Abstract
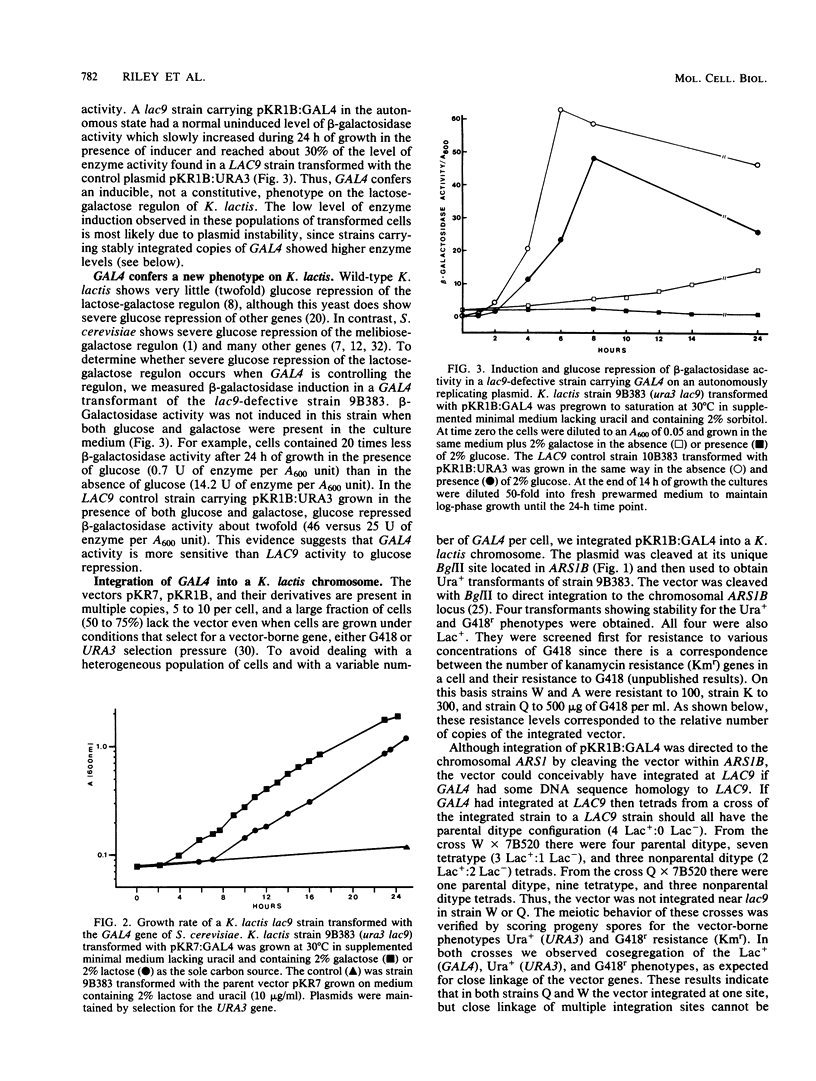
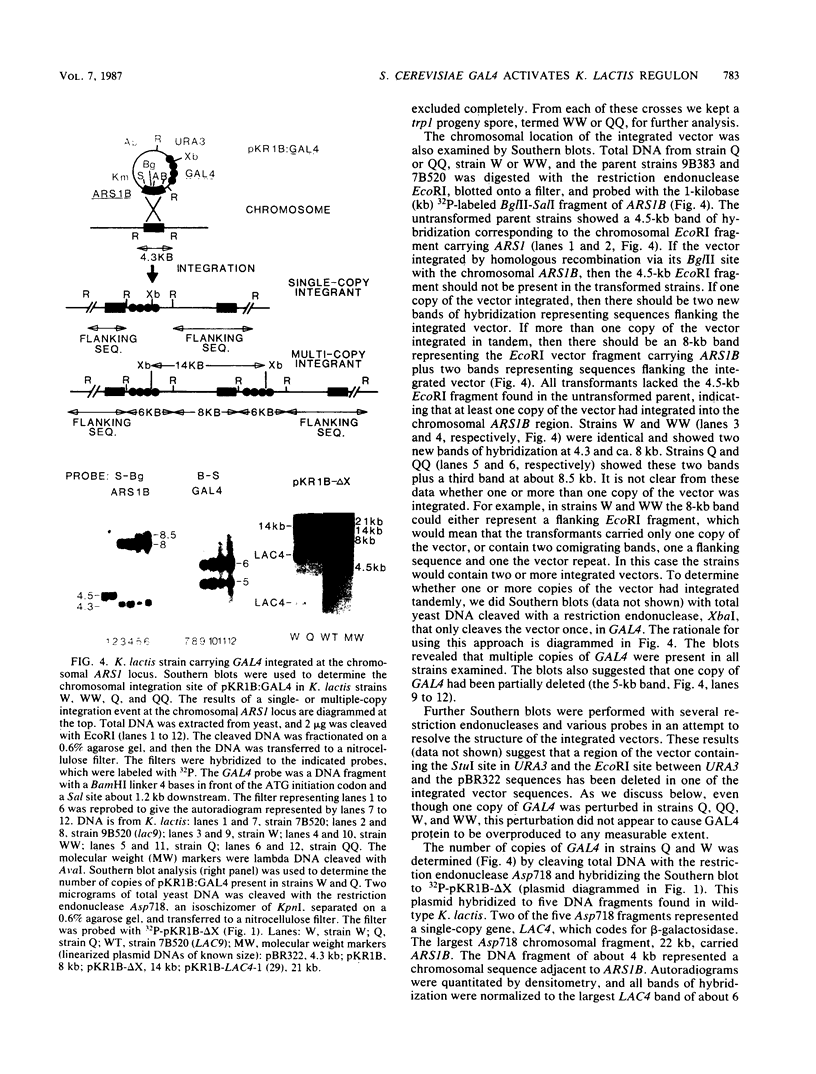
A Kluyveromyces lactis mutant defective in lac9 cannot induce beta-galactosidase or galactokinase activity and is unable to grow on lactose or galactose. When this strain was transformed with the GAL4 positive regulatory gene of Saccharomyces cerevisiae it was able to grow on lactose or galactose as the sole carbon source. Transformants bearing GAL4 exhibited a 4.5-h generation time on galactose or lactose, versus 24 h for the nontransformed lac9 strain. A K. lactis lac9 strain bearing two integrated copies of GAL4 showed 3.5-fold induction of beta-galactosidase activity and 1.8-fold induction of galactokinase activity compared with 15.6-fold and 4.4-fold induction, respectively, for the LAC9 wild-type strain. In transformants bearing 10 integrated copies of GAL4, the induced level of beta-galactosidase was nearly as high as in the LAC9 wild-type strain. In addition to restoring lactose and galactose gene expression, GAL4 in K. lactis lac9 mutant cells conferred a new phenotype, severe glucose repression of lactose and galactose-inducible enzymes. Glucose repressed beta-galactosidase activity 35- to 74-fold and galactokinase activity 14- to 31-fold in GAL4 transformants, compared with the 2-fold glucose repression exhibited in the LAC9 wild-type strain. The S. cerevisiae MEL1 gene was repressed fourfold by glucose in LAC9 cells. In contrast, the MEL1 gene in a GAL4 lac9 strain was repressed 20-fold by glucose. These results indicate that the GAL4 and LAC9 proteins activate transcription in a similar manner. However, either the LAC9 or GAL4 gene or a product of these genes responds differently to glucose in K. lactis.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams B. G. Induction of galactokinase in Saccharomyces cerevisiae: kinetics of induction and glucose effects. J Bacteriol. 1972 Aug;111(2):308–315. doi: 10.1128/jb.111.2.308-315.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel J., Mortimer R. Genetic order of the galactose structural genes in Saccharomyces cerevisiae. J Bacteriol. 1971 Oct;108(1):179–183. doi: 10.1128/jb.108.1.179-183.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci U S A. 1985 Jan;82(1):43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig K. D., Dahlems U., Das S., Hollenberg C. P. Analysis of a eukaryotic beta-galactosidase gene: the N-terminal end of the yeast Kluyveromyces lactis protein shows homology to the Escherichia coli lacZ gene product. Nucleic Acids Res. 1984 Mar 12;12(5):2327–2341. doi: 10.1093/nar/12.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOUGLAS H. C., HAWTHORNE D. C. ENZYMATIC EXPRESSION AND GENETIC LINKAGE OF GENES CONTROLLING GALACTOSE UTILIZATION IN SACCHAROMYCES. Genetics. 1964 May;49:837–844. doi: 10.1093/genetics/49.5.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Denis C. L., Ciriacy M., Young E. T. A positive regulatory gene is required for accumulation of the functional messenger RNA for the glucose-repressible alcohol dehydrogenase from Saccharomyces cerevisiae. J Mol Biol. 1981 Jun 5;148(4):355–368. doi: 10.1016/0022-2836(81)90181-9. [DOI] [PubMed] [Google Scholar]
- Dickson R. C., Markin J. S. Physiological studies of beta-galactosidase induction in Kluyveromyces lactis. J Bacteriol. 1980 Jun;142(3):777–785. doi: 10.1128/jb.142.3.777-785.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Sheetz R. M., Lacy L. R. Genetic regulation: yeast mutants constitutive for beta-galactosidase activity have an increased level of beta-galactosidase messenger ribonucleic acid. Mol Cell Biol. 1981 Nov;1(11):1048–1056. doi: 10.1128/mcb.1.11.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas H. C., Hawthorne C. D. Uninducible mutants in the gal i locus of Saccharomyces cerevisiae. J Bacteriol. 1972 Mar;109(3):1139–1143. doi: 10.1128/jb.109.3.1139-1143.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federoff H. J., Eccleshall T. R., Marmur J. Carbon catabolite repression of maltase synthesis in Saccharomyces carlsbergensis. J Bacteriol. 1983 Oct;156(1):301–307. doi: 10.1128/jb.156.1.301-307.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Kikuchi Y., Nogi Y., Fukasawa T. Regulation of expression of the galactose gene cluster in Saccharomyces cerevisiae. Isolation and characterization of the regulatory gene GAL4. Mol Gen Genet. 1983;191(1):31–38. doi: 10.1007/BF00330886. [DOI] [PubMed] [Google Scholar]
- Johnston S. A., Hopper J. E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew O. M., Douglas H. C. Genetic co-regulation of galactose and melibiose utilization in Saccharomyces. J Bacteriol. 1976 Jan;125(1):33–41. doi: 10.1128/jb.125.1.33-41.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou S. C., Christensen M. S., Cirillo V. P. Galactose transport in Saccharomyces cerevisiae. II. Characteristics of galactose uptake and exchange in galactokinaseless cells. J Bacteriol. 1970 Sep;103(3):671–678. doi: 10.1128/jb.103.3.671-678.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Isolation and preliminary characterization of the GAL4 gene, a positive regulator of transcription in yeast. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6827–6831. doi: 10.1073/pnas.79.22.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Hopper J. E. The relationship of regulatory proteins and DNase I hypersensitive sites in the yeast GAL1-10 genes. Nucleic Acids Res. 1985 Dec 9;13(23):8409–8423. doi: 10.1093/nar/13.23.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Yoshimatsu T., Oshima Y. Recessive mutations conferring resistance to carbon catabolite repression of galactokinase synthesis in Saccharomyces cerevisiae. J Bacteriol. 1983 Mar;153(3):1405–1414. doi: 10.1128/jb.153.3.1405-1414.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post-Beittenmiller M. A., Hamilton R. W., Hopper J. E. Regulation of basal and induced levels of the MEL1 transcript in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jul;4(7):1238–1245. doi: 10.1128/mcb.4.7.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M. I., Dickson R. C. Genetic and biochemical characterization of the galactose gene cluster in Kluyveromyces lactis. J Bacteriol. 1984 May;158(2):705–712. doi: 10.1128/jb.158.2.705-712.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sheetz R. M., Dickson R. C. Lac4 is the structural gene for beta-galactosidase in Kluyveromyces lactis. Genetics. 1981 Aug;98(4):729–745. doi: 10.1093/genetics/98.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz R. M., Dickson R. C. Mutations affecting synthesis of beta-galactosidase activity in the yeast Kluyveromyces lactis. Genetics. 1980 Aug;95(4):877–890. doi: 10.1093/genetics/95.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekrishna K., Dickson R. C. Construction of strains of Saccharomyces cerevisiae that grow on lactose. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7909–7913. doi: 10.1073/pnas.82.23.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekrishna K., Webster T. D., Dickson R. C. Transformation of Kluyveromyces lactis with the kanamycin (G418) resistance gene of Tn903. Gene. 1984 Apr;28(1):73–81. doi: 10.1016/0378-1119(84)90089-1. [DOI] [PubMed] [Google Scholar]
- Torchia T. E., Hamilton R. W., Cano C. L., Hopper J. E. Disruption of regulatory gene GAL80 in Saccharomyces cerevisiae: effects on carbon-controlled regulation of the galactose/melibiose pathway genes. Mol Cell Biol. 1984 Aug;4(8):1521–1527. doi: 10.1128/mcb.4.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitomer R. S., Nichols D. L. Kinetics of glucose repression of yeast cytochrome c. J Bacteriol. 1978 Jul;135(1):39–44. doi: 10.1128/jb.135.1.39-44.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]