difficult to model precisely. In our device, the cell is pushed in a first narrow channel, C1, designed in such a way that the deformed cell plugs the channel (Figure 1a-ii). Considering that C1 has a width W smaller than the cell diameter, its height H1 should be chosen so that the deformed cell touches all sides of channel C1. Considering volume conservation, a spherical cell of initial diameter Di squeezed in a slit of width W is roughly deformed into a “puck” of thickness W and diameter Dpu
| (2) |
C1 should, therefore, be designed with a height H1 smaller than Dpu in order to limit leakage around the puck. The force acting on a cell in C1 is thus dominated by pressure drop and can be determined as described in the section “Pressure force in the presence of gutters”. After the first constriction C1, a narrower channel C2 with the same width W but a smaller height H2 is used to perform the quantitative force vs. deformation measurement during the displacement of the cell from C1 to C2 (Figures 1a-iii, 1a-iv). Other advantages of this double constriction design are to provide a cell deformation that is 2 dimensional and localized (with a small difference between H1 and H2), which significantly simplifies the estimate of the rheological properties of the cell.
Figure 1.
Device rationale and fabrication. (a) Sketch representing the passage of a cell in a double constriction device. (i) Spherical cell blocked at the entrance of a rectangular constriction. Red arrows illustrate flow leaks around the cell in the channel corners. (ii) The cell is squeezed in constriction C1. The deformed cell fills the cross-section of the channel and plugs the passage of the fluid around the cell. (iii) Cell deformation during the penetration into constriction C2, L is the projection length of the cell in C2. (iv) End of the stage of cell entrance in constriction C2. (b) Design of the microfluidic device showing the inputs E1 and E2 for medium and cell suspensions and the output EO. The junction Ji divides the flow between the analysis circuit (red dashed box), and a bypass (black dashed box). Ji is used to select and isolate cells of interest. The dark circle localizes the portion of the device zoomed in (c). The scale bar corresponds to 500 μm. (c) Design of the double constriction device, showing the entrance and exit zones (grey), the first constriction C1 of cross-section W × H1 = 6 × 11 μm (green) and the second constriction C2 of cross-section W × H2 = 6 × 8.5 μm (red). The entrance channel leading to C1 has a radius of curvature Rcu of 150 μm. All channels except C2 have a targeted height of 17 μm. Theblack disk has a diameter of 14 μm like the cells used in the experiments. The scale bar corresponds to 100 μm. (d)SEMmicrograph of the resin mould of double constriction zone (the scale bar corresponds to 20 μm).
Setup used for single cell experiment
The circuit to select, to isolate, and to manipulate single cells (Figure 1b) has already been described by Gabriele et al.54 The microfluidic chip has two inputs: E1 is connected to a reservoir of pure cell culture medium R1, E2 is connected to a reservoir R2 containing the suspension of cells to analyze. An output EO is finally connected to a reservoir RO. The fluids injected in entries E1 and E2 converge at junction JC into a single channel up to junction Ji, where the flow is split between the analysis circuit, AC (red dashed box in Figure 1b), and a bypass (black dashed box in Figure 1b). The junction Ji is an important element of the device. Indeed, it is characterized by a highly anisotropic flow division in favor of the bypass route, which fulfils four functions: passive filtering of submicron particles from the AC, selection of single cells to introduce in the AC, finer precision control in the AC through a pressure division, and maintenance of a constant pressure drop ΔPAC across the AC with or without any cell in AC.
The device is built by superposing two layers of resins of thickness 8.5 μm on the whole design, except on C2 which is only coated with one layer of 8.5 μm. THP-1 cells of mean diameter 14 μm move freely with the flow in the whole device except in the constrictions, since the height of the ceiling is HD = 17 μm and the width of the channels is larger than 20 μm. This design insures that cells are not damaged or perturbed by interactions with channel walls before they enter the double constriction section. To manipulate cells, the macroscopic pressure drop across the device, ΔPext, is changed by a simple actuation of the height of reservoirs R1 relatively to R2 and RO on a centimeter range, which allows us to reach a precision of a few Pascal for the local pressure driving the cells. Cell selection and isolation are performed at junction Ji, and the force-deformation measurement is performed upon cell passage from constrictions C1 to C2.
The double constriction
To avoid excessive stress on cells before the force-deformation measurement, it is important to introduce the cells inside C1 as gently as possible. The geometry of the entrance is known to influence the entry time of the cell,55 a larger radius of curvature Rcu facilitating the penetration of the cells. The entrance channel toward C1 is, therefore, designed with a large curvature Rcu of 150 μm (Figure 1c). The typical diameter of the cells is Di = 14 μm, which leads to a puck diameter Dpu = 17.5 μm for channels of width W = 6 μm. In practice, the superposition of two resin layers on patterns of small area results into a cumulated thickness smaller than expected. The height of C1 is thus of 11 μm instead of twice 8.5 μm (Figure 1d). At the end of the process, C1 and C2 have the same width W = 6 μm, and heights of, respectively, H1 = 11 μm and H2 = 8.5 μm. The small height difference between C1 and C2 is important in the following, since it allows to assume small and localized deformations in the modeling of the force-deformation process.
RESULTS AND DISCUSSION
Cell entry in a rectangular constriction with reduced leakages around the cell
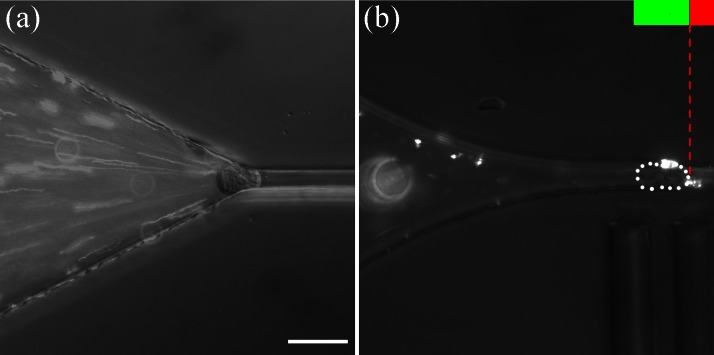
Figure 2 presents a sequence of micrographs of a THP-1 cell of diameter 17 μm entering successively C1 and C2 at a constant external applied pressure of ΔPext = 400 Pa, which corresponds to a pressure drop across the constriction of 160 Pa. Due to the large radius of curvature of the connection, the penetration of cells in C1 is significantly facilitated as compared to entries in constrictions with sharp funnel-shaped connections.5, 6 In order to visualize possible leaks around a cell in C1, we added in the fluid fluorescent nanoparticles of diameter 200 nm. We compare in Figure 3 the flow conditions in a “single constriction” device with funneled entry, which has been used in previous works,5, 6, 10 and in the new “double constriction” device. A force vs. deformation experiment begins when the cell has reached the constriction entrance after the converging neck of the funnel in the case of a “single constriction” device, and the entrance of C2 for the double constriction device. At this point, the flow around the cell is still important in the “single constriction” device, whereas it apparently vanishes in the “double constriction” device. The path followed by fluorescent particles while the image is captured is indeed much shorter in the second situation. This observation validates the double constriction design approach to limit leakage in channel corners.
Figure 2.
Cell force vs. deformation experiment in a double constriction. Series of micrographs showing THP-1 cells of diameter Di = 17 μm (a) flowing freely with the flow in the entrance zone of C1, (b) squeezed during entry in C1, (c) stopped in C1 at the entrance of C2, (d) and (e) transferring from C1 to C2, and (f) after complete entry in C2. The green and red bars in (a) indicate the position along the axis of the transition between constrictions C1 and C2, the black arrow in (a) shows the direction of the fluid flow in the whole experiment, and the yellow arrows point to the back and front edges of the cell. The scale bar corresponds to 20 μm (enhanced online).
Figure 3.
Reduction of flow leaks around a deformed cell in a double constriction. Micrographs showing the fluid flow, materialized by fluorescent nanoparticles, when a cell has just arrived at the entrance of (a) the constriction of a simple constriction device5 of size 4 × 16 μm, and (b) the constriction C2 of a double constriction device. The flow is stopped by the cell in (b) but not (a). White dashed lines underline the edges of the cell. The green and red bars in (b) indicate the position along the axis of the transition between constrictions C1 and C2. The scale bar corresponds to 10 μm.
In terms of orders of magnitude, a typical pressure drop measured across the constriction is ΔPAC = 160 Pa (ΔPext = 400 Pa), which leads to a force on the order of 10−8 N for a cell of 6 × 11 μm2 cross section that would perfectly clog the channel. The fluid velocity V leading to the same pressure in the case of a free channel would be on the order of 500 μm/s. The corresponding Stokes forces acting on the cell (3πηDiV) would thus be of order 10−10 N. This estimate explains why, with a single constriction device, stiff cells remain blocked for a long time at the entrance and then suddenly enter with a large speed. This sharp transition can be explained by the non-linear increase of the force acting on the cell upon cell deformation. Indeed, the driving force starts from an extremely low value when weak deformation of the cell leads to important leaks of the carrier fluid. However, this force increases by two orders of magnitude when the cell is deformed and plugs the channel. From our experience, a single constriction device is not appropriate to perform reproducible and quantitative measurements with stiff cells. Conversely, the double constriction device allows an accurate comparison of cell properties in a wide range of stiffness, which has allowed us to investigate pathologies involving the plugging of lung microvasculature by stiffened leukocytes.56 In the present work, we demonstrate how this device can also be used to infer quantitative rheological analysis on single cells.
Quantitative force vs. deformation experiment
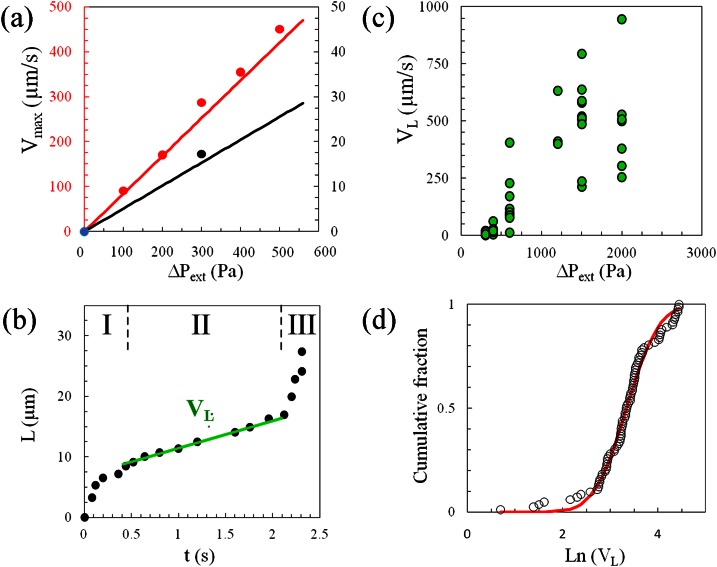
The quantitative measurement of the force acting on a cell entering C2 requires the absolute control of the local pressure in the device. Flow and pressure through the device can be deduced from a lumped element modeling of the microfluidic circuit applying Kirchhoff's and Ohm's laws, as classically done with electrical circuits. Each channel resistance is calculated using the solution of Stokes' law in a rectangular channel, whose dimensions are measured through optical profilometry. The details of the derivation for our circuitry are reported by Gabriele et al.54 In Figure 4a, we compare the predicted maximum fluid velocities Vmax in two positions of the channels with experimental data obtained from tracking the displacement of fluorescent nanoparticles. The good agreement (within 8%) between these data (without fitting parameter) validates our modeling of the local pressure drops in the circuit.
Figure 4.
Measurement with a double construction device. (a) Comparison of experimental (dots) and computed (solid line) of the maximum fluid velocities Vmax at two positions in the device, before the bifurcation Ji (red data) and in the analysis circuit (black data) at positions indicated by, respectively, red and black arrows in Fig. 1b. (b) Cell projection length in C2, L, versus time, t, of a THP-1 cell of diameter 14 μm at an applied pressure ΔPext = 400 Pa. Stages I, II, and III correspond, respectively, to the entry of the cell front in C2, the transfer of the cell body from constriction C1 to C2, and the final entry of the rear of the cell. The slope in stage II yields the velocity of the projection length, VL. (c) Velocity of the projection length in stage II, VL, against the external applied pressure ΔPext. (d) Cumulative fraction of cells vs. VL at ΔPext = 400 Pa of normal THP-1 cells (dots) and fit (black line) with a log-normal distribution in semi-log representation.
Concerning cell deformation, we first verified that the volume of the cell is constant as the cell penetrates into the constriction. For instance, the cell in Figure 2 has initially a spherical shape of diameter Di = 14 ± 0.5 μm and is transformed into a block of length L1 = 18 μm, height H1 = 11 μm, and width W = 6 μm in C1, and L2 = 22 μm and H2 = 8.5 μm in C2. The volume remains, therefore, within 1400 ± 150 μm3. The characterization of the deformation of a cell penetrating in C2 is based on the measurement of the extension of the cell in C2, L, as a function of time (Figure 4b). The curves L(t) present three stages. In the transient first and third stages, the fast increase of L corresponds to the respective entry of the leading and receding edges of the cell in the section C2. The intermediate regime corresponds to the penetration of the main body of the cell. In this stage, L(t) is linear, which allows us to define the corresponding steady velocity, VL. The steadiness of VL for a given cell indicates that the dissipation induced by the deformation of the cell is constant. VL is, therefore, a qualitative indicator of a cell rheological dissipation modulus and appears roughly proportional to the external applied pressure (Figure 4c). At a given pressure, the data obtained for a population of cells present a distribution that can be nicely fitted by a log-normal function (Figure 4d). A log-normal distribution is consistent with the literature data for other living cells, obtained with other rheometric techniques.39
This distribution provides a qualitative characterization of the deformability of a cell population. A careful analysis is nevertheless required to extract more quantitative insights. Indeed, the absolute force acting on a cell entering into C2 results not only from the pressure drop across the cell but also from the hydrodynamic friction with walls. Several parameters should thus be considered before estimating this force: (1) the pressure drop changes upstream and downstream of the cell as a function of the velocity VL, (2) the pressure drop dependency with residual flow along the corners of the channel, and (3) the thickness of the lubrication films squeezed between the cell and the channel walls. All the above issues involve complex and intricate hydrodynamic phenomena that will be presented successively in the following paragraphs.
Cell-channel interface: Lubrication film and “gutters” in channel corners
The thin glass wall on the microfluidic chip allows RICM to be implemented to the experimental setup. RICM observations of cells traveling in constriction C2 can be used to determine the topography of the cell membrane in the “contact zone” corresponding to the lower channel (Figure 5a). The darkest regions on the upper and lower parts of the pictures correspond to the glass/PDMS interface, whereas the bright stripe in between corresponds to the glass/water interface inside the channel. The “contact zone” between the cell and the glass surface appears as a dark region in the channel. The grey levels correspond to different distances between the cell and the substrate, darker zones corresponding to thinner lubrication films.
Figure 5.
Effect of the size of the cell and of the contact zone on the cell flow. (a) RICM imaging of the cell-wall interface of different THP-1 cells at the end of constriction C2 moving at a steady-state velocity showing: (i) a flat homogeneous contact zone without measurable gutters along the corners, (ii) a contact zone with gutters of width 1.5 μm along the corner, and (iii) an inhomogeneous contact zone. The scale bar corresponds to 10 μm. (b) Cell velocity in C2, VC2, measured 500 μm away from the entrance of C2, normalized by the mean fluid velocity in C2 in the absence of cells, VMF at the same applied pressure versus the initial diameter of the cell, Di, for different cells and applied pressures. (c) Cell-to-wall distance in the contact zone, h, estimated from RICM images of cells traveling in C2, versus the cell diameter, Di, for different cells and applied pressures. (d) Distance h versus cell velocity in C2, VC2, for cells of diameters limited to a range of 13–15 μm. Plots (b)–(d) correspond to pooled data at different ΔPext in the range of 300-2000 Pa (enhanced online).
Contact zones present different patterns from one cell to the other. In Figure 5a-i, the contact zone is homogeneous, indicating that the cell membrane is flat in the vicinity of the walls. The distance from the cell to the surface is on the order of 20 nm in this particular example, but typically ranges from 20 to 80 nm for different cells at different applied pressures. The bright ring around the contact zone indicates a larger distance, which corresponds to the concavity of the edge of the cell. In Figure 5a (see movie), the contour of the contact zone has a higher radius of curvature at the front of the cell than at the rear, which is reminiscent of the shape of floppy vesicles forced into constrictions.57 Interestingly, the contact zone may occupy the whole apparent channel width, at least within the resolution of the pictures (1 pixel corresponds to 0.18 μm), which confirms that cells can plug efficiently the cross-section of the channels. However, efficient plugging is not systematic and Figure 5a-ii presents an example with a gutter between the side of the contact zone and the channel corner. Gutters have a maximum width G on the order of 1.5 μm and are more frequent with smaller cells. Finally, Figure 5a-iii presents an example of a contact zone with an irregular topography. This phenomenon concerns about half of cells and is not correlated with any obvious external aspect of the cells. It may arise from inner mechanical inhomogeneities leading to inhomogeneous pressures on walls. A deep understanding of the shape of the gutters and of the contact zone properties as a function of the cells characteristics would deserve much further investigations but is beyond the scope of this paper. In the following, we take advantage of the geometrical characterization of gutters and contact zones to model and analyze the hydrodynamic behavior of cells entering constrictions.
Effect of cell size on the passage through the constriction
Since our method relies on the deformation of polydisperse cells moving through a constriction of fixed geometry, it is important to investigate the influence of the size of the cells in our measurements. We first consider the velocity of the cell in C2, after the complete entry of the cell in C2. We report in Figure 5b the cell velocity 500 μm downstream of C2 entrance, VC2, normalized by the mean fluid velocity in C2 at the same applied pressure but in the absence of the cell, VMF. The ratio VC2/VMF is found to depend strongly on the size of the cell: it is larger than 1 for the smallest cells, decreases with the cell size, and reaches a plateau on the order of 0.3 for cells of diameters larger than 15 μm. The fact that small cells travel faster than the average fluid velocity in the absence of cells can be explained by the position of cells in the centre of the channel where the flow is faster than in the vicinity of the walls. Conversely, large cells tend to clog more efficiently the constriction, like a “piston.” Since the ratio VC2/VMF is significantly smaller than 1, we can deduce that “piston”-cells are slowed down by friction against the walls.58 Additional insights on friction can finally be extracted from RICM images, which shows that the mean cell-to-wall distance h in the contact zone depends strongly on cell size for cells traveling in C2 (Figure 5c). The distance h is found to be smaller for larger cells, indicating a higher pressure against the wall. These observations show the importance to select cells of comparable size to investigate the dependence of friction with the cells' velocity. Indeed, data with cells of diameters limited to the range of 13–15 μm (Figure 5d) show that h is hardly dependent on velocity. This result is further discussed in the friction force section.
Pressure force in the presence of gutters
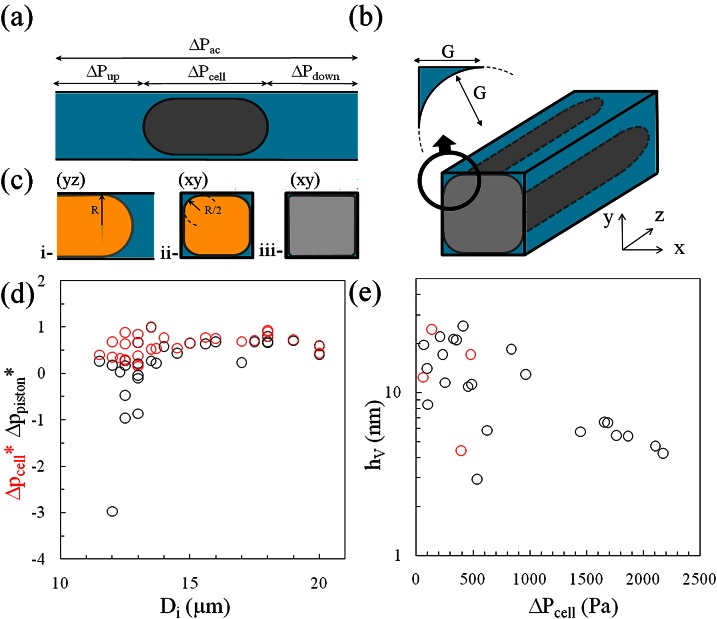
In our setup, fluid flows are precisely controlled through the pressure drop across the device, ΔPext, imposed externally with columns of fluid. However, the relevant parameter to derive the cell force vs. deformation data is the local pressure drop across the cell, ΔPcell. Indeed, the resulting pressure force, Fpressure, acting on a cell moving along a constriction is given by
| (3) |
where Scell is the cross section of the cell in the constriction. Let us estimate ΔPcell from ΔPext. The pressure drop across the analysis circuit, ΔPac, is not significantly affected by the presence of a cell in the analysis circuit, as a result of the large asymmetry of flow rate at the Ji junction.54 ΔPac is, therefore, stable and proportional to ΔPext. We now consider the pressure drop in the analysis circuit in the presence of a cell. As sketched in Figure 6a, ΔPac can be decomposed into the pressure across the cell ΔPcell, and the pressure drops upstream and downstream of the cell
| (4) |
where Rup and Rdown are the hydrodynamic resistances of the analysis circuit upstream and downstream of the cell and can be directly derived from the geometry of the channels. The only unknown parameter in Eq. 4 is the flow rate Q, which is not directly measured but can be inferred from the velocity and the shape of the cell in the constriction. In the absence of leaks through the gutters, the cell indeed acts as a tight piston and Q is equal to the cell velocity, Vcell, times cell cross-section Scell. The local pressure acting on cells in the absence of gutters, ΔPpiston, can been expressed as
| (5) |
where W and H are the width and height of the channel. Due to the possible leakage of the fluid along the gutters, Q is actually the resultant of two contributions, the flow rate due to the piston-cell, Vcell Scell, and the sum of the flow rates in each gutter, Qgutter. The cross-section of gutters in each corner is delimited by the channel walls and by the surface of the cell, which is approximately an arc of circle (Figure 6b), with a radius of curvature G. We solve numerically Stokes equation in this geometry with no slip boundary conditions on the walls (due to the high contrast of viscosity with the solution, the cell is considered as a rigid wall) within the limit where the velocity of the cell is negligible in comparison with the average velocity in the gutter. We obtain the hydrodynamic resistance of gutters, Rgutter
| (6) |
where the length of the gutters, Lgutter, and the radius of curvature, G, are experimentally determined from RICM pictures, G being equal to the width of the non-contact zone in corners (Figure 6b). Still within the limit of a slow cell, the global flow rate can be expressed as:
| (7) |
Combining Eqs. 4, 7 finally allows us to express the local pressure drop across a cell moving in a constriction with flow leakage in gutters, to ΔPcell, as
| (8) |
where all parameters are experimentally available. The influence of gutters on the local pressure across a cell appears clearly in Figure 6d, where ΔPcell and ΔPpiston (both normalized by ΔPAC) are reported for cells of different diameters and for ΔPAC ranging from 100 to 1000 Pa. Equation 8 corrects efficiently the irrelevant negative pressure drops obtained for the smallest cells with Eq. 5. However, the correction becomes negligible for cells larger than 14 μm. With the present device, gutter effects on the pressure force Fpressure can, therefore, be neglected for cells of diameter larger than 14 μm.
Figure 6.
Pressure and friction force. (a) Sketch showing the distribution of pressure drop in the analysis circuit with a single cell in the channel. (b) Sketch of the contact zone around a cell squeezed in a channel. Inset is a zoom on the cross-section in the plane (xy) of the gutter. The width of the gutter, G, which is also the radius of curvature of the cell along the corner, can be directly measured in RICM pictures on the interval between the edges of the channel and the contact zone of the cell. (c) Comparison of the section of a bubble or a liquid droplet (yellow) obstructing a channel of side 2R with the case of a living cell (grey). Section plane is (yz) in (i) and (xy) (ii) and (iii). The tip of droplet or bubble has a spherical cap shape with a radius close to R leading to rounded sections with radii R/2 in corners. From our observations, corners of cells are much sharper. (d) ΔPcell* (red circles) and ΔPpiston* (black circles) correspond to ΔPcell and ΔPpiston calculated with Eqs. 8, 5 and normalized by ΔPac. Data are reported against the diameter of the cells, Di, and correspond to a range of ΔPac between 100 and 1000 Pa. (e) Thickness of lubrication film, hv, estimated from the ratio of Ffriction and Fpressure as calculated using Eqs. 3, 10. The red dots correspond to data where ΔPpiston < 0 and ΔPcell > 0, which supports further the validity of Eq. 8.
Note that the latter result is not trivial at the light of the literature on deformable objects (bubbles and drops) traveling in channels of rectangular cross-section.59, 60, 61, 62, 63, 64, 65 Wong et al.63, 64 have, for instance, shown how changing a channel cross section from circular to polygonal modifies drastically the fluid flow in the presence of bubbles, because the lubricating layer present in the gutters is orders of magnitude thicker than the thin layer observed in a cylindrical channel.58 In a cylindrical channel, a bubble is indeed more difficult to move because of the large drag exerted in the homogeneous lubrication film. Conversely, in a rectangular channel, the liquid can flow along the leaky corners with a speed of one order of magnitude larger than the speed of the bubble. However, these conclusions for bubbles do not apply straightforwardly to cells, which adopt different conformations than bubbles and drops in a constriction. Indeed, the shapes of both front and rear ends of a quasi-static drop or bubble in a square constriction of side 2 R are hemispherical shapes with a radius close to R. According to Laplace law, the curvature of the bubble/drop in channel corners is, therefore, R/2 (with a minor correction66). Gutters for bubbles or drops are, therefore, much wider than gutters observed with cells (Figure 6c). Indeed, interfacial tension of bubbles/drops is in the range of 10–70 mN/m, whereas the cell cortical tension is typically 10−2 mN/m.67 This difference by three orders of magnitude can explain why surface tension plays a much lesser role in dictating the shape of cells as compared to drops and bubbles.
Friction force and lubrication film
For a highly viscous object like a cell, the main dissipation due to the cell motion relative to the channel walls arises from shearing the lubricating film separating the moving object from the cell walls. This assumption is valid as long as viscous dissipation in the film is lower than it would be in within the cell in the absence of lubrication68
| (9) |
With hcell = 5 μm, hfilm = 10 nm, ηfilm = 10−3 Pa s, and ηcell = 100 Pa s,67 the ratio of Eq. 9 is equal to 200. In our case, the friction force, Ffriction, can thus be written as
| (10) |
where h is the average thickness, Sc the area of the contact zone on the lower channel wall measured from RICM pictures, and 2(1 + H2/W) is a multiplicative factor which accounts for the contact zone corresponding to the three other interfaces. From a theoretical point of view, the thickness of the lubricating layer is expected to rely on a balance between viscous dissipation and interfacial forces and to follow Bretherton's law69, 58
| (11) |
which should be valid while surface forces remain negligible (typically for h > 100 nm). However, our experimental results in Figure 5c indicate a thickness on the order of 70 nm practically independent from the cell velocity. Equation 11 is, therefore, not valid in our case. The thickness of the lubricating layer may, however, be inferred from the balance between Ffriction and Fpressure in a steady regime, using Eqs. 3, 10 with the data for the cell velocity in C2. This approach yields a thickness for the lubrication film in a range of 5-30 nm (Figure 6e), which is significantly smaller than the data measured by RICM. Nevertheless, the gap between the glass and the cell membrane is not only occupied by a free flowing film layer but also by the polymer and glycocalyx layers coating the glass and cell surfaces.70 Our optical measurements may, therefore, overestimate the actual thickness of the lubricating film. However, both approaches agree that the lubrication film is too thin to exclude the effect of surface forces and consider only a hydrodynamical approach. A deeper characterization of the complex cell/wall friction mechanics is a difficult task and is beyond the scope of this paper. For our present purpose, our analysis of the balance of Ffriction with Fpressure allows us to get the magnitude of the friction force on a cell in a constriction versus its velocity, which can be used to interpret force–deformation experiments.
Small deformation and steady state regime
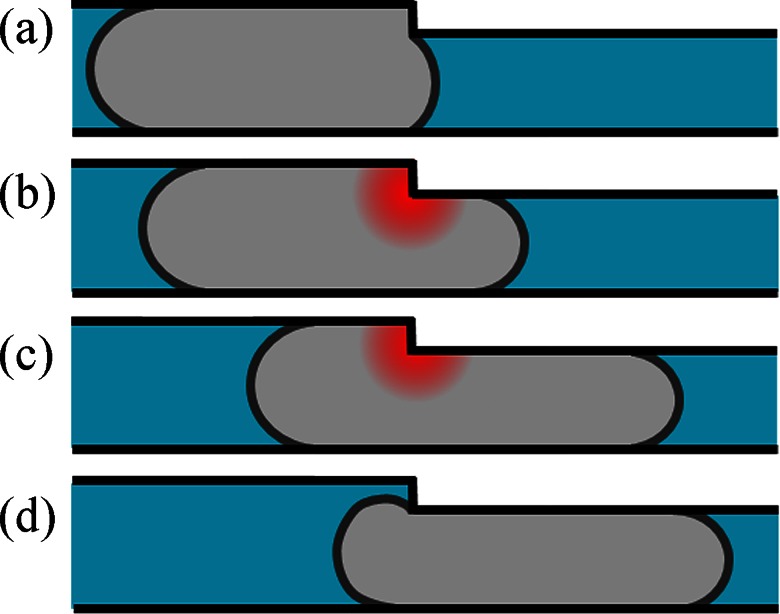
We now focus to the phase where a cell enters at constant velocity in the second constriction C2 (Figure 4, stage II). We would expect that the dissipation induced by the step is dominant (Figures 7b, 7c). Since the step is relatively small (2 μm for a height on the order of 10 μm and a cell length around 20 μm), most of the dissipation should be localized and invariant during the whole transfer of the body of the cell from constrictions 1 to 2. However, we find that the velocity of the cell during this stage, VL, is not negligible as compared to the velocity of the cell travelling freely in the constriction 2, VC2. In other words, the friction force Ffriction cannot be neglected when compared to the force exerted by the step Fstep. To the best of our knowledge, this additional friction force has never been taken into account in previous works.
Figure 7.
Constant viscous dissipation in the vicinity of the step. Sketch representing the different stages of the cell entry from constrictions C1 to C2, (a) stage I, (b) and (c) stage II, and (d) stage III. During stage II, the viscous dissipation due to cell deformation is constant and localized in the vicinity of step (red shadow).
Note finally that the friction force should in principle be different in C2 and C1. Nevertheless, since the cross-sections of both constrictions are very close we neglect this slight difference in force. The linear regime observed in stage II confirms this approximation.
Apparent cell loss modulus
We have shown that the cell deformation in double constrictions can be monitored with a quantitative control of friction and fluid leaks between cell and channel walls. We describe how to extract rheological properties of single cells. The only rheological data accessible in the literature have been performed with the micropipette technique. Interestingly, this technique is very similar to the present double constriction method. In most of these works, cells are considered as viscous liquid droplets surrounded by an elastic membrane.36 The elastic behavior influences mostly the threshold pressure to deform single cells through the constrictions, whereas for small deformations, pressure dissipation within the cell cortex was deemed negligible.71 We thus propose to model the cell as a mostly viscous blob and to extract an apparent viscosity. This approach has the advantage to provide data that are directly comparable to previous micropipette data. However, recent experiments suggest that a more appropriate model should include visco-elasticity.39 Nevertheless, the loss modulus (viscous dissipation) is found to be dominant for frequency solicitations larger than 100 Hz. In our data, with VL > 200 μm/s and step height of 2 μm, the typical frequency rate is larger than 100 Hz and cell can, therefore, reasonably be considered as mostly viscous.
To determine the viscous dissipation imposed by the step during the quasi-static transfer of cells from C1 to C2 (Figure 4, stage II), we have simulated with the finite element software freefem++ the flow of a viscous fluid filling entirely the double constriction with boundary conditions of total slip at the channel walls. The selection of this condition was motivated by the presence of a lubrication layer between the cell and the walls of the channel. The contribution of the step in the global pressure drop has been finally fitted by the third order polynomial function
| (12) |
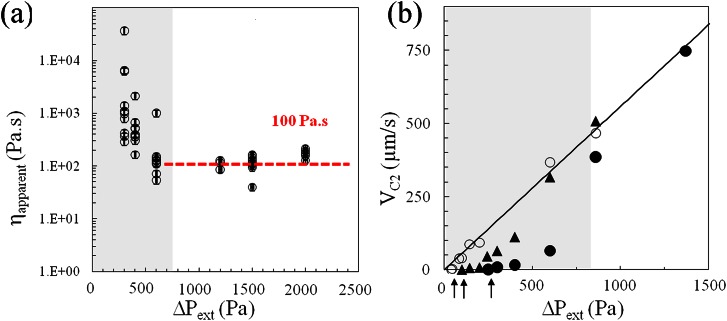
where u2 is the velocity of the viscous fluid in C2 during its passage of the step and = H1−H2 is the step in height. This expression provides an estimate of the pressure with an accuracy of 2%. If friction is neglected, an apparent viscosity of the cell, ηapparent, can be derived directly from Eq. 12 using ΔPcell in place of ΔPstep and VL in place of u2. However, since friction during the cell entry in C2 is not negligible, the applied pressure ΔPcell is dissipated both in the deformation induced by the step and in friction. We assume, as discussed further below, that the friction force is proportional to the cell velocity. The contribution of the step on the pressure drop is then given by ΔPstep = ΔPcell.(1−VL/VC2), where VC2 is the cell velocity after completed entry in C2. Figure 8a reports the estimation of the apparent viscosity for different cells at different applied pressures, where ΔPstep has been corrected. We observe a plateau around ηapparent = 100 Pa s at large pressures, which is remarkably consistent with the data from the literature for THP-1 cells72 and other leukocytes36, 37, 67 The apparent viscosity ηapparent also tends to slightly increase with the applied pressure (or the solicitation rate), which is consistent with recent characterization of cell rheological behavior.73, 74 These results validate our microfluidic method and our analysis to extract quantitative information from the passage of a cell in a microfluidic constriction at high-applied pressures.
Figure 8.
Measurement of the cell loss modulus with a double constriction setup. (a) Apparent cell viscosity, ηapparent measured for different cells versus applied pressure. The grey zone corresponds to the range of pressure where friction is abnormally high. (b) Velocity of a cell in C2, VC2, versus applied pressure ΔPext for 3 different cells of diameter Di = 15.5 μm (•) and 13.5 μm (▲,○). The solid line is a guide to the eye for the linear behavior at high pressure. The black arrows indicate the threshold of pressure required for moving the cell. The grey area underlines the range of pressures, where VC2 is abnormally lower than the linear dependence.
Non-linear friction vs. velocity
At low applied pressures, our analysis of ηapparent in Figure 8a yields a diverging cell viscosity, while the loss modulus of living cell is expected to decrease for lower rates of deformation. To understand this non-physical behavior, we conducted precise measurements of the cell velocity VC2 versus pressure for individual cells (Figure 8b). In the regime of high pressure, we observe that VC2 increases linearly with pressure, as assumed in our analysis of ηapparent. However, in the regime of low pressure, the evolution of the friction force is strongly non-linear: VC2 decreases more rapidly than the linear behavior observed at high pressures, and we find a threshold in pressure before initiating a movement of the cell (between ΔPext = 100 and 300 Pa from one cell to another). Since our analysis is based on linear friction, our estimate of ηapparent can only be valid in the regime of high pressures, ΔPext > 800 Pa (grey area in Figure 8). Altogether, our results confirm that friction can play a significant role during the passage of the cell in constrictions, although this effect has been largely overlooked in the literature. However, we have shown that friction effects in the linear regime of high cell velocities can be efficiently taken into account to assess quantitatively the apparent loss modulus of individual cells from their deformation during their passage in a constriction.
Final protocol to measure cell loss modulus with a double constriction
In this paper, the precise investigation of friction and gutter effects has necessitated a series of measurements with multiple techniques including interference microscopy. Finally, rheological measurements with the double constriction can, however, be assessed from a rather simple protocol. Only two parameters shall be recorded by video-microscopy: the kinetics of the projection length in C2, L(t), and the cell velocity in C2 after completed entry, VC2. The velocity of the extension of the cell VL is extracted from the linear regime of L(t). The effective pressure drop across the cells, ΔPcell, is determined from a standard pressure drop calculation in a circuit of resistive channels, considering that cells behave as non-leaky pistons. This latter assumption is valid for cells sufficiently deformed in C1, i.e., in our case, for cells of diameter larger than 14 μm. Finally, Eq. 11 with u2 = VL and ΔPstep = ΔPcell.(1 − VL/VC2) yields the apparent cell viscosity in the plateau region at high pressure. This protocol has been optimized for THP-1 cells. To extend the method to other cellular systems, appropriate adjustments may have to be developed. The geometry of the double constriction should be adapted to the average size of the cells of interest. Moreover, the experimental conditions where gutters are negligible and friction is linear should be determined.
CONCLUSION
We have designed a novel double constriction device to measure in line the loss modulus of circulating cells in a microfluidic chip, by monitoring the deformation kinetics of a cell when passing a small step. The squeezing of a cell in a first constriction limits flow leaks along the angular edges of the channels and the deformation is measured in a second constriction. Optical interferometry of the cell/wall interface and steady-state cell velocities in straight channels have been used to estimate the effective pressure and friction forces acting on the cell during the passage of the step by taking into account residual flow leaks through the corners of the channels. Since the thickness of lubrication films is on the order of tens of nanometers, friction is not purely hydrodynamic but also involves surface forces' effects. Moreover, friction is not negligible as compared to pressure forces and has to be accounted for in rheological measurements. The dependence of friction with velocity is not linear for low velocities and we measure a threshold in pressure to initiate the motion of the cell. In the linear friction regime at high velocity, the measurement of the velocity of the cell exiting the step can be used to estimate friction during step passage. The loss modulus of individual cells can finally been determined quantitatively. One advantage of this novel cell micro-rheometer relies on the possible integration of multiple analysis devices for single-cells. For instance, the surface of the flat channels permits a precise investigation of cell-surface interactions in micro-channels using optical microscopy techniques for surface investigations (RICM, Total Internal Reflection fluorescence microscopy (TIRF), Surface Enhanced Ellipsometric Contrast microscopy (Wet-SEEC)75). The method is adapted to probe cells of high rigidity and can be useful for the study of pathologies such as acute respiratory distress syndrome, cancer, sepsis, or pneumonia. We hope it will inspire future tools for medical analysis.
ACKNOWLEDGMENTS
The authors are grateful to P. Bongrand, F. Gallet, A. Asnacios, and M. Sokol for helpful discussions. They also thank Rhodia Company (LOF, Pessac, France), as well as I. Ozerov and F. Bedu from Cinam (CNRS, Marseilles, France) for technical support with microfabrication. P.P.'s Ph.D. grant was supported by Région PACA and the company CAPSUM-SAS.
References
- Looney M. R., Thornton E. E., Sen D., Lamm W. J., Glenny R. W., and Krummel M., Nat. Methods 8, 91–96 (2011). 10.1038/nmeth.1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. and Sun Y., Micro Nano Lett. 6, 327–331 (2011). 10.1049/mnl.2011.0010 [DOI] [Google Scholar]
- Hou H. W., Lee W. C., Leong M. C., Sonam S., Vedula S. R. K., and Lim C. T., Cell. Mol. Bioeng. 4, 591–602 (2011). 10.1007/s12195-011-0209-4 [DOI] [Google Scholar]
- Nishino M., Tanaka H., Ogura H., Inoue Y., Koh T., Fujita K., and Sugimoto H., J. Trauma 59, 1425–1431 (2005). 10.1097/01.ta.0000197356.83144.72 [DOI] [PubMed] [Google Scholar]
- Gabriele S., Benoliel A. M., Bongrand P., and Theodoly O., Biophys. J. 96, 4308–4318 (2009). 10.1016/j.bpj.2009.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preira P., Leoni T., Valignat M.-P., Lellouch A., Robert P., Forel J.-M., Papazian L., Dumenil G., Bongrand P., and Théodoly O., Int. J. Nanotechnol. 9, 529–547 (2012). 10.1504/IJNT.2012.045340 [DOI] [Google Scholar]
- Rosenbluth M. J., Lam W. A., and Fletcher D. A., Lab Chip 8, 1062–1070 (2008). 10.1039/b802931h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw K. C., Manimaran M., Francis E. H., and Swaminathan S., Microvasc. Res. 72, 153–160 (2006). 10.1016/j.mvr.2006.06.003 [DOI] [PubMed] [Google Scholar]
- Yap B. and Kamm R. D., J. Appl. Physiol. 98, 1930–1939 (2005). 10.1152/japplphysiol.01226.2004 [DOI] [PubMed] [Google Scholar]
- Shelby J. P., White J., Ganesan K., Rathod P. K., and Chiu D. T., Proc. Natl. Acad. Sci. U.S.A 100, 14618–14622 (2003). 10.1073/pnas.2433968100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins R. J., Piel M., Faure-Andre G., Lennon-Dumenil A. M., Joanny J. F., Prost J., and Voituriez R., Phys. Rev. Lett. 102, 058103 (2009). 10.1103/PhysRevLett.102.058103 [DOI] [PubMed] [Google Scholar]
- Jacobelli J., Friedman R. S., Conti M. A., Lennon-Dumenil A. M., Piel M., Sorensen C. M., Adelstein R. S., and Krummel M. F., Nat. Immun. 11, 953–961 (2010). 10.1038/ni.1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw K. C., Manimaran M., Tay F. E. H., and Swaminathan S., J. Phys.: Conf. Ser. 34, 747–751 (2006). 10.1088/1742-6596/34/1/123 [DOI] [Google Scholar]
- Van der Meer A. D., Poot A. A., Duits M. H. G., Feijen J., and Vermes I., J. Biomed. Biotechnol. 2009, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E. W. K. and Simmons C. A., Lab Chip 10, 143–160 (2010). 10.1039/b913390a [DOI] [PubMed] [Google Scholar]
- Song J. W., Gu W., Futai N., Warner K. A., Nor J. E., and Takayama S., Anal. Chem. 77, 3993–3999 (2005). 10.1021/ac050131o [DOI] [PubMed] [Google Scholar]
- Tkachenko E., Gutierrez E., Ginsberg M. H., and Groisman A., Lab Chip 9, 1085–1095 (2009). 10.1039/b812184b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan O. F. and Sefton M. V., Biomed. Microdevices 13, 69–87 (2011). 10.1007/s10544-010-9472-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meer A. D., Poot A. A., Feijen J., and Vermes I., Biomicrofluidics 4, 1–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. W., Cavnar S. P., Walker A. C., Luker K. E., Gupta M., Tung Y. C., Luker G. D., and Takayama S., PLoS ONE 4(6), e5756 (2009). 10.1371/journal.pone.0005756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J. B., Wu L., Wu J. Z., Zheng Y. H., Zhao H., Jin Q. H., and Zhao J. L., Lab Chip 9, 3118–3125 (2009). 10.1039/b909312e [DOI] [PubMed] [Google Scholar]
- McFaul S. M., Lin B. K., and Ma H., Lab Chip 12, 2369–2376 (2012). 10.1039/c2lc21045b [DOI] [PubMed] [Google Scholar]
- Vanapalli S. A., Duits M. H. G., and Mugele F., Biomicrofluidics 3, 012006 (2009). 10.1063/1.3067820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preira P., Grandné V., Forel J.-M., Gabriele S., Camara M., and Theodoly O., Lab Chip 13, 161–170 (2013). 10.1039/c2lc40847c [DOI] [PubMed] [Google Scholar]
- Huh D., Matthews B. D., Mammoto A., Montoya-Zavala M., Yuan Hsin H., and Ingber D. E., Science 328, 1662–1668 (2010). 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther A., Yasotharan S., Vagaon A., Lochovsky C., Pinto S., Yang J., Lau C., Voigtlaender-Bolz J., and Bolz S.-S., Lab Chip 10, 2341–2349 (2010). 10.1039/c004675b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abkarian M., Faivre M., and Stone H. A., Proc. Natl. Acad. Sci. U.S.A. 103, 538–542 (2006). 10.1073/pnas.0507171102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada K., Sekisuka E., Oshio C., and Minamitani H., Microvasc. Res. 61, 231–239 (2001). 10.1006/mvre.2001.2307 [DOI] [PubMed] [Google Scholar]
- Rand R. P. and Burton A. C., Biophys. J. 4, 115–135 (1964). 10.1016/S0006-3495(64)86773-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E. A., Biophys. J. 13, 941–954 (1973). 10.1016/S0006-3495(73)86036-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien S., Blood Cells 3, 71–99 (1977). [Google Scholar]
- Lichtman M. A., J. Clin. Invest. 52, 350–358 (1973). 10.1172/JCI107191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. E. and Myers K. A., J. Reticuloendothel. Soc. 18, 337–345 (1975). [PubMed] [Google Scholar]
- Schmid-Schonbein G. W., Shih Y. Y., and Chien S., Blood 56, 866–875 (1980). [PubMed] [Google Scholar]
- Schmid-Schonbein G. W., Paul Sung K.-L., Tozere H., Skalak R., and Chien S., Biophys. J. 36, 243–256 (1981). 10.1016/S0006-3495(81)84726-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E. and Kukan B., Blood 64, 1028–1035 (1984); available at http://bloodjournal.hematologylibrary.org/content/64/5/1028.abstract%7d. [PubMed] [Google Scholar]
- Needham D. and Hochmuth R. M., J. Biomech. Eng. 112, 269–276 (1990). 10.1115/1.2891184 [DOI] [PubMed] [Google Scholar]
- Richelme F., Benoliel A. M., and Bongrand P., Cell Motil. Cytoskeleton 45(2), 93–105 (2000). [DOI] [PubMed] [Google Scholar]
- Balland M., Desprat N., Icard D., Fereol S., Asnacios A., Browaeys J., Henon S., and Gallet F., Phys. Rev. E 74, 021911 (2006). 10.1103/PhysRevE.74.021911 [DOI] [PubMed] [Google Scholar]
- Herricks T., Antia M., and Rathod P. K., Cell. Microbiol. 11, 1340–1353 (2009). 10.1111/j.1462-5822.2009.01334.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford S. C., Derganc J., Shevkoplyas S. S., Yoshida T., and Bitensky M. W., Br. J. Haematol. 135, 395–404 (2006). 10.1111/j.1365-2141.2006.06279.x [DOI] [PubMed] [Google Scholar]
- Khan Z. S. and Vanapalli S. A., Biomicrofluidics 7, 011806 (2013). 10.1063/1.4774310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang E., Shin S.-J., Lee K. H., and Lee S.-H., Lab Chip 10, 1856–1861 (2010). 10.1039/c002695f [DOI] [PubMed] [Google Scholar]
- Fiddes L. K., Raz N., Srigunapalan S., Tumarkan E., Simmons C. A., Wheeler A. R., and Kumacheva E., Biomaterials 31, 3459–3464 (2010). 10.1016/j.biomaterials.2010.01.082 [DOI] [PubMed] [Google Scholar]
- Zhou C., Yue P., and Feng J., Ann. Biomed. Eng. 35, 776–780 (2007). 10.1007/s10439-007-9286-x [DOI] [PubMed] [Google Scholar]
- Leong F. Y., Li Q., Lim C. T., and Chiam K. H., Biomech. Model. Mechanobiol. 10, 755–766 (2011). 10.1007/s10237-010-0271-1 [DOI] [PubMed] [Google Scholar]
- Guo Q., Park S., and Ma H. S., Lab Chip 12, 2687–2695 (2012). 10.1039/c2lc40205j [DOI] [PubMed] [Google Scholar]
- Walter N., Micoulet A., Seufferlein T., and Spatz J. P., BioInterphases 6, 117–125 (2011). 10.1116/1.3625258 [DOI] [PubMed] [Google Scholar]
- Xia Y. and Whitesides G. M., Angew. Chem. 37, 550–575 (1998). [DOI] [PubMed] [Google Scholar]
- Tsuchiya S., Yamabe M., Yamaguchi Y., Kobayashi Y., T. Konno, and K. Tada. Int. J. Cancer 26, 171–176 (1980). 10.1002/ijc.2910260208 [DOI] [PubMed] [Google Scholar]
- Vitte J., Benoliel A.-M., Eymeric P., Bongrand P., and Pierres A., Biophys. J. 86, 4059–4074 (2004). 10.1529/biophysj.103.038778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., and Stuurman N., “ Computer control of microscopes using μmanager,” Curr. Protoc. Mol. Biol. 92, 14.20.1–14.20.17 (2010). 10.1002/0471142727.mb1420s92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoly O., Huang Z.-H., and Valignat M.-P., Langmuir 26, 1940–1948 (2010). 10.1021/la902504y [DOI] [PubMed] [Google Scholar]
- Gabriele S., Versaevel M., Preira P., and Theodoly O., Lab Chip 10, 1459–1467 (2010). 10.1039/c002257h [DOI] [PubMed] [Google Scholar]
- Bathe M., Shirai A., Doerschuk C. M., and Kamm R. D., Biophys. J. 83, 1917–1933 (2002). 10.1016/S0006-3495(02)73955-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preira P., Forel J.-M., Robert P., Xeridat F., Brissy O., Bongrand P., Papazian L., and Theodoly O., “Leukocyte stiffening property of plasma in early ARDS revealed by a microfluidic single-cell study- role of cytokines and protection with antibodies” (unpublished). [DOI] [PMC free article] [PubMed]
- Vitkova V., Mader M., and Podgorski T., Europhys. Lett. 68, 398–404 (2004). 10.1209/epl/i2004-10211-9 [DOI] [Google Scholar]
- Baroud C. N., Gallaire F., and Dangla R., Lab Chip 10, 2032–2045 (2010). 10.1039/c001191f [DOI] [PubMed] [Google Scholar]
- Bico J. and Quéré D., J. Colloid Interface Sci. 247, 162–166 (2002). 10.1006/jcis.2001.8106 [DOI] [PubMed] [Google Scholar]
- Kolb W. B. and Cerro R., J. Colloid Interface Sci. 159, 302–311 (1993). 10.1006/jcis.1993.1327 [DOI] [Google Scholar]
- Thulasidas T. C., Abraham M. A., and Cerro R. L., “ Bubble-train flow in capillaries of circular and square cross section,” Chem. Eng. Sci. 50, 183–199 (1995). 10.1016/0009-2509(94)00225-G [DOI] [Google Scholar]
- Ransohoff T. C. and Radke C. J., J. Colloid Interface Sci. 121, 392–401 (1988). 10.1016/0021-9797(88)90442-0 [DOI] [Google Scholar]
- Wong H., Radke C. J., and Morris S., J. Fluid Mech. 292, 71–94 (1995). 10.1017/S0022112095001443 [DOI] [Google Scholar]
- Wong H., Radke C. J., and Morris S., J. Fluid Mech. 292, 95–110 (1995). 10.1017/S0022112095001455 [DOI] [Google Scholar]
- Vanapalli S. A., Banpurkar A. G., van den Ende D., Duits M. H. G., and Mugele F., Lab Chip 9, 982–990 (2009). 10.1039/b815002h [DOI] [PubMed] [Google Scholar]
- Dong M. and Chatziz I., J. Colloid Interface Sci. 172, 278–288 (1995). 10.1006/jcis.1995.1253 [DOI] [Google Scholar]
- Hochmuth R. M., J. Biomech. 33, 15–22 (2000). 10.1016/S0021-9290(99)00175-X [DOI] [PubMed] [Google Scholar]
- Bico J. and Quéré D., J. Fluid Mech. 467, 101–127 (2002). 10.1017/S002211200200126X [DOI] [Google Scholar]
- Bretherton F. P., J. Fluid Mech. 10, 166–188 (1961). 10.1017/S0022112061000160 [DOI] [Google Scholar]
- Damiano E. R., Microvasc. Res. 55, 77–91 (1998). 10.1006/mvre.1997.2052 [DOI] [PubMed] [Google Scholar]
- Yeung A. and Evans E., Biophys. J. 56, 139–149 (1989). 10.1016/S0006-3495(89)82659-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richelme F., Benoliel A. M., and Bongrand P., Exp. Biol. Online 2(5), 1 (1997). [Google Scholar]
- Trepat X., Linhong D., An A. S., Navajas D., Tschumperlin D. J., Gerthoffer W. T., Butler J. P., and Fredberg J. J., Nature 447, 592–595 (2007). 10.1038/nature05824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabry B., Maksym G. N., J. Butler, Glogauer M., Navajas D., and Fredberg J., Phys. Rev. Lett. 87, 148102 (2001). 10.1103/PhysRevLett.87.148102 [DOI] [PubMed] [Google Scholar]
- Ducret A., Valignat M. P., Mouhamar F., Mignot T., and Theodoly O., Proc. Natl. Acad. Sci. U.S.A. 109(25), 10036-10041 (2012). 10.1073/pnas.1120979109 [DOI] [PMC free article] [PubMed] [Google Scholar]