Abstract
Background
Cold acclimation in woody perennials is a metabolically intensive process, but coincides with environmental conditions that are not conducive to the generation of energy through photosynthesis. While the negative effects of low temperatures on the photosynthetic apparatus during winter have been well studied, less is known about how this is reflected at the level of gene and metabolite expression, nor how the plant generates primary metabolites needed for adaptive processes during autumn.
Results
The MapMan tool revealed enrichment of the expression of genes related to mitochondrial function, antioxidant and associated regulatory activity, while changes in metabolite levels over the time course were consistent with the gene expression patterns observed. Genes related to thylakoid function were down-regulated as expected, with the exception of plastid targeted specific antioxidant gene products such as thylakoid-bound ascorbate peroxidase, components of the reactive oxygen species scavenging cycle, and the plastid terminal oxidase. In contrast, the conventional and alternative mitochondrial electron transport chains, the tricarboxylic acid cycle, and redox-associated proteins providing reactive oxygen species scavenging generated by electron transport chains functioning at low temperatures were all active.
Conclusions
A regulatory mechanism linking thylakoid-bound ascorbate peroxidase action with “chloroplast dormancy” is proposed. Most importantly, the energy and substrates required for the substantial metabolic remodeling that is a hallmark of freezing acclimation could be provided by heterotrophic metabolism.
Keywords: Microarray, Sitka spruce, Carbon metabolism, Cell walls, Adaptation mechanisms, Visualization
Background
The transition from active growth to dormancy in woody perennials of the temperate and boreal regions involves molecular, cellular, and whole-plant responses. Triggered by gradual decreases in temperature and day length, this remodeling reflects a complex array of adaptations to the physical and metabolic stress imposed by freezing temperatures. Substantial changes in the transcriptome and metabolome accompany this transition and enable the plant to avoid cellular damage resulting from direct thermal effects of freezing temperatures on macromolecules, extracellular ice formation, and the generation of reactive oxygen species (ROS). In deciduous angiosperms and evergreen gymnosperms, the light- and hormone-regulated transcriptional and metabolic changes associated with cambial and vegetative bud dormancy and freezing tolerance are initiated by prolonged nights long prior to the actual freezing [1]–[7].
Because most conifers retain their leaves (needles) during winter, protection from freezing has to extend to these photosynthetic organs. The photosynthetic apparatus is prone to oxidative damage as temperatures drop due to energetic and metabolic imbalance [8]. This imbalance is a result of the temperature dependency of metabolic processes, including photochemistry. Light is absorbed by photosystems regardless of the temperature, but enzymes are inhibited at low temperatures, resulting in over-reduction of the photosystems and photoinhibition. High-energy electrons produced in excess reduce molecular oxygen to generate ROS causing photosystem damage [9]. In evergreen conifers, such oxygen reduction in a Mehler-type reaction is coupled to antioxidant defenses and represents an important mechanism of dissipating excess energy absorbed by photosystem I at low temperatures [8,10].
Photosynthetic acclimation to low temperatures and freezing in evergreen gymnosperms also involves other mechanisms, including the re-localization of chloroplasts, reorganization and aggregation of photosystem antennae, chlororespiration, and non-photochemical quenching leading to conversion of absorbed light energy to heat rather than to reductant for CO2 fixation and growth [8,11]–[14]. Lodgepole pine (Pinus contorta) was shown to reduce the antenna size and the number of reaction centers in photosystem II during winter hardening to minimize light absorption [15]. Additional mechanisms that facilitate the transition from energy harvesting to energy dissipation that occurs with winter hardening include changes in abundance of specific thylakoid proteins [16], non-photochemical quenching via the xanthophyll cycle, and antenna protonation via thylakoid pH changes to dissipate the excess energy to heat [8]. A second, zeaxanthin-independent, quenching mechanism has also been described, involving charge recombination between photosystem II reaction center components [14,17]. Photosystem I is less sensitive to low temperatures than photosystem II and supports xanthophyll-mediated non-photochemical quenching, while maintaining active cyclic electron transport for ATP synthesis at low temperatures [8,11,18]. Chlororespiration and cyclic electron transport enable the dissipation of overproduced excitation energy via alternative electron acceptors, including the terminal oxidase in the plastid (PTOX), a bi-functional protein involved in carotenoid biosynthesis and oxidation of plastoquinol produced by over-reduced electron transport chain (ETC.) [19]–[23].
Potential excess NADPH produced as a result of linear photosynthetic electron transport in the almost complete absence of carbon fixation in the stroma can also be dissipated with or without altering ATP synthesis through the action of the alternate ETC. in mitochondria via operation of the plastidic malate/oxaloacetate shuttle [24]. The alternate mitochondrial ETC. also provides means of preventing ROS formation during mitochondrial aerobic respiration [25]. If ROS scavenging does not occur rapidly enough, under such conditions as extreme biotic or abiotic stresses including low temperatures, damage to mitochondrial and other cellular components and even programmed cell death may occur [26]–[30]. ROS production can also be prevented by lowering O2 levels and/or redirecting respiratory electron flux to other substrates. These alternate routes for electrons during respiration involve alternative oxidases, external and internal alternative NAD(P)H dehydrogenases, and uncoupling proteins, that are especially active upon imposition of abiotic stress that leads to ROS production [31]–[34]. These enzymes play a key role in plant acclimation and tolerance to low temperatures by preventing ROS formation when the traditional route involving complexes III and IV is inhibited [35]–[38]. These alternate routes bring about a lowering of ROS levels by maintaining active electron flow and preventing over-reduction of electron transport components, or redox imbalance [29,33,34,39,40].
Alternative oxidases compete for electrons with cytochrome c oxidase (complex III), using oxygen as a substrate. In addition to maintaining mitochondrial electron flow, they can also prevent ROS production directly by lowering the levels of oxygen, which can react with electrons to generate ROS, up to 60% during abiotic stresses [34,41,42]. Rotenone-insensitive alternative internal and external NAD(P)H dehydrogenases interfere with mitochondrial proton transport along with alternative oxidases [33,34,43]–[45]. These dehydrogenases bypass complex I, while alternative oxidases compete for the electrons with complex III, thus preventing a total of three protons from being transported from the matrix to the inter-membrane space by NADH dehydrogenase and cytochrome c reductase and oxidase [33,34]. Unlike alternative oxidases and dehydrogenases, uncoupling proteins diminish the electrochemical potential directly by transporting protons back to the matrix with the result that less ATP is synthesized. Uncoupling of the ETC. from oxidative phosphorylation stimulates fluxes through the ETC. and subsequently substrate oxidation during carbon and energy imbalance that occurs during a variety of stresses. As such, uncoupling proteins are also involved in controlling ROS levels and promoting tolerance of plants to stresses [31]–[34].
The transition from energy harvesting to dissipation of the photosynthetic apparatus in needles that takes place during winter hardening is a major adaptive response. However, freezing acclimation also involves a wide range of other cellular adaptations, which requires a substantial source of metabolic precursors and chemical energy that, given the circumstances, clearly cannot originate from linear photosynthetic electron transport and CO2 fixation. For example, detailed bioinformatic analyses of transcriptomic and metabolic remodeling during winter hardening in Sitka spruce (Picea sitchensis) revealed the presence of highly complex regulatory networks involving ethylene-mediated signaling, active trafficking of cell wall carbohydrates through the endomembrane system, and significant remodeling of cell walls [46]. Here we implement systems biology tools to further interrogate these datasets, which involve five time points sampled from late summer to early winter [47]. Specifically, the current paper focuses on adaptation of the photosynthetic apparatus to low temperature, with the focus on whole processes relevant to energy metabolism from the global perspective of systems biology rather than individual genes.
Results
Photosynthesis and respiration
The vast majority of responsive genes associated with chloroplast function were down-regulated after the time point 2 (TP2) and remained in that state for the duration of the time course (Figure 1 and Additional file 1: Table S1). Notable exceptions were PTOX/IMMUTANS, a nuclear gene encoding a plastid terminal oxidase, and Psb, a chloroplast gene encoding the CP47 subunit of the reaction center of Photosystem II (Figure 1), in addition to the plastid-targeted antioxidant gene products discussed below (Figure 2). In contrast, respiration-related genes were up-regulated (Figures 3 and 4). For example, Atmg00516, which encodes a subunit of mitochondrial NAD(P)H dehydrogenase (complex I), showed a consistent about 3-fold up-regulation throughout the time-course, and other components of the ETC. exhibited low, but consistent up-regulation, and clustered with the genes encoding alternative mitochondrial ETC. components. However, there were also corresponding sets of genes involved in respiration (both nucleus- and mitochondrion-encoded) that were down-regulated (Figures 3 and 4).
Figure 1.
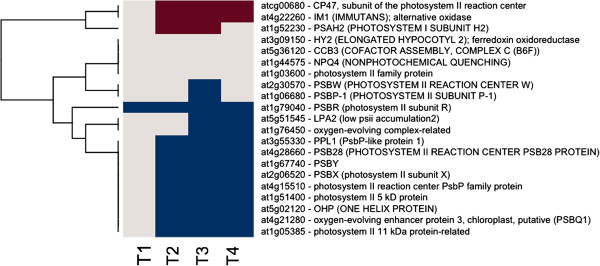
Photosynthetic Gene Expression. The gene expression levels across the rows are hierarchically clustered. Magenta cells represent significant positive gene expression changes, blue cells represent significant negative changes, and off-white cells represent no change or no statistically significant change. The level of significance used was at P ≤ 0.05.
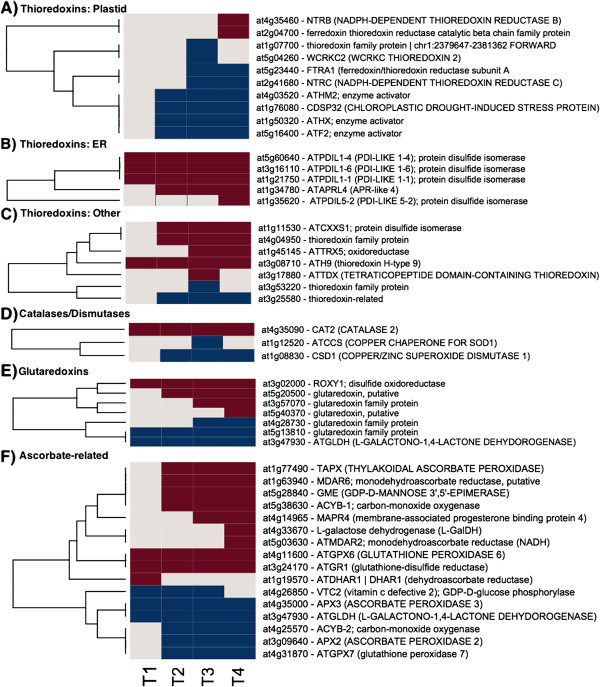
Figure 2.
Antioxidant Gene Expression. The gene expression levels across the rows are hierarchically clustered. Magenta cells represent significant positive expression changes, blue cells represent significant negative changes, and off white cells represent no change or no statistically significant change. The level of significance used was at P ≤ 0.05. Genes and metabolites are grouped according to MapMan bin or MapMan bin combined with cellular location. The figure is divided into panels: A, thioredoxins transcripts associated with plastids, B, thioredoxins associated with the endoplasmic reticulum (ER), C, thioredoxins transcripts with unknown or locations other than plastids or endoplasmic reticulum, D, catalases and dismutases transcripts, E, glutaredoxin Transcripts, and F, ascorbate-related transcripts.
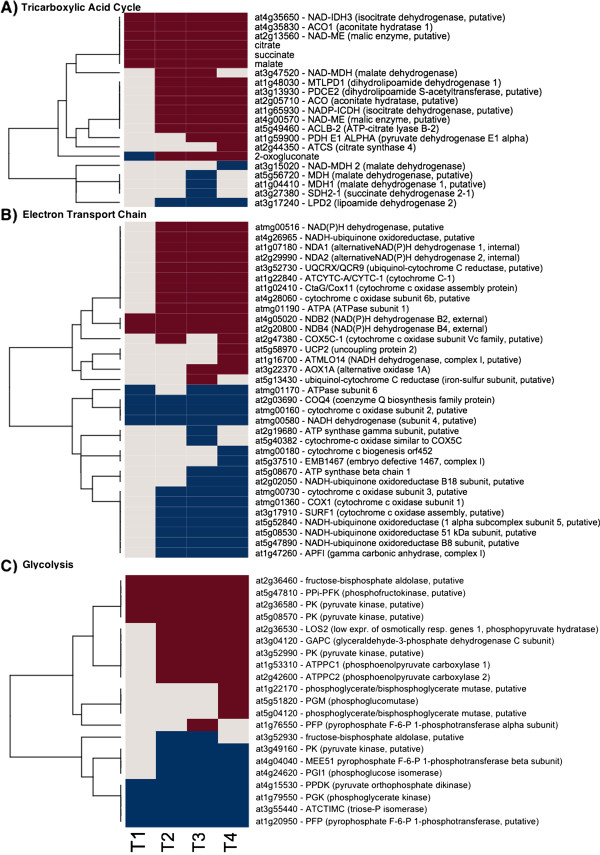
Figure 3.
Tricarboxylic Acid Cycle, ETC., and Glycolysis Gene Expression and Metabolite Changes. The gene expression levels across the rows are hierarchically clustered. Magenta cells represent significant positive expression changes, blue cells represent significant negative changes, and off white cells represent no change or no statistically significant change. The level of significance used was at P ≤ 0.05. The figure is divided into panels: A, transcripts associated with the Tricarboxylic Acid Cycle, B, ETC. transcripts, and C, glycolysis related transcripts.
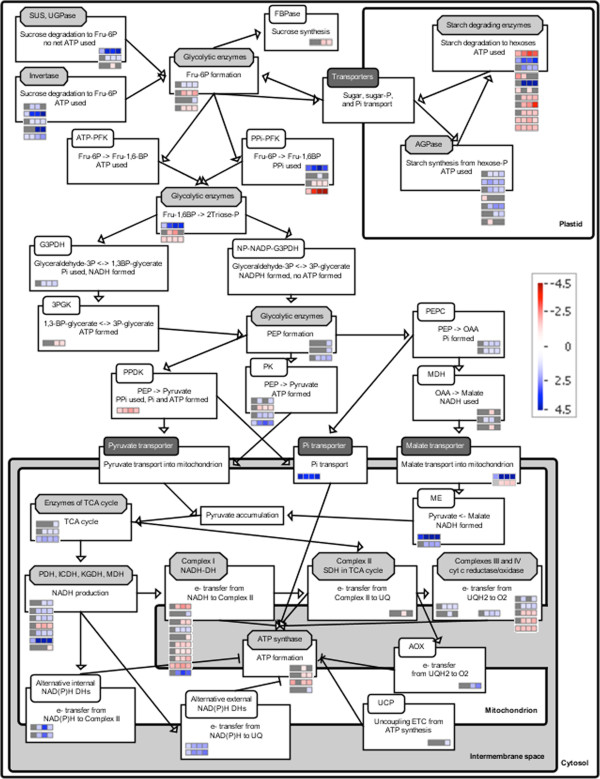
Figure 4.
SBGN Pathway of heterotrophic metabolism and changes in expression of metabolic genes in Sitka spruce needles during winter hardening. Metabolic pathways were drawn using the BEACON tool to represent activities as boxed reactions with associated temporal changes in expression of relevant genes (rows in colored bins/small boxes) in TP1 through TP4 obtained from MapMan. Gray boxes indicate that the change in gene expression was not statistically significant, while blue and red indicate a statistically significant up- and down-regulation, respectively (P ≤ 0.05). Arrows connecting the activities indicate positive influence to generate a metabolic network, while “T” represents an inhibitory effect, e.g., components of the alternative ETC. in mitochondria interfere with adenosine triphosphate (ATP) synthesis. Each activity is associated with an enzyme (white rounded rectangle), enzyme complex (light gray octagon), or transporter (dark gray rounded rectangle). Abbreviations: 3PGK, 3-phosphoglycerate kinase; AGPase, ADP-glucose pyrophosphorylase; AOX, alternative oxidase; cyt, cytochrome; DH, dehydrogenase; FBPase, fructose-1,6-bisphosphatase; Fru, fructose; G3PDH, glyceraldehydes-3-phosphate DH; ICDH, isocitrate DH; KGDH, α-ketoglutarate DH; MDH, malate DH; ME, malic enzyme; NAD(P), oxidized nicotinamide adenine dinucleotide (phosphate); NAD(P)H, reduced nicotinamide adenine dinucleotide (phosphate); NP, non-phosphorylating; P, phosphate; PEP, phosphoenolpyruvate; PEPC, PEP carboxylase; PDH, pyruvate DH; PFK, phosphofructokinase; PK, pyruvate kinase; Pi, inorganic phosphate; PPi, pyrophosphate; PPDK, pyruvate phosphate dikinase; SDH, succinate DH; SUS, sucrose synthase; TCA, tricarboxylic acid cycle; UCP, uncoupling protein; UGPase, UDP-glucose pyrophosphorylase; UQ/UQH2, oxidized/reduced ubiquinone.
Antioxidant function
Genes encoding proteins related to antioxidant activity in various organelles responded in diverse ways during the time course (Figure 2 and Additional file 2: Table S2). Massive increases in expression levels of peroxisomal catalase 2 were the most striking results in this overall category. Increases in the expression of genes encoding differentially targeted thioredoxin-related proteins were also observed, including three genes encoding protein disulfide isomerases (PDI gene family) targeted to the ER, and genes homologous to AtERO, which interacts with PDI in the ER. Genes encoding proteins that function in the ubiquitous ROS-scavenging ascorbate/glutathione pathway showed varying responses. Glutathione reductase 1, encoding a gene product targeted to the peroxisome and the cytosol, was up-regulated. Two monodehydroascorbate reductases, which generate ascorbate from monodehydroascorbate substrates, were up-regulated. One of these proteins is targeted to the chloroplast stroma. Two genes encoding ascorbate peroxidases were also up-regulated. After CAT2, thylakoid-associated L-ascorbate peroxidase was among the strongest up-regulated antoxidant genes. Re-reduction of ascorbate using reduced glutathione as a substrate was evident by the up-regulation of a dehydroascorbatereductase (subcellular location undefined), as well as an intrinsic trans-membrane cytochrome b561, and a membrane-associated heme binding protein that may be involved in intra-vesicular reduction of monodehydroascorbate. Within the glutaredoxin category, a spruce homolog of the disulfide oxidoreductase ROXY1, which has a regulatory role in angiosperm flower development, was highly up-regulated at the later time points. Notably up-regulated early in the time course were several genes involved in ascorbate biosynthesis, including an L-galactose dehydrogenase, which catalyzes the conversion of L-galactose to L-galactono-1,4-lactone, and a GDP-mannose 3,5-epimerase, which catalyzes an important step in the ascorbate biosynthetic pathway – the conversion of GDP-d-mannose to GDP-l-galactose.
The Automated Layout Pipeline for Inferred NEtworks (ALPINE) pipeline was used to visualize documented “guilt by association” relationships for the thylakoid-bound ascorbate peroxidase (tAPX) (Table 1). The “guilt by association” relationships are imported from GeneMania and drawn from a large dataset containing functional associations to tAPX. These functional associations include protein and genetic interactions, pathways, co-expression, co-localization and protein domain similarity. The ALPINE visualization integrates the functional associations of genes, Gene Ontology (GO) annotations, and subcellular location data into a single unified view. This view is filtered by user supplied gene expression data to return only gene associations obtained from the input. This ability to exclusively analyze a particular dataset and return only the gene associations of interest and significance is unique to ALPINE and to our knowledge, there are no bioinformatics tools available with the ALPINE’s useful and unique combination of functionalities and capabilities.This strategy revealed clues concerning antioxidant-related events in winter hardening that are not apparent using other singular strategies. Of the 28 genes that were found to be associated with tAPX, all but five were down-regulated, and all but one gene encoded a plastid-targeted protein. The plastid-targeted down-regulated genes fell into several categories, for example, photosystem II, chloroplast-based protein synthesis, protein folding, and RNA regulation of transcription. In contrast, a gene encoding a member of the rhodanase family was up-regulated, as was a plastid-targeted, NADP-linked oxidoreductase. Taken together, tAPX may be associated with the regulatory mechanism that brings about the virtual cessation of many chloroplast activities that occur during winter hardening.
Table 1.
Gene association network generated by querying thylakoid ascorbate peroxidase (tAPX, AT1G77490) using the ALPINE tool
| AT1G77490 |
thylakoidal ascorbate |
Plastid |
PlastidRedox.Ascorbate and Glutathione.Ascorbate |
3.41 |
1.51E-04 |
| AT4G39970 |
Haloacid dehalogenase‒like hydrolase (HAD) superfamily |
Plastid |
Not Assigned. No Ontology |
2.53 |
1.15E-03 |
| AT5G51820 |
phosphoglucomutase |
Plastid |
Glycolysis.Plastid Branch.Phosphoglucomutase |
1.5 |
4.98E-02 |
| AT4G27700 |
Rhodanese/Cell cycle control phosphatase superfamily |
Plastid |
Misc.Rhodanese |
1.48 |
8.16E-03 |
| AT1G04420 |
NAD(P)‒linked oxidoreductase superfamily protein |
Plastid |
Minor CHO Metabolism.Others |
1.47 |
4.78E-04 |
| AT5G27290 |
AT5G27290unknown protein |
Plastid |
Not Assigned.Unknown |
1.36 |
4.71E-02 |
| AT5G65220 |
AT5G65220Ribosomal L29 family protein |
Plastid |
Protein.Synthesis.Ribosomal Protein.Prokaryotic. |
−1.32 |
4.09E-02 |
| AT4G21280 |
AT4G21280photosystem II subunit QA |
Plastid |
PS.Lightreaction.Photosystem II.PSII Polypeptide Subunits |
−1.33 |
1.45E-02 |
| AT1G32550 |
2Fe‒2S ferredoxin‒like superfamily protein |
N/A |
Misc.Other Ferredoxins and Rieske Domain |
−1.33 |
4.95E-02 |
| AT1G76450 |
Photosystem II reaction center PsbP family protein |
Plastid |
PS.Lightreaction.Photosystem II.PSII Polypeptide Subunits |
−1.37 |
1.34E-02 |
| AT2G43560 |
FKBP‒like peptidyl‒prolyl cis‒trans isomerase family protein |
Plastid |
Protein.Folding |
−1.37 |
1.54E-02 |
| AT3G52150 |
RNA‒binding (RRM/RBD/RNP motifs) family protein |
Plastid |
RNA.Regulation of Transcription.Unclassified |
−1.41 |
2.59E-02 |
| AT3G18890 |
NAD(P)‒binding Rossmann‒fold superfamily protein |
Plastid |
Signalling.Light |
−1.45 |
2.02E-02 |
| AT3G51510 |
unknown protein |
Plastid |
Not Assigned.Unknown |
−1.46 |
1.54E-02 |
| AT1G48350 |
Ribosomal L18p/L5e family protein |
Plastid |
Protein.Synthesis.Ribosomal Protein.Prokaryotic.Chloroplast |
−1.46 |
1.54E-02 |
| AT1G43670 |
Inositol monophosphatase family protein |
Membrane |
Metabolism.Synthesis.Sucrose.F |
−1.51 |
3.97E-02 |
| AT1G75690 |
DnaJ/Hsp40 cysteine‒rich domain superfamily protein |
Plastid |
Protein.Folding |
−1.6 |
4.19E-02 |
| AT5G14910 |
Heavy metal transport/detoxification |
Plastid |
Not Assigned.No Ontology |
−1.62 |
3.44E-02 |
| AT1G21350 |
Thioredoxin super family protein |
Plastid |
Not Assigned.Unknown |
−1.63 |
1.33E-02 |
| AT5G13510 |
Ribosomal protein L10 family protein |
Plastid |
Protein.Synthesis.Ribosomal Protein.Prokaryotic. |
−1.69 |
3.43E-05 |
| AT3G48420 |
Haloacid dehalogenase‒like hydrolase (HAD) superfamily |
Plastid |
Not Assigned.Unknown |
−1.7 |
2.50E-02 |
| AT1G16080 |
unknown protein |
Plastid |
Not Assigned.Unknown |
−1.73 |
4.36E-03 |
| AT4G09650 |
ATP SYNTHASE DELTA‒SUBUNIT; hydrogen ion |
Plastid |
Not Assigned.Unknown |
−1.9 |
3.44E-04 |
| AT3G54210 |
Ribosomal protein L17 family protein |
Plastid |
Protein.Synthesis.Ribosomal Protein.Prokaryotic.Chloroplast. |
−1.92 |
4.44E-04 |
| AT3G61870 |
unknown protein |
Plastid |
Not Assigned.Unknown |
−2.06 |
2.71E-03 |
| AT5G52970 |
thylakoid lumen 15.0 kDa protein |
Plastid |
Not Assigned.No Ontology |
−2.06 |
4.12E-04 |
| AT4G01310 |
Ribosomal L5P family protein |
Plastid |
Protein.Synthesis.Ribosomal Protein.Prokaryotic. |
−2.42 |
8.36E-04 |
| AT5G42070 | unknown protein | Plastid | Not Assigned.Unknown | −3.01 | 2.08E-05 |
All associations were found to be co-expression associations, and all 28 genes in the network demonstrate a significant (q <0.05) expression value in T5 of the dataset.
Alternative ETC
Components of the alternative electron transport are known to interfere with mitochondrial ATP production. Two external alternative NAD(P)H dehydrogenases showed a consistent up-regulation in response to freezing temperatures across the time series, while two internal ones from TP2 to TP4 (Figures 3 and 4). Alternative oxidase A1 gene (At3g22370 in Arabidopsis) was up-regulated at TP3 and 4, while an ortholog of uncoupling protein 2 (At5g58970) at TP4 only (Figures 3 and 4).
Carbon and reductant sources for respiration
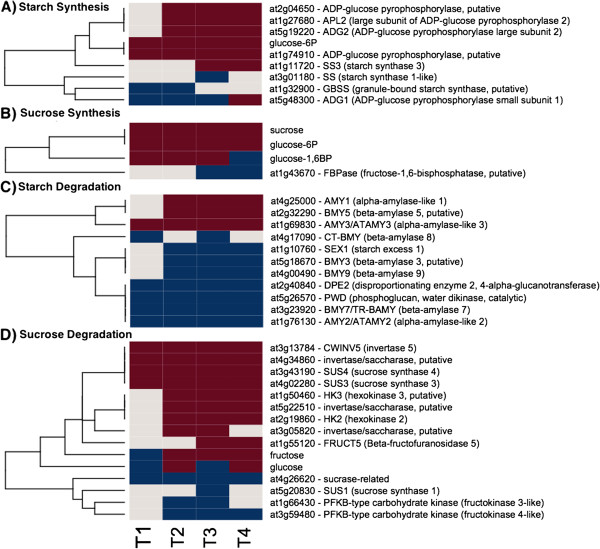
To determine the source of reductant for mitochondrial electron transport, we assessed changes in gene expression of various genes involved in central carbon metabolic pathways that could provide these substrates (Additional file 3: Table S3). The only potentially active pathways involved sugars. Transcript levels of four invertase genes, which are involved in starch degradation, increased, including a cell wall invertase, whose expression level increased through TP4 (Figures 4 and 5). A homolog of At4g34860, a cytosolic invertase, showed large increases through TP4. The expression of homologs to members of the sucrose synthase (SUS) gene family showed a somewhat analogous pattern with SUS4 exhibiting a substantial increase through TP4. SUS3 also showed an increase, albeit a more modest one, which peaked between TP3 and TP4. In contrast, SUS1 showed a decrease in TP3 and no significant changes at other TPs. However, sucrose levels remained constant and above baseline levels during freezing acclimation. More generally, genes encoding starch biosynthesis enzymes were moderately up-regulated, while those associated with degradation were down-regulated (Figures 4 and 5). ADP glucose pyrophosphorylase (AGPase) involved in starch biosynthesis is a heteromer of two large and two small subunits. APL1 and 2, two of the three genes encoding the large AGPase subunits responded and showed increases in the expression from TP2 to TP4. A gene encoding the small subunit of AGPase (APS1) showed a decrease in expression at TP1, 2 and 3 and an increase in TP4. One gene encoding starch synthase showed an initial decrease in TP1 and 2, while the other responsive starch synthase gene was down-regulated at TP3 (Figures 4 and 5).
Figure 5.
Starch and Sucrose Metabolism Gene Expression and Metabolite Changes. The gene expression levels across the rows are hierarchically clustered. Magenta cells represent significant positive expression changes, blue cells represent significant negative changes, and off white cells represent no change or no statistically significant change. The level of significance used was at P ≤ 0.05. The figure is divided into panels: A, transcripts associated with starch synthesis, B, sucrose synthesis, C, starch degradation, and D, sucrose degradation.
Most genes encoding glycolytic enzymes did not have considerably altered levels of relevant transcripts in response to freezing (Figures 3 and 4). However, some glycolytic genes showed a more substantial up- or down-regulation, including the pyrophosphate-dependent phosphofructokinase 2 (At5g47810) that showed a maximal 5.2-fold increase and a constant up-regulation. The same step in glycolysis can also be catalyzed by another pyrophosphate-dependent enzyme, fructose-6-P 1-phosphotransferase and the corresponding α - subunit (At1g20950) was correspondingly down-regulated (a maximal 5.2-fold decrease). Fructose-1,6-bisP aldolase (At2g36460) transcript levels increased consistently (~4-fold) in response at all TPs. In Arabidopsis, At5g08570 encodes a protein similar to cytosolic pyruvate kinase, and the corresponding gene in Sitka spruce showed a 3-fold increase in transcript levels. Transcript levels of gluconeogenetic pyruvate pyrophosphate dikinase (At4g15530) decreased at TP1 and remained low throughout the time course. Phosphoenolpyruvatecarboxykinase (encoded by At4g37870 in Arabidopsis) transcript gradually decreased 4.4 fold through TP4.
Dihydrolipoamide dehydrogenase 1 (LPD1, At1g48030), the E3 subunit 1 of pyruvate dehydrogenase, was moderately (up to 1.8-fold), but significantly up-regulated from TP2 to TP4 (Figures 3 and 4). In contrast, LPD2 (At3g17240), the E3 subunit 2 of pyruvate dehydrogenase, was down-regulated about 1.8-fold between TP2 and TP4. The steady-state transcript levels for other subunits of pyruvate dehydrogenase remained unchanged during freezing. This was true for most of the transcripts encoding the remaining enzymes of the TCA cycle, though they tended to be slightly above the baseline levels (typically less than a 1.5-fold change) throughout the time course. A putative cytosolic NADP-dependent isocitrate dehydrogenase (At1g65930) showed the same pattern and degree of increases in gene expression as LPD1. A putative regulatory subunit of the mitochondrial NAD-dependent isocitrate dehydrogenase III (At4g35650) increased 2-fold in TP1 and showed a stable 4.4-fold increase in transcript levels in TP2 through TP4. Malic enzyme gene (At4g00570) showed only a moderate 2.2-fold up-regulation in response to freezing, but the consistent 9- to 11-fold induction of the At2g13560-encoded NAD-dependent malic enzyme transcript levels in TP2, 3, and 4 was quite striking. Citrate and succinate levels were moderately increased in a stable manner, while malate levels gradually increased up to 2.9 fold during freezing (Figure 4).
Discussion and conclusions
Temperate and boreal conifers must survive harsh winters while retaining their photosynthetic tissue, and have evolved intricate mechanisms to maintain a certain level of active metabolism, signaling, and protective processes at low temperatures in their needles. The comprehensiveness of our data afforded a global view of winter hardening as it is reflected in numerous processes. The use of bioinformatics tools enabled the visualization of these changes in a cellular context. The results point to numerous photoprotective processes that are active during winter, serve to confirm previous reports, and provide new insights into the activity of genes associated with chloroplast function, energy metabolism, antioxidant processes, and possible regulatory mechanisms.
Genes related to energy quenching mechanisms including a spruce PsbS homolog and plastid terminal oxidase PTOX increased their expression, which is in an agreement with previous reports [10]. PsbS activity is associated with changes in photosystem II that occur as part of the photoprotective mechanisms that are set in place as the temperature drops, while PTOX represents an important component of the dissipation of excess light energy in photosynthesis as an alternative electron acceptor and O2-utilizing enzyme helping to prevent ROS formation in hardening conifer needles [10]. In Ranunculus glacialis, an alpine plant growing in high altitudes at high light and low temperatures, PTOX protein levels correlated with the use of alternative ETC. and enabled high fluxes through photosynthetic ETC. despite the excess of light energy. PTOX was implicated to serve as a safety valve for the over-reduction of photosystems during high light and low temperatures in alpine plants [48]. PTOX has also been implicated in increasing the flux through the linear ETC. and subsequent increased tolerance of photosystems to high light in cold-hardened Arabidopsis thaliana[49]. This result suggests that the energy dissipation mechanisms set in motion within the chloroplast during winter hardening were effective. One manifestation of this may have been a relatively low level of ROS generation as the temperature dropped. However, redox effects on cellular responses to the environment are not confined to a simple containment of ROS. Much data point to regulatory roles for specific antioxidant proteins in plant cells under stress. Several of these antioxidant, regulatory genes were up-regulated over the winter hardening time course, including catalase CAT2, glutaredoxin ROXY1, and glutathione reductase GR1. Catalases regulate the levels of hydrogen peroxide, which is an important signaling molecule in plant stress responses [50,51]. ROXY1encodes a glutaredoxin with a documented regulatory role in development in Arabidopsis [52]–[54], while GR1exerts a regulatory effect on responses to cold stress, an effect that cannot be replaced by GR2, or thioredoxins [55].
The ROS scavenging pathway may have been operational in the chloroplast as winter hardening proceeded, since other components, such as mondehydroascorbate reductase, were also up-regulated (Table 1). However, it is also possible that tAPX was acting in a different/additional capacity to protect the photosynthetic machinery. Duan et al. (2012) have shown that tAPX plays a specific role in alleviating the photosystem I and II-associated photoinhibition that occurs with chilling stress in Arabidopsis [56], and spruce tAPX may play an analogous role in the chloroplasts of hardening needles. Interestingly, Maruta et al. (2010) provide evidence for a specific regulatory role for tAPX in chloroplast responses to stress, resonant with the role shown for GR1 [57]. In the case of tAPX and the winter hardening data, the guilt by association analysis revealed an association of tAPX exclusively with plastid-targeted gene products, in which the great majority of those responsive genes were down-regulated. It is possible that tAPX plays an important role in the adaptation mechanism that results in the onset of chloroplast dormancy in conifer needles. Taken together, these results suggest specific regulatory roles for tAPX, ROXY1, and GR1 in the adaptation mechanism underlying winter hardening.
Freezing temperatures impose severe stress on the mitochondrial ETC. A set of genes encoding components of the respiratory ETC. were strongly or mildly up-regulated, suggesting that the mitochondrial ETC. is active. Mitochondrial aerobic respiration is a source of ROS formed through the reduction of molecular oxygen by electrons coming from complexes I and III of the ETC. [25]. The ROS production can be prevented by redirecting respiratory electron flux to other substrates by involving the major players of the alternate ETC. including alternative oxidases, external and internal alternative NAD(P)H dehydrogenases, and uncoupling proteins [31]–[34]. Genes encoding all these components of the alternate ETC. were up-regulated during the time course, suggesting that these alternate pathways are active, along with the conventional ETC., during freezing acclimation. Up-regulation of genes encoding the components of the alternate ETC. is typical during cold acclimation in plants in general and is accompanied with metabolic changes. Tobacco plants subjected to cold showed an increase in alternative oxidase levels and sugar content compared to the transgenic plants with suppressed expression of this enzyme [58]. These transgenic plants also showed an increased lipid peroxidation at normal, but not at low, temperatures. Alternative oxidases are regulated at the transcriptional level by abscisic acid insensitive 4 (ABI4) transcription factor and are activated by pyruvate [59]–[61]. Accumulation of pyruvate as the final product of glycolysis indicates an imbalance between glycolytic fluxes and the mitochondrial ETC. Pyruvate then activates alternative oxidases to stimulate flux through the mitochondrial ETC. to keep pace with glycolysis [34,62,63]. Suppressing the external NAD(P)H dehydrogenase NDB4 resulted in increased levels of NDB2 and alternative oxidase 1A in tobacco, which lead to increased salt tolerance and improved growth [45].
Glycolysis and the TCA cycle as sources of carbon, ATP, and reductant
Avoiding ROS production comes at a price, as every alternate route leads to a decreased ATP production by mitochondrial ATP synthase. Both normal and alternate ETCs were active based on our data, which brings a question of the potential source(s) of reductant for these ETCs and source(s) of ATP that would supplement ATP lost due to the potentially high activities of the alternate routes for electrons. Most glycolytic genes did not show major steady-state transcript differences during freezing acclimation, though persistent up-regulation was observed, suggesting that glycolysis was active. In plants, glycolysis is regulated at the transcriptional and allosteric levels and in a complex way due to metabolic flexibility [33,34,64]. In situations when ATP cannot be easily produced in heterotrophic systems (e.g., during hypoxia, chilling, or phosphate starvation), pyrophosphate- instead of ATP-utilizing enzyme forms are used to drive glycolysis [33,34,65,66]. One of the genes encoding phosphofructokinase was up-regulated up to 5.2-fold during winter hardening. Up-regulation of a pyrophosphate-dependent phosphofructokinase isoform suggests the possibility of conservation of ATP during freezing acclimation, similar to phosphate starvation stress. To maximize ATP production, alternate glycolytic steps involving substrate-level phosphorylation are preferentially utilized. The conversion of glyceraldehyde-3P to 3-phosphoglycerate can be done in two ways by: (i) non-phosphorylating glyceraldehyde-3P dehydrogenase in a single step to only produce NADPH and (ii) phosphoglycerate kinase and phosphorylating glyceraldehyde-3P dehydrogenase in two steps involving NADH and ATP production [34]. The phosphorylating glyceraldehyde-3P dehydrogenase was moderately, but consistently up-regulated, while the kinase showed a consistent, less than 1.5-fold down-regulation, suggesting that the two-step reaction producing both the reductant and ATP could be active. ATP production by substrate-level phosphorylation appears to be active in general – three transcripts coding for pyruvate kinase showed varying, but consistent up-regulation. Pyruvate kinase catalyzes a key step regulating glycolysis and pyruvate is a known allosteric regulator of a number of glycolytic enzymes including pyruvate kinase, activity of which is induced during hypoxia when glycolysis is the major source of ATP [33,34]. Based on these observations, it appears that glycolytic routes conserving or producing ATP are preferentially used. This is in an agreement with the global activation of alternative ETC. routes. As such, glycolysis could be a source of ATP supplementing lower ATP levels obtained from respiration and could help with high-energy cofactor balancing.
In conventional glycolysis, pyruvate kinase converts phosphoenolpyruvate to pyruvate and ATP is made. However, mitochondria could oxidize substrates other than pyruvate, such as malate [33], during winter hardening. In this scenario, the pyruvate kinase step is bypassed by three consecutive steps with no net NADH and ATP production or consumption (Figure 4). However, this bypass allows NADH from the cytosol to be shuttled inside mitochondria where it can be used by the conventional or alternative ETCs. First, cytosolic phosphoenolpyruvate is carboxylated to oxaloacetate by phosphoenolpyruvate carboxylase, a highly regulated enzyme whose activity increases during phosphate starvation [67]–[70]. Second, oxaloacetate is reduced to malate by cytosolic NADH-utilizing malate dehydrogenase, and malate is transported to mitochondria. The dicarboxylate/tricarboxylate transporter DTC encoded by the Arabidopsis At5g19760 gene ortholog that is capable of transporting malate to mitochondria [71] showed a consistent 5-fold up-regulation. Third, malate either enters the TCA cycle or is decarboxylated to pyruvate by NAD-dependent malic enzyme in mitochondria. Most genes encoding the enzymes of the TCA cycle showed a moderate up-regulation or no significant changes in their steady-state transcript levels, suggesting that the TCA cycle is operational. For example, a putative regulatory subunit of the mitochondrial NAD-dependent isocitrate dehydrogenase III (At4g35650) [72,73] showed a stable 4.4-fold increase in transcript levels in TP2 through TP4. Malic enzyme has been implicated in generating NADH and pyruvate needed for lipid synthesis in heterotrophic and photoheterotrophic systems such as plant seeds [74]–[81].
The physiological significance of this bypass in most in vivo situations is not well understood [33]. However, existing metabolomic and transcriptomic data [82] suggest that the bypass is operational (along with the conventional pyruvate kinase step) during freezing acclimation. Malate levels increased significantly at TP1 and reached the levels that were nearly 3-fold higher than baseline later in the time course. Two genes encoding phosphoenolpyruvate carboxylase and one gene encoding malic enzyme were moderately up-regulated in response to freezing acclimation, while cytosolic malate dehydrogenase transcript levels were not significantly different in all TPs. However, the Sitka spruce homolog of the Arabidopsis At2g13560-encoded NAD-dependent malic enzyme 1, which is allosterically activated by fumarate [83], showed a 9- to 11-fold up-regulation in its transcript levels during freezing acclimation and could drive significant fluxes through this bypass. Low temperature stress was also shown to induce the expression of a gene encoding malic enzyme in maize [24]. To drive efficient conventional and alternative ETCs during winter hardening, this bypass could: (i) shuttle NADH from cytosol to mitochondria, (ii) bring additional carbon from glycolysis to the TCA cycle to stimulate alternative and conventional mitochondrial ETCs, and (iii) generate phosphate for ATP synthesis.
Potential sources of hexoses for glycolysis
The reaction catalyzed by invertase requires 2 ATP molecules to generate fructose-6P, while the net NTP consumption for the sucrose synthase pathway is zero and the latter is up-regulated during hypoxia to save ATP [84]. Both invertases and sucrose synthases showed significant increases in the corresponding transcript levels in Sitka spruce needles during freezing acclimation. However, steady-state sucrose levels did not change significantly. So, if there were an increase in sucrose degradation to provide glucose and fructose for glycolysis, there would be a corresponding increase in sucrose supply to maintain unchanging sucrose levels. Because photosynthetic carbon fixation is close to inactive, one can hypothesize that sucrose would have to be transported from organs that have a great potential to store it or made from other, more condensed form of carbohydrate such as starch that can easily be converted to sucrose.
Starch can accumulate transiently in photosynthetically active cells or be stored for a prolonged period of time in tubers and seeds [85]–[89]. This makes starch an excellent candidate as a storage compound that could be mobilized and used as a source of hexoses during winter hardening. However, the majority of genes associated with starch degradation were down-regulated, while those involved in its biosynthesis were up-regulated, which would make starch present in needles a seemingly unlikely storage form of carbon. Regulation of starch metabolism is complex, as it involves several levels of regulation, including transcriptional, protein phosphorylation, allosteric, and redox regulation, to contribute to the balance between photosynthate production and demand in source and sink tissues [87,90]–[92] and as such, transcript levels alone cannot provide enough information regarding the direction of starch metabolism during winter hardening. Changes in starch levels were not measured during freezing acclimation in Sitka spruce needles [82], but Fisher &Holl [93] showed that starch content progressively decreased in Scots pine needles during the winter, suggesting that this polysaccharide could possibly serve as a storage compound for the winter hardening processes also in Sitka spruce needles despite the up-regulation in starch synthesis genes. However, an increase in starch synthesis was also observed in Arabidopsis leaves exposed to prolonged moderate osmotic stress [94] that lead to similar global gene expression responses involving cell wall remodeling, vesicular transport, and hormonal signaling as in winter hardening [46], but the reason for these increases in starch synthesis remains to be elucidated.
Addressing the question of the storage form of carbon in needles during winter hardening is quite complicated. Starch is a potential candidate, but conifers also accumulate di and triacylglycerols as storage compounds in the needles [93]. Unfortunately these compounds were not measured in Sitka spruce needles during winter hardening [82], but both types of lipids showed a similar trend as their levels gradually increased during fall and winter in Scots pine needles, followed by a rapid decrease in spring prior to bud break, suggesting that lipid degradation provides additional carbon and energy for new buds before photosynthesis is fully operational [93]. It is unclear whether these lipids can also be used as a source of energy during winter hardening. Starch levels decreasing and lipid levels increasing in needles during the winter is consistent with the hypothesis that starch rather than lipids are substrates for respiration and active cell-wall remodeling during winter. In addition, degradation of oligo and polysaccharides is more energy effective for producing hexoses for substantial cell-wall remodeling than potential gluconeogenesis (down-regulated during winter hardening) from lipids. A possibility also exists that soluble and insoluble sugars are stored in other parts of the trees and then transported in the form of sucrose to needles to help maintaining active metabolism during prolonged freezing. All these compounds including di and triacylglycerols are present in the sapwood of Scots pine [93], but detailed source-sink relationship studies are needed to confirm the possibility of sucrose transport to needles in winter.
Methods
Plant material, microarray analysis, and metabolite profiling
Plants and gene expression data for this study are described in [47]. Sitka spruce plants originating from Prince Rupert, British Columbia, Canada, were grown in an outdoor, raised-bed garden at Vancouver, British Columbia, Canada. In the fourth growth season (2004), needle tissue from plants were sampled for RNA extraction on August 30, October 18, November 22, December 1, and December 13. Expression changes associated with these time points are referred to as TP1 for the ratio of October 18:August 30, TP2 for the ratio of November 22:August 30, and so on. Microarray analysis was performed using a 21.8K spruce cDNA microarray. The slides were scanned and quantified using ImaGene software (BioDiscovery, Inc., El Segundo, CA), and subsequently normalized using variance stabilizing normalization. Fold-changes and probability values for each cDNA clone were calculated using a linear model in which the normalized intensity values in the Cy3 and Cy5 channels were adjusted for dye and array effects [47]. In addition to transcriptome analysis, data on soluble metabolites quantified by gas chromatography–mass spectrometry (GC-MS) were taken from [82] for the same samples [47]. All statistical analyses were carried out using the R statistical package (http://www.r-project.org).
Metabolic mapman
The MapMan tool enables the classification, statistical analysis, and visualization of transcripts and metabolites into hierarchical categories (known as bins) without the redundancies encountered when using other ontologies [95]. A modification to MapMan that provides a view of multiple instances of a bin simultaneously, e.g., to enable visualization of time courses, was implemented.
BEACON
Beacon is a software and database system designed for the construction, editing, annotation, and storage of signal transduction pathways. For the representation of pathway entities, Beacon implements a graphical language known as the Activity Flow (AF) language, which is part of a set of standardized graphical languages defined in the Systems Biology Graphical Notation project [96]. The AF language portrays pathways at the level of biological activities, phenotypes, and perturbations, with a particular focus on the influences that these entities have on one another in a given pathway. The language is of particular use in the representation of signal transduction pathways due to the flexibility inherent in these viewpoints of biological entities. The Beacon Pathway Editor, specifically, refers to the graphical user interface component for the construction, editing, and annotation of signal transduction pathways using the AF graphical language. A beta version of the Beacon Pathway Editor has been released to a select group of users and is expected to be available to the public in the fall of this year. Once completed, the Beacon Pathway Editor will be extended to allow for the simulation of user-constructed pathways, enabling the possibility for “what if” analyses by manipulating the states of biological activities and perturbations in a given pathway. The database used to store Beacon pathways is also in development, with one eventual goal being the creation of an inference engine to infer new connections among pathway entities based on the information contained in all stored pathways.
ALPINE
The Automated Layout Pipeline for Inferred NEtworks (ALPINE) tool is an in-house project that is currently in the early stages of development. The purpose of the tool is to make the generation of inferred gene association networks, the filtering of those networks to include only those genes that are significant in a user-provided gene expression data set, and the layout of those networks based on subcellular localization annotations an entirely automated process. At present, the tool is implemented as a plugin for the Cytoscape [97] visualization environment to act as an intermediary between the existing GeneMANIA and Mosaic Cytoscape plugins. The GeneMANIA plugin allows users to enter a set of query genes and to generate an inferred gene association network for a variety of association types (physical interactions, genetic interactions, co-expression associations, and more) [98]. The Mosaic plugin allows users to partition, color, and layout existing networks based on GO annotations and other network attributes [99]. The ALPINE plugin provides a graphical user interface that allows the user to set the required parameters for network inference via GeneMANIA and a network layout via Mosaic. Additionally, the user interface allows the user to load a gene expression data set, and to filter the generated network to include only those genes with significant expression values in a minimum number of time points specified by the user. The ultimate result is that, once the user has entered the required parameters, the tasks of network inference, filtering, and layout are handled automatically by the ALPINE tool.
For the networks generated using ALPINE in the current paper, we required a greater specificity in the subcellular localizations annotated to each gene than those provided by the Mosaic portion of ALPINE. In order to achieve a greater specificity, the resulting Cytoscape networks were uploaded to the Arabidopsis Interactions Viewer [100], one of many available tools included in the Bio-Array Resource for Plant Biology collection, where more specific subcellular localizations, based on data from the Arabidopsis Subcellular Database, were annotated to each gene and a layout was performed to group genes based on subcellular localizations. Optional use of the Arabidopsis Interactions Viewer for subcellular localization annotations is currently not included in the ALPINE tool; however, it is possible that the tool may include such an option before it is released for public use.
Abbreviations
ABI: Abscisic acid insensitive; ADP: Adenosine diphosphate; AF: Activity flow; AGPase: ADP glucose pyrophosphorylase; ALPINE: Automated layout pipeline for inferred networks; ATP: Adenosine triphosphate; cDNA: Complementary deoxyribonucleic acid; DTC: Dicarboxylate/tricarboxylate transporter; ER: Endoplasmic reticulum; ETC: Electron transport chain; GC-MS: Gas chromatography–mass spectrometry; GDP: Guanosine diphosphate; GR: Glutathione reductase; GO: Gene ontology; LPD: Dihydrolipoamide dehydrogenase; NAD(P)H: Reduced nicotinamide adenine dinucleotide (phosphate); NTP: Nucleotide triphosphate; PDI: Protein disulfide isomerases; PTOX: Plastidic terminal oxidase; RNA: Ribonucleic acid; ROS: Reactive oxygen species; SUS: Sucrose synthase; tAPX: Thylakoid-bound ascorbate peroxidase; TCA: Citric acid cycle; TP: Time point.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
EC, RG, JAH, and LSH developed the concept of this computational study. EC, RG, and JAH interpreted and discussed biological findings and concepts of the computationally derived results. EC prepared Figure 4. CK, HS, and EM performed all computational analyses under the supervision of RG and LSH and prepared all other figures and tables. All authors reviewed and approved the final manuscript.
Supplementary Material
TCA and Electron Transport. Raw data file showing MapMan bin, Arabidopsis thaliana homolog ID, annotations, time point expression, and location.
Antioxidants. Raw data file showing MapMan bin, Arabidopsis thaliana homolog ID, annotations, time point expression, and location.
Starch and sucrose metabolism. Raw data file showing MapMan bin, Arabidopsis thaliana homolog ID, annotations, time point expression, and location.
Contributor Information
Eva Collakova, Email: collakov@vt.edu.
Curtis Klumas, Email: curtisk@vt.edu.
Haktan Suren, Email: hsuren@vt.edu.
Elijah Myers, Email: esm2310@vt.edu.
Lenwood S Heath, Email: heath@vt.edu.
Jason A Holliday, Email: jah1@vt.edu.
Ruth Grene, Email: grene@vt.edu.
Acknowledgements
This work was supported by NSF ABI1062472.
References
- Ruttink T, Arend M, Morreel K, Storme V, Rombauts S, Fromm J, Bhalerao RP, Boerjan W, Rohde A. A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell. 2007;19(8):2370–2390. doi: 10.1105/tpc.107.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druart N, Johansson A, Baba K, Schrader J, Sjodin A, Bhalerao RR, Resman L, Trygg J, Moritz T, Bhalerao RP. Environmental and hormonal regulation of the activity-dormancy cycle in the cambial meristem involves stage-specific modulation of transcriptional and metabolic networks. Plant J. 2007;50(4):557–573. doi: 10.1111/j.1365-313X.2007.03077.x. [DOI] [PubMed] [Google Scholar]
- Joosen RVL, Lammers M, Balk PA, Bronnum P, Konings M, Perks M, Stattin E, Van Wordragen MF, van der Geest AHM. Correlating gene expression to physiological parameters and environmental conditions during cold acclimation of Pinus sylvestris, identification of molecular markers using cDNA microarrays. Tree Physiol. 2006;26(10):1297–1313. doi: 10.1093/treephys/26.10.1297. [DOI] [PubMed] [Google Scholar]
- Mohamed R, Wang CT, Ma C, Shevchenko O, Dye SJ, Puzey JR, Etherington E, Sheng XY, Meilan R, Strauss SH. Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. 2010;62(4):674–688. doi: 10.1111/j.1365-313X.2010.04185.x. [DOI] [PubMed] [Google Scholar]
- Ruonala R, Rinne PLH, Kangasjarvi J, van der Schoot C. CENL1 expression in the rib meristem affects stem elongation and the transition to dormancy in Populus. Plant Cell. 2008;20(1):59–74. doi: 10.1105/tpc.107.056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez LMG, El Kayal W, Ju CJT, Allen CCG, King-Jones S, Cooke JEK. Integrated transcriptomic and proteomic profiling of white spruce stems during the transition from active growth to dormancy. Plant Cell Environ. 2012;35(4):682–701. doi: 10.1111/j.1365-3040.2011.02444.x. [DOI] [PubMed] [Google Scholar]
- Bohlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312(5776):1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- Oquist G, Huner NPA. Photosynthesis of overwintering evergreen plants. Annu Rev Plant Biol. 2003;54:329–355. doi: 10.1146/annurev.arplant.54.072402.115741. [DOI] [PubMed] [Google Scholar]
- Scheller HV, Haldrup A. Photoinhibition of photosystem I. Planta. 2005;221(1):5–8. doi: 10.1007/s00425-005-1507-7. [DOI] [PubMed] [Google Scholar]
- Savitch LV, Ivanov AG, Krol M, Sprott DP, Oquist G, Huner NPA. Regulation of energy partitioning and alternative electron transport pathways during cold acclimation of lodgepole pine is oxygen dependent. Plant Cell Physiol. 2010;51(9):1555–1570. doi: 10.1093/pcp/pcq101. [DOI] [PubMed] [Google Scholar]
- Adams WW, Zarter CR, Ebbert V, Demmig-Adams B. Photoprotective strategies of overwintering evergreens. Bioscience. 2004;54(1):41–49. doi: 10.1641/0006-3568(2004)054[0041:PSOOE]2.0.CO;2. [DOI] [Google Scholar]
- Sveshnikov D, Ensminger I, Ivanov AG, Campbell D, Lloyd J, Funk C, Huner NPA, Oquist G. Excitation energy partitioning and quenching during cold acclimation in Scots pine. Tree Physiol. 2006;26(3):325–336. doi: 10.1093/treephys/26.3.325. [DOI] [PubMed] [Google Scholar]
- Tanaka A. Photosynthetic activity in winter needles of the evergreen tree Taxus cuspidata at low temperatures. Tree Physiol. 2007;27(5):641–648. doi: 10.1093/treephys/27.5.641. [DOI] [PubMed] [Google Scholar]
- Ivanov AG, Sane PV, Hurry V, Oquist G, Huner NPA. Photosystem II reaction centre quenching: mechanisms and physiological role. Photosynth Res. 2008;98(1–3):565–574. doi: 10.1007/s11120-008-9365-3. [DOI] [PubMed] [Google Scholar]
- Savitch LV, Leonardos ED, Krol M, Jansson S, Grodzinski B, Huner NPA, Oquist G. Two different strategies for light utilization in photosynthesis in relation to growth and cold acclimation. Plant Cell Environ. 2002;25(6):761–771. doi: 10.1046/j.1365-3040.2002.00861.x. [DOI] [Google Scholar]
- Verhoeven A, Osmolak A, Morales P, Crow J. Seasonal changes in abundance and phosphorylation status of photosynthetic proteins in eastern white pine and balsam fir. Tree Physiol. 2009;29(3):361–374. doi: 10.1093/treephys/tpn031. [DOI] [PubMed] [Google Scholar]
- Ivanov AG, Sane PV, Krol M, Gray GR, Balseris A, Savitch LV, Oquist G, Huner NPA. Acclimation to temperature and irradiance modulates PSII charge recombination. FEBS Lett. 2006;580(11):2797–2802. doi: 10.1016/j.febslet.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Ivanov AG, Krol M, Sveshnikov D, Malmberg G, Gardestrom P, Hurry V, Oquist G, Huner NPA. Characterization of the photosynthetic apparatus in cortical bark chlorenchyma of Scots pine. Planta. 2006;223(6):1165–1177. doi: 10.1007/s00425-005-0164-1. [DOI] [PubMed] [Google Scholar]
- Carol P, Stevenson D, Bisanz C, Breitenbach J, Sandmann G, Mache R, Coupland G, Kuntz M. Mutations in the Arabidopsis gene immutans cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell. 1999;11(1):57–68. doi: 10.1105/tpc.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DY, Wright DA, Wetzel C, Voytas DF, Rodermel S. The immutans variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell. 1999;11(1):43–55. doi: 10.1105/tpc.11.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier G, Cournac L. Chlororespiration. Annu Rev Plant Biol. 2002;53:523–550. doi: 10.1146/annurev.arplant.53.100301.135242. [DOI] [PubMed] [Google Scholar]
- Sun X, Wen T. Physiological roles of plastid terminal oxidase in plant stress responses. Journal Biosci. 2011;36(5):951–956. doi: 10.1007/s12038-011-9161-7. [DOI] [PubMed] [Google Scholar]
- Rumeau D, Peltier G, Cournac L. Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 2007;30(9):1041–1051. doi: 10.1111/j.1365-3040.2007.01675.x. [DOI] [PubMed] [Google Scholar]
- Weber AP, Linka N. Connecting the plastid: transporters of the plastid envelope and their role in linking plastidial with cytosolic metabolism. Annu Rev Plant Biol. 2011;62:53–77. doi: 10.1146/annurev-arplant-042110-103903. [DOI] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48(12):909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Robson CA, Vanlerberghe GC. Transgenic plant cells lacking mitochondrial alternative oxidase have increased susceptibility to mitochondria-dependent and -independent pathways of programmed cell death. Plant Physiol. 2002;129(4):1908–1920. doi: 10.1104/pp.004853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VK, Wood SM, Knowles VL, Plaxton WC. Phosphite accelerates programmed cell death in phosphate-starved oilseed rape (Brassica napus) suspension cell cultures. Planta. 2003;218(2):233–239. doi: 10.1007/s00425-003-1088-2. [DOI] [PubMed] [Google Scholar]
- Andronis EA, Roubelakis-Angelakis KA. Short-term salinity stress in tobacco plants leads to the onset of animal-like PCD hallmarks in planta in contrast to long-term stress. Planta. 2010;231(2):437–448. doi: 10.1007/s00425-009-1060-x. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Mittler R. Reactive oxygen species and temperature stresses: A delicate balance between signaling and destruction. Physiol Plant. 2006;126(1):45–51. doi: 10.1111/j.0031-9317.2005.00582.x. [DOI] [Google Scholar]
- Krishnamurthy KV, Krishnaraj R, Chozhavendan R, Christopher FS. The programme of cell death in plants and animals - A comparison. Current Sci. 2000;79(9):1169–1181. [Google Scholar]
- Sweetlove LJ, Lytovchenko A, Morgan M, Nunes-Nesi A, Taylor NL, Baxter CJ, Eickmeier I, Fernie AR. Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc Nat Acad Sci US Am. 2006;103(51):19587–19592. doi: 10.1073/pnas.0607751103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira FTS, Sassaki FT, Maia IG. Arabidopsis thaliana Uncoupling Proteins (AtUCPs): insights into gene expression during development and stress response and epigenetic regulation. J Bioenerg Biomembr. 2011;43(1):71–79. doi: 10.1007/s10863-011-9336-9. [DOI] [PubMed] [Google Scholar]
- Plaxton WC, Podesta FE. The functional organization and control of plant respiration. Critical Rev Plant Sci. 2006;25(2):159–198. doi: 10.1080/07352680600563876. [DOI] [Google Scholar]
- van Dongen JT, Gupta KJ, Ramirez-Aguilar SJ, Araujo WL, Nunes-Nesi A, Fernie AR. Regulation of respiration in plants: A role for alternative metabolic pathways. J Plant Physiol. 2011;168(12):1434–1443. doi: 10.1016/j.jplph.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Fiorani F, Umbach AL, Siedow JN. The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature. A study of Arabidopsis AOX1a transgenic plants. Plant Physiol. 2005;139(4):1795–1805. doi: 10.1104/pp.105.070789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugie A, Naydenov N, Mizuno N, Nakamura C, Takumi S. Overexpression of wheat alternative oxidase gene Waox1a alters respiration capacity and response to reactive oxygen species under low temperature in transgenic Arabidopsis. Genes Gen Sys. 2006;81(5):349–354. doi: 10.1266/ggs.81.349. [DOI] [PubMed] [Google Scholar]
- Matos AR, Hourton-Cabassa C, Cicek D, Reze N, Arrabaca JD, Zachowski A, Moreau F. Alternative oxidase involvement in cold stress response of Arabidopsis thaliana fad2 and FAD3+ cell suspensions altered in membrane lipid composition. Plant Cell Physiol. 2007;48(6):856–865. doi: 10.1093/pcp/pcm061. [DOI] [PubMed] [Google Scholar]
- Wang HH, Huang JJ, Liang XL, Bi YR. Involvement of hydrogen peroxide, calcium, and ethylene in the induction of the alternative pathway in chilling-stressed Arabidopsis callus. Planta. 2012;235(1):53–67. doi: 10.1007/s00425-011-1488-7. [DOI] [PubMed] [Google Scholar]
- Popov VN. Possible role of free oxidation processes in the regulation of reactive oxygen species production in plant mitochondria. Biochem Soc Transact. 2003;31:1316–1317. doi: 10.1042/BST0311316. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012;35(2):259–270. doi: 10.1111/j.1365-3040.2011.02336.x. [DOI] [PubMed] [Google Scholar]
- Cvetkovska M, Vanlerberghe GC. Alternative oxidase modulates leaf mitochondrial concentrations of superoxide and nitric oxide. New Phytol. 2012;195(1):32–39. doi: 10.1111/j.1469-8137.2012.04166.x. [DOI] [PubMed] [Google Scholar]
- Liao YWK, Shi K, Fu LJ, Zhang S, Li X, Dong DK, Jiang YP, Zhou YH, Xia XJ, Liang WS. The reduction of reactive oxygen species formation by mitochondrial alternative respiration in tomato basal defense against TMV infection. Planta. 2012;235(2):225–238. doi: 10.1007/s00425-011-1483-z. [DOI] [PubMed] [Google Scholar]
- Elhafez D, Murcha MW, Clifton R, Soole KL, Day DA, Whelan J. Characterization of mitochondrial alternative NAD(P)H dehydrogenases in arabidopsis: Intraorganelle location and expression. Plant Cell Physiol. 2006;47(1):43–54. doi: 10.1093/pcp/pci221. [DOI] [PubMed] [Google Scholar]
- Moore CS, Cook-Johnson RJ, Rudhe C, Whelan J, Day DA, Wiskich JT, Soole KL. Identification of AtNDI1, an internal non-phosphorylating NAD(P)H dehydrogenase in arabidopsis mitochondria. Plant Physiol. 2003;133(4):1968–1978. doi: 10.1104/pp.103.029363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Barthet M, Melino V, Smith P, Day D, Soole K. Alterations in the Mitochondrial Alternative NAD(P)H Dehydrogenase NDB4 Lead to Changes in Mitochondrial Electron Transport Chain Composition, Plant Growth and Response to Oxidative Stress. Plant Cell Physiol. 2011;52(7):1222–1237. doi: 10.1093/pcp/pcr073. [DOI] [PubMed] [Google Scholar]
- Grene R, Klumas C, Suren H, Yang K, Collakova E, Myers ES, Heath LS, Holiday J. Mining and visualization of microarray and metabolomic data reveal extensive cell wall remodeling during winter hardening in Sitka spruce (Picea sitchensis) Plant Sci: Front; 2012. p. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday JA, Ralph SG, White R, Bohlmann J, Aitken SN. Global monitoring of autumn gene expression within and among phenotypically divergent populations of Sitka spruce (Picea sitchensis) New Phytol. 2008;178(1):103–122. doi: 10.1111/j.1469-8137.2007.02346.x. [DOI] [PubMed] [Google Scholar]
- Laureau C, DEP R, Latouche G, Moreno-Chacon M, Finazzi G, Kuntz M, Cornic G, Streb P. Plastid terminal oxidase (PTOX) has the potential to act as a safety valve for excess excitation energy in the alpine plant species Ranunculus glacialis L. Plant Cell Environ. 2013. In press. [DOI] [PubMed]
- Ivanov AG, Rosso D, Savitch LV, Stachula P, Rosembert M, Oquist G, Hurry V, Huner NP. Implications of alternative electron sinks in increased resistance of PSII and PSI photochemistry to high light stress in cold-acclimated Arabidopsis thaliana. Photosynth Res. 2012;113(1–3):191–206. doi: 10.1007/s11120-012-9769-y. [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F, Noctor G. Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J Exp Bot. 2010;61(15):4197–4220. doi: 10.1093/jxb/erq282. [DOI] [PubMed] [Google Scholar]
- Jannat R, Uraji M, Hossain MA, Islam MM, Nakamura Y, Mori IC, Murata Y. Catalases negatively regulate methyl jasrnonate signaling in guard cells. J Plant Physiol. 2012;169(10):1012–1016. doi: 10.1016/j.jplph.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Xing SP, Rosso MG, Zachgo S. ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development. 2005;132(7):1555–1565. doi: 10.1242/dev.01725. [DOI] [PubMed] [Google Scholar]
- Xing S, Zachgo S. ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 2008;53(5):790–801. doi: 10.1111/j.1365-313X.2007.03375.x. [DOI] [PubMed] [Google Scholar]
- Li S, Lauri A, Ziemann M, Busch A, Bhave M, Zachgo S. Nuclear Activity of ROXY1, a Glutaredoxin Interacting with TGA Factors, Is Required for Petal Development in Arabidopsis thaliana. Plant Cell. 2009;21(2):429–441. doi: 10.1105/tpc.108.064477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A, Hager J, Chaouch S, Queval G, Han Y, Taconnat L, Saindrenan P, Gouia H, Issakidis-Bourguet E, Renou JP. Arabidopsis GLUTATHIONE REDUCTASE1 Plays a Crucial Role in Leaf Responses to Intracellular Hydrogen Peroxide and in Ensuring Appropriate Gene Expression through Both Salicylic Acid and Jasmonic Acid Signaling Pathways. Plant Physiol. 2010;153(3):1144–1160. doi: 10.1104/pp.110.153767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan M, Feng HL, Wang LY, Li D, Meng QW. Overexpression of thylakoidal ascorbate peroxidase shows enhanced resistance to chilling stress in tomato. J Plant Physiol. 2012;169(9):867–877. doi: 10.1016/j.jplph.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Maruta T, Tanouchi A, Tamoi M, Yabuta Y, Yoshimura K, Ishikawa T, Shigeoka S. Arabidopsis Chloroplastic Ascorbate Peroxidase Isoenzymes Play a Dual Role in Photoprotection and Gene Regulation under Photooxidative Stress. Plant Cell Physiol. 2010;51(2):190–200. doi: 10.1093/pcp/pcp177. [DOI] [PubMed] [Google Scholar]
- Wang J, Rajakulendran N, Amirsadeghi S, Vanlerberghe GC. Impact of mitochondrial alternative oxidase expression on the response of Nicotiana tabacum to cold temperature. Physiol Plant. 2011;142(4):339–351. doi: 10.1111/j.1399-3054.2011.01471.x. [DOI] [PubMed] [Google Scholar]
- Djajanegara I, Holtzapffel R, Finnegan PM, Hoefnagel MHN, Berthold DA, Wiskich JT, Day DA. A single amino acid change in the plant alternative oxidase alters the specificity of organic acid activation. FEBS Lett. 1999;454(3):220–224. doi: 10.1016/S0014-5793(99)00808-X. [DOI] [PubMed] [Google Scholar]
- Giraud E, Van Aken O, Ho LHM, Whelan J. The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 2009;150(3):1286–1296. doi: 10.1104/pp.109.139782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach AL, Gonzalez-Meler MA, Sweet CR, Siedow JN. Activation of the plant mitochondrial alternative oxidase: insights from site-directed mutagenesis. Biochimica Et Biophysica Acta-Bioenergetics. 2002;1554(1–2):118–128. doi: 10.1016/s0005-2728(02)00219-0. [DOI] [PubMed] [Google Scholar]
- Oliver SN, Lunn JE, Urbanczyk-Wochniak E, Lytovchenko A, Van Dongen JT, Faix B, Schmalzlin E, Fernie AR, Geigenberger P. Decreased expression of cytosolic pyruvate kinase in potato tubers leads to a decline in pyruvate resulting in an in Vivo repression of the alternative oxidase. Plant Physiol. 2008;148(3):1640–1654. doi: 10.1104/pp.108.126516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabalza A, Van Dongen JT, Froehlich A, Oliver SN, Faix B, Gupta KJ, Schmalzlin E, Igal M, Orcaray L, Royuela M. Regulation of respiration and fermentation to control the plant internal oxygen concentration. Plant Physiol. 2009;149(2):1087–1098. doi: 10.1104/pp.108.129288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136(1):2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Sonnewald U, Blemelt S. Characterisation of the ATP-dependent phosphofructokinase gene family from Arabidopsis thaliana. FEBS Lett. 2007;581(13):2401–2410. doi: 10.1016/j.febslet.2007.04.060. [DOI] [PubMed] [Google Scholar]
- Winkler C, Delvos B, Martin W, Henze K. Purification, microsequencing and cloning of spinach ATP-dependent phosphofructokinase link sequence and function for the plant enzyme. FEBS J. 2007;274(2):429–438. doi: 10.1111/j.1742-4658.2006.05590.x. [DOI] [PubMed] [Google Scholar]
- Chollet R, Vidal J, Oleary MH. Phosphoenolpyruvate carboxylase: A ubiquitous, highly regulated enzyme in plants. Ann Rev Plant Physiol Plant Mol Biol. 1996;47:273–298. doi: 10.1146/annurev.arplant.47.1.273. [DOI] [PubMed] [Google Scholar]
- Moraes TF, Plaxton WC. Purification and characterization of phosphoenolpyruvate carboxylase from Brassica napus (rapeseed) suspension cell cultures - Implications for phosphoenolpyruvate carboxylase regulation during phosphate starvation, and the integration of glycolysis with nitrogen assimilation. Eur J Biochem. 2000;267(14):4465–4476. doi: 10.1046/j.1432-1327.2000.01495.x. [DOI] [PubMed] [Google Scholar]
- O’Leary B, Park J, Plaxton WC. The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem J. 2011;436:15–34. doi: 10.1042/BJ20110078. [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Thomas M, Vidal J. From enzyme activity to plant biotechnology: 30 years of research on phosphoenolpyruvate carboxylase. Plant Physiol Biochem. 2003;41(6–7):533–539. [Google Scholar]
- Picault N, Palmieri L, Pisano I, Hodges M, Palmieri F. Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria - Bacterial expression, reconstitution, functional characterization, and tissue distribution. J Biol Chem. 2002;277(27):24204–24211. doi: 10.1074/jbc.M202702200. [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell. 2004;16(1):241–256. doi: 10.1105/tpc.016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Behal RH, Oliver DJ. Characterization of a mutation in the IDH-II subunit of the NAD(+)-dependent isocitrate dehydrogenase from Arabidopsis thaliana. Plant Sci. 2004;166(4):983–988. doi: 10.1016/j.plantsci.2003.12.012. [DOI] [Google Scholar]
- Alonso AP, Goffman FD, Ohlrogge JB, Shachar-Hill Y. Carbon conversion efficiency and central metabolic fluxes in developing sunflower (Helianthus annuus L.) embryos. Plant J. 2007;52(2):296–308. doi: 10.1111/j.1365-313X.2007.03235.x. [DOI] [PubMed] [Google Scholar]
- Alonso AP, Dale VL, Shachar-Hill Y. Understanding fatty acid synthesis in developing maize embryos using metabolic flux analysis. Metab Eng. 2010;12(5):488–497. doi: 10.1016/j.ymben.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Allen DK, Ohlrogge JB, Shachar-Hill Y. The role of light in soybean seed filling metabolism. Plant J. 2009;58(2):220–234. doi: 10.1111/j.1365-313X.2008.03771.x. [DOI] [PubMed] [Google Scholar]
- Allen DK, Libourel IGL, Shachar-Hill Y. Metabolic flux analysis in plants: coping with complexity. Plant Cell Environ. 2009;32(9):1241–1257. doi: 10.1111/j.1365-3040.2009.01992.x. [DOI] [PubMed] [Google Scholar]
- Junker BH, Lonien J, Heady LE, Rogers A, Schwender J. Parallel determination of enzyme activities and in vivo fluxes in Brassica napus embryos grown on organic or inorganic nitrogen source. Phytochem. 2007;68(16–18):2232–2242. doi: 10.1016/j.phytochem.2007.03.032. [DOI] [PubMed] [Google Scholar]
- Lonien J, Schwender J. Analysis of metabolic flux phenotypes for two arabidopsis mutants with severe impairment in seed storage lipid synthesis. Plant Physiol. 2009;151(3):1617–1634. doi: 10.1104/pp.109.144121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VV, Sriram G, Fulton DB, Zhou R, Westgate ME, Shanks JV. Metabolic flux maps comparing the effect of temperature on protein and oil biosynthesis in developing soybean cotyledons. Plant Cell Environ. 2008;31(4):506–517. doi: 10.1111/j.1365-3040.2008.01781.x. [DOI] [PubMed] [Google Scholar]
- Sriram G, Iyer VV, Fulton DB, Shanks JV. Identification of hexose hydrolysis products in metabolic flux analytes: A case study of levulinic acid in plant protein hydrolysate. Metab Eng. 2007;9(5–6):442–451. doi: 10.1016/j.ymben.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Dauwe R, Holliday JA, Aitken SN, Mansfield SD. Metabolic dynamics during autumn cold acclimation within and among populations of Sitka spruce (Picea sitchensis) New Phytol. 2012;194(1):192–205. doi: 10.1111/j.1469-8137.2011.04027.x. [DOI] [PubMed] [Google Scholar]
- Tronconi MA, Wheeler MCG, Drincovich ME, Andreo CS. Differential fumarate binding to Arabidopsis NAD(+)-malic enzymes 1 and-2 produces an opposite activity modulation. Biochimie. 2012;94(6):1421–1430. doi: 10.1016/j.biochi.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Bologa KL, Fernie AR, Leisse A, Loureiro ME, Geigenberger P. A bypass of sucrose synthase leads to low internal oxygen and impaired metabolic performance in growing potato tubers. Plant Physiol. 2003;132(4):2058–2072. doi: 10.1104/pp.103.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM. The diurnal metabolism of leaf starch. Biochem J. 2007;401:13–28. doi: 10.1042/BJ20061393. [DOI] [PubMed] [Google Scholar]
- Weise SE, Van Wijk KJ, Sharkey TD. The role of transitory starch in C-3, CAM, and C-4 metabolism and opportunities for engineering leaf starch accumulation. J Exp Bot. 2011;62(9):3109–3118. doi: 10.1093/jxb/err035. [DOI] [PubMed] [Google Scholar]
- Geigenberger P. Regulation of Starch Biosynthesis in Response to a Fluctuating Environment. Plant Physiol. 2011;155(4):1566–1577. doi: 10.1104/pp.110.170399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelia D, Zeeman SC. Progress in Arabidopsis starch research and potential biotechnological applications. Curr Opin Biotechnol. 2011;22(2):271–280. doi: 10.1016/j.copbio.2010.11.014. [DOI] [PubMed] [Google Scholar]
- Jeon JS, Ryoo N, Hahn TR, Walia H, Nakamura Y. Starch biosynthesis in cereal endosperm. Plant Physiol Biochem. 2010;48(6):383–392. doi: 10.1016/j.plaphy.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Kotting O, Kossmann J, Zeeman SC, Lloyd JR. Regulation of starch metabolism: the age of enlightenment? Curr Opin Biotechnol. 2010;13(3):321–329. doi: 10.1016/j.pbi.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Stitt M, Lunn J, Usadel B. Arabidopsis and primary photosynthetic metabolism - more than the icing on the cake. Plant J Cell Mol Biol. 2010;61(6):1067–1091. doi: 10.1111/j.1365-313X.2010.04142.x. [DOI] [PubMed] [Google Scholar]
- Smith AM, Stitt M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007;30(9):1126–1149. doi: 10.1111/j.1365-3040.2007.01708.x. [DOI] [PubMed] [Google Scholar]
- Fischer C, Holl W. Food reserves of scots pine (pinus-sylvestris l) .1. Seasonal-changes in the carbohydrate and fat reserves of pine needles. Trees-Struct Funct. 1991;5(4):187–195. doi: 10.1007/BF00227524. [DOI] [Google Scholar]
- Skirycz A, De Bodt S, Obata T, De Clercq I, Claeys H, De Rycke R, Andriankaja M, Van Aken O, Van Breusegem F, Fernie AR. Developmental stage specificity and the role of mitochondrial metabolism in the response of arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol. 2010;152(1):226–244. doi: 10.1104/pp.109.148965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species. Maize. Plant Cell Environ. 2009;32(9):1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- Le Novere N, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Demir E, Wegner K, Aladjem MI, Wimalaratne SM. The systems biology graphical notation. Nature Biotechnol. 2009;27(8):735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Gen Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26(22):2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Hanspers K, Kuchinsky A, Salomonis N, Xu D, Pico AR. Mosaic: making biological sense of complex networks. Bioinformatics. 2012;28(14):1943–1944. doi: 10.1093/bioinformatics/bts278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler-Lee J, O’Toole N, Ammar R, Provart NJ, Millar AH, Geisler M. A predicted interactome for Arabidopsis. Plant Physiol. 2007;145(2):317–329. doi: 10.1104/pp.107.103465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TCA and Electron Transport. Raw data file showing MapMan bin, Arabidopsis thaliana homolog ID, annotations, time point expression, and location.
Antioxidants. Raw data file showing MapMan bin, Arabidopsis thaliana homolog ID, annotations, time point expression, and location.
Starch and sucrose metabolism. Raw data file showing MapMan bin, Arabidopsis thaliana homolog ID, annotations, time point expression, and location.