Abstract
FACT (facilitates chromatin transcription) is a histone chaperone that promotes chromatin recovery during transcription, with additional roles in cell differentiation. Although several models of the action of FACT during transcription have been proposed, they remain to be experimentally evaluated. Here we show that human FACT (hFACT) facilitates transcription through chromatin and promotes nucleosome recovery in vitro. FACT action depends on the presence of histone H2A/H2B dimers in the nucleosome. Kinetic analysis suggests that hFACT decreases the lifetime of nonproductive RNA polymerase II (Pol II)–nucleosome complexes and facilitates the formation of productive complexes containing nucleosomal DNA partially uncoiled from the octamer. Taken together, our data suggest that hFACT interacts with DNA-binding surfaces of H2A/H2B dimers, facilitating uncoiling of DNA from the histone octamer. Thus, hFACT–H2A/H2B interactions play a key role in overcoming the nucleosomal barrier by Pol II and promoting nucleosome survival during transcription.
Keywords: elongation, dynamics, DNA uncoiling, mechanism
FACT (facilitates chromatin transcription) is the transcription and replication factor (1, 2) involved in cell differentiation (3), and is also an important target for anticancer drugs (4). Human FACT (hFACT) is a heterodimer protein complex composed of two subunits [suppressor of Ty 16 homolog (Spt16) and structure specific recognition protein 1 (SSRP1)] that has histone chaperone activity (5, 6). FACT stimulates transcript elongation through nucleosomes in vitro (2, 6). In vivo, FACT colocalizes with RNA polymerase II (Pol II) and displays similar kinetics of recruitment and chromosome tracking (7, 8). FACT is also essential for maintenance of chromatin structure during transcript elongation by Pol II (8–11).
Different models have been proposed to describe the mechanism of FACT’s action. FACT can induce global accessibility of nucleosomal DNA without histone H2A/H2B displacement (12, 13) and thus can facilitate action of processive enzymes on DNA. On the other hand, it has been suggested that FACT destabilizes nucleosomes by facilitating dissociation of histone H2A/H2B dimer from nucleosomes, thereby facilitating transcription through chromatin (6). The mechanism of FACT action during transcription remains to be experimentally evaluated.
In this study, we systematically examined the effect of FACT on transcription through a nucleosome by Pol II in vitro. Our results show that FACT alleviates nucleosomal pausing, and that the presence of H2A/H2B dimers is required for FACT action. Kinetic studies suggest that the alternating FACT–dimer interactions result in an increased rate of conversion from the nonproductive to productive Pol II–nucleosome complexes and thus facilitate transcription and nucleosome survival.
Results
H2A/H2B Dimers Mediate FACT-Dependent Transcription Through a Nucleosome.
In our experiments, we used yeast Pol II and DNA fragments bearing single nucleosomes assembled on DNA sequences having high affinity for the histone octamer, which can precisely position nucleosomes (14, 15). This experimental system faithfully recapitulates many key characteristics of the mechanism of transcription through chromatin in vivo (16–18), as well as the effect of FACT during Pol II transcription through the nucleosomes (6, 14).
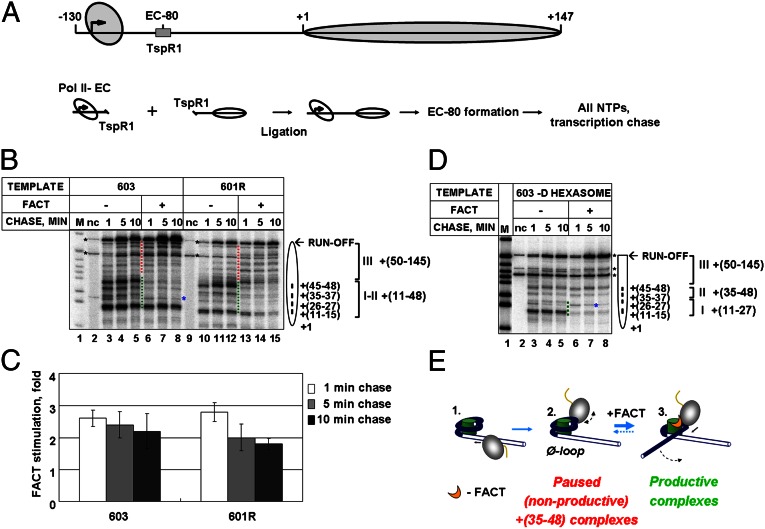
To map the regions of nucleosomal pausing affected by hFACT, we reconstituted nucleosomes on 603 and 601R positioning sequences, allowing efficient transcription through the nucleosomes (14). Pol II elongation complexes (ECs) were assembled using synthetic DNA and RNA oligonucleotides and ligated to DNA fragments containing nucleosomes (Fig. 1A). The nascent RNA was pulse-labeled, and the EC-80 was stalled 80 bp upstream of the promoter–proximal nucleosomal boundary (the number indicates the distance of the active center of the enzyme from this boundary). Then transcription was continued at 150 mM KCl in the presence or absence of hFACT.
Fig. 1.
FACT facilitates transcription through nucleosomes: The role of the promoter-distal H2A/H2B dimer. (A) (Upper) Nucleosomal template for Pol II transcription. (Lower) Experimental approach. (B) 603 and 601R nucleosomes were transcribed by Pol II at 150 mM KCl in the absence or presence of FACT, followed by analysis of pulse-labeled RNA by denaturing PAGE. Pol II pausing regions that are positively or negatively affected by FACT are indicated by green and red dotted lines, respectively. Blue and black asterisks indicate DNA-specific pause and labeled DNA fragments, respectively. nc, no chase. (C) Quantitation of the run-off transcripts (Fig. 1B). Averages from three experiments and SDs are shown. (D) The promoter-distal (D) dimer is required for FACT-dependent transcription through the +(35–48) region. (E) The proposed mechanism of FACT-dependent relief of nucleosomal +(35–48) pausing.
On both nucleosomal templates, major pauses were observed at positions +(11–15), +(26, 27), +(35–37), and +(45–48) bp from the promoter–proximal nucleosome boundary (regions I and II; Fig. 1B and Fig. S1). In the presence of hFACT, all four major pauses were partially relieved, with the lowest effect on the +(11–15) region. Pol II pausing within the +(50–145) region was increased in the presence of hFACT (region III; Fig. 1B and Fig. S1), although this effect is less pronounced in the case of 603 template and could be explained in part by more efficient Pol II progression past the early pauses in the presence of FACT. hFACT similarly affects nucleosomal pausing and the yield of the run-off transcripts on both templates (Fig. 1C).
hFACT is a histone H2A/H2B chaperone (6) and strongly relieves the +(35–48) pausing that is caused in part by the presence of promoter-distal H2A/H2B histone dimer (d-dimer) (15). Therefore, the effect of presence of the d-dimer on hFACT-dependent transcription by Pol II was studied. 603 nucleosomes missing the d-dimer [i.e., 603-D hexasome (15)] were transcribed in the presence or absence of hFACT. In contrast with 603 nucleosomes, the +(11–27) pausing in the hexasomes was relieved more efficiently, whereas the +(35–48) pausing remained unaffected by hFACT (Fig. 1D, Fig. S1C, and Fig. S2). hFACT did not affect transcription through the H3/H4 tetramer (Fig. S3), suggesting that the presence of at least one H2A/H2B dimer is required for hFACT action.
How can the presence of the d-dimer affect the +(35–48) pausing that occurs when the active center of the enzyme is more than 60 bp upstream of the dimer-bound DNA region? It was shown that the +(35–48) pausing is relieved due to uncoiling of the promoter-distal end of nucleosomal DNA from the d-dimer (15). Alternatively, the +(35–48) pausing could be relieved by the removal of the d-dimer from DNA (15). Therefore, to relieve the +(35–48) pausing, FACT could either facilitate removal of the d-dimer from the nucleosome or induce partial uncoiling of promoter-distal DNA end from the octamer. Given that removal of the dimer does not fully eliminate the +(35–48) pausing, and that the presence of the dimer is required for hFACT action (Fig. 1D), the data indicate that hFACT facilitates uncoiling of the nucleosomal DNA from the octamer. We propose that hFACT, like the histone chaperone nucleosome assembly protein 1 (Nap1) (19), facilitates DNA uncoiling by competing with DNA for the positively charged DNA-binding site on the d-dimer (Fig. 1E and Discussion). As expected, after removal of promoter-proximal dimer (P-dimer) FACT efficiently relieved the remaining pausing in the +45 region (Fig. S4).
How can the d-dimer negatively affect relief of the earlier +(11–27) pausing by FACT? This pausing is caused by strong DNA–histone interactions immediately downstream of the transcribing Pol II (20, 21), most likely impeding polymerase-induced uncoiling of nucleosomal DNA from the promoter-proximal H2A/H2B dimer (22). At the same time, the promoter-distal DNA end is not uncoiled before Pol II transcribes past the +35 region (15, 21). Thus, we propose that hFACT affects +(11–27) pausing by competing with DNA for binding to the P-dimer, thereby facilitating DNA uncoiling. In this case, the observed inhibitory effect of the distal dimer on hFACT action is explained by competition between the P- and d-dimers for hFACT binding. In agreement with this idea, an increase in hFACT concentration results in further relief of the +(11–27) pausing.
In summary, the data suggest that hFACT facilitates transcription through the regions I and II by alternatively interacting with H2A/H2B dimers and competing with DNA for binding to the dimers within the histone octamer (Fig. 1E). This model of FACT action makes several predictions. First, because histone–DNA interactions are the primary FACT targets, FACT is expected to facilitate transcription through the nucleosome by a different RNA polymerase (RNAP) that uses a similar mechanism of transcription through chromatin, such as Escherichia coli RNAP (15, 23). Second, regions of nucleosomal DNA that have high affinities to core histones and prevent uncoiling of nucleosomal DNA from the octamer (15) may interfere with FACT action. Third, FACT is expected to increase the rate of conversion from paused (nonproductive) to productive complexes (Fig. 1E) or to decrease the rate of the reverse reaction (i.e., formation of nonproductive complexes). Finally, because intranucleosomal binding of H2A/H2B dimers depends on the presence of nucleosomal DNA, FACT could prevent dissociation of H2A/H2B dimers from the nucleosome by substituting for the DNA that is uncoiled from the dimers during transcription.
To evaluate the model’s first prediction, we transcribed the 603 nucleosomes by E. coli RNAP in the absence or presence of hFACT (Fig. S5). The data suggest that hFACT similarly affects the transcription by different polymerases sharing the Pol II-specific mechanism of transcription through chromatin. This in turn suggests that histone–DNA interactions are the primary targets during hFACT action.
To evaluate whether the DNA regions that prevent uncoiling of promoter-distal nucleosomal DNA from the octamer may interfere with FACT action, we studied the effect of nucleosomal sequences with high affinity to core histones [polar barrier sequences (PBS) (24)] on hFACT action (Fig. S6). These data, together with our previous observations, suggests that PBS sequence limits DNA uncoiling from the octamer and has a dominant effect, preventing further hFACT-dependent DNA uncoiling and transcription.
Disruption of the Dimer–Tetramer Interface Is Not Required for FACT Action.
Our working model (Fig. 1E) suggests that FACT targets the DNA-binding surface of the H2A/H2B dimer(s). An alternative model suggests that FACT can facilitate dissociation of the histone H2A/H2B dimer from nucleosomes and thus facilitate transcription through chromatin (6). According to the former model, disruption or stabilization of the protein–protein interaction interface between each H2A/H2B dimer and the H3/H4 tetramer (e.g., by mutations or protein–protein cross-linking) would not be expected to strongly affect FACT action, but an effect would be predicted by the dimer displacement model.
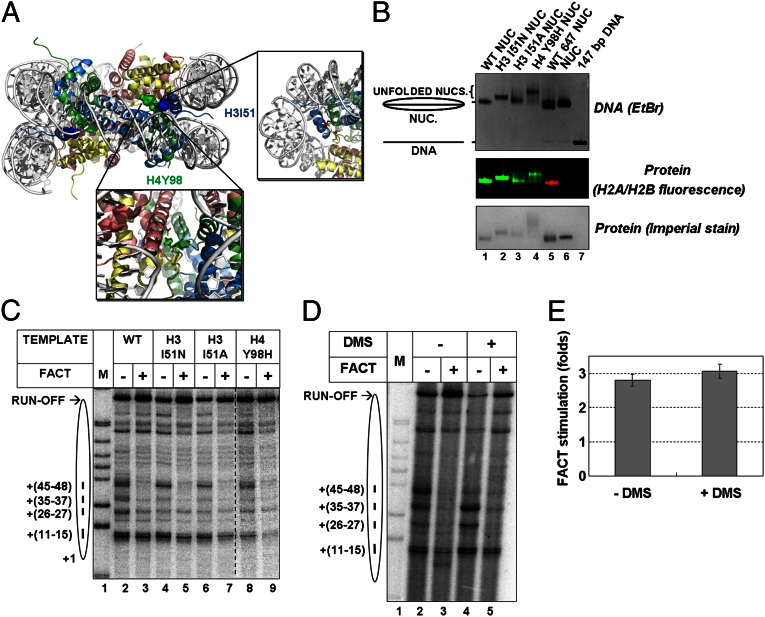
To explore this, we first studied the effect of different mutations in core histones designed to weaken dimer–tetramer interactions (H3-I51N, H3-I51A, and H4-Y98H; Fig. 2A) on dimer–tetramer interactions, overall conformation of nucleosomes, the ability of mutant histones to form histone–histone complexes and nucleosomes, and FACT-dependent transcription. Nucleosomes were prepared from individually refolded H2A/H2B dimers (fluorescently labeled on H2B) and WT or mutant (H3-H4)2 tetramers and were analyzed by native PAGE (Fig. 2B). All combinations of WT and mutant histones (containing H3-I51N, H3-I51A, or H4-Y98H mutations) form stable nucleosomes on the 147-bp 601 DNA fragment. As expected from its role in stabilizing the interaction of the H3 αN helix with the H2A-docking domain, a mutation of isoleucine H3I51 to glutamate generates slower migrating nucleosomes, indicative of a more open nucleosomal structure. The replacement of tyrosine H4Y98 with histidine results in more reduced mobility, suggesting more severe structural distortions in the nucleosome. The nondisruptive replacement of H3I51 with alanine produces only subtle effects. All mutant nucleosomes remain intact during gel electrophoresis (Fig. 2B). The data suggest that at least the H3-I51N and H4-Y98H mutations destabilize the dimer–tetramer interface and induce a change in nucleosome conformation, dictating their slower mobilities in the native gel.
Fig. 2.
Disruption of dimer–tetramer interface is not required for FACT action. (A) Contacting residues in the dimer–tetramer interfaces of the core histone octamer. H2A, H2B, H3, and H4 are shown in yellow, red, blue and green, respectively. (B) 147-bp 601 DNA fragments were assembled into nucleosomes using WT and mutant (H3-I51N, H3-I51A, or H4-Y98H) histones. A point mutant of H2B (H2BT112C) was labeled with Alexa Fluor 488. Nucleosomes were run on 5% native PAGE, scanned on a Typhoon Imager at a wavelength of 520 nm (H2A/H2B fluorescence), and then stained with ethidium bromide (for DNA) or Imperial stain (for protein). (C) Effect of mutations disrupting the dimer–tetramer interface on transcription through chromatin in the absence or presence of FACT, as shown by analysis of pulse-labeled RNA by denaturing PAGE. (D) Effect of cross-linking of histone octamer by DMS on transcription through the nucleosome in the absence or presence of FACT, as shown by analysis of pulse-labeled RNA by denaturing PAGE. (E) The effect of FACT on transcription of intact and cross-linked nucleosomes. Averages from three experiments and SDs are shown.
Presence of each of the mutations results in partial relief of the +26 and +97 pauses at 150 mM KCl and in a stronger +45 pausing at 40 mM KCl (Fig. S7). The overall effect of mutations on nucleosome traversal is moderately positive, with a minor increase in the yield of run-off transcripts at 150 mM KCl (Fig. 2C and Fig. S7). Only the H4-Y98H mutation considerably affected hFACT action, resulting in an ∼25% decrease in the magnitude of transcription stimulation by hFACT, most likely related to the significantly higher level of transcription in the absence of hFACT (Fig. 2C). More efficient transcription in the absence of hFACT could occur owing to easier uncoiling of nucleosomal DNA from the mutant octamer. Although the H3-I51N and H3-I51A mutations clearly affect nucleosome conformation (Fig. 2B) and have a detectable effect on transcription through the nucleosome in the absence of hFACT (Fig. S7), they do not have a strong affect on hFACT action (Fig. 2C).
We next cross-linked core histones in the nucleosome by dimethyl suberimidate (DMS). DMS does not change the charge of histones and thus does not perturb the nucleosome structure. The histone octamer was cross-linked either after or before nucleosome assembly; similar data were obtained in both cases (Fig. 2 and Fig. S8). All histones in the octamer were cross-linked by DMS (Fig. S8B); cross-linking did not affect the mobility of the nucleosomes in a native gel (Fig. S8 A and C). Transcription of intact and cross-linked 603 nucleosomes by Pol II in the absence of FACT produced similar pausing patterns, with exception of the +45 pause, which was reduced on cross-linked templates (Fig. 2D and Fig. S8D). FACT similarly affected pausing on intact and cross-linked nucleosomes (Fig. 2D and Fig. S8D), increasing the yield of run-off transcripts by approximately threefold (Fig. 2E). The data show that cross-linking affects FACT action only minimally, as predicted by the model (Fig. 1E).
In summary, our data indicate that mutations destabilizing the dimer–tetramer interface and cross-linking of all histones in the transcribed nucleosomes minimally affect hFACT action during transcription, suggesting that hFACT does not target the dimer–tetramer interaction interface.
hFACT Facilitates Formation of Productive ECs.
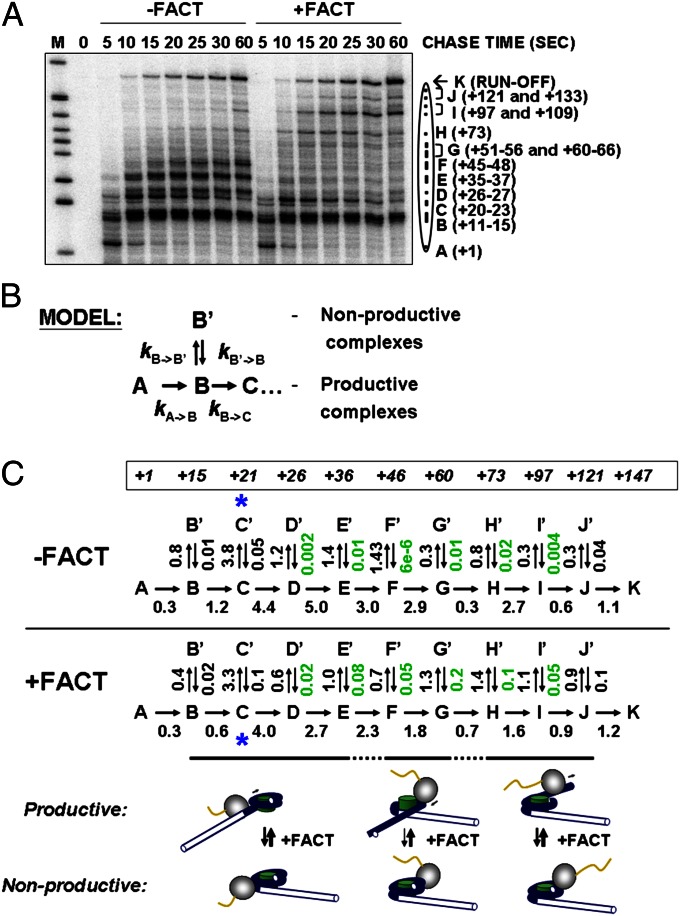
To evaluate the effect of hFACT on the rate of conversion from paused (nonproductive) complexes to productive complexes (e.g., Fig. 1E), we analyzed the kinetics of transcription through the 603 nucleosome in the presence or absence of hFACT (Fig. 3). Time courses of transcription through the nucleosome by Pol II were evaluated (Fig. 3A). Pol II pausing was quantified, and the resulting data were plotted against time and fit to a sequential multistep model (Fig. 3B) (25).
Fig. 3.
FACT facilitates formation of productive intranucleosomal Pol II complexes. (A) 603 nucleosomes were transcribed by Pol II at 150 mM KCl in the absence or presence of FACT for indicated time intervals, followed by analysis of pulse-labeled RNA by denaturing PAGE. The intranucleosomal pauses (from A to K) were quantified using a Cyclone Phosphor Imager (PerkinElmer). (B) The quantified data were analyzed using an elongation model that produces a good fit of the experimental data to the calculated curves (Fig. S10). (C) KinTek Kinetic Explorer software (25) was used to determine the rate constants of each step of transcription through the nucleosome. Averages from three experiments are shown. The rate constants positively affected by FACT by more than sevenfold are shown in green. Expected complexes formed at each region (15) are shown.
During transcription through chromatin by Pol II, multiple nonproductive complexes containing stalled and arrested ECs are formed (26, 27). Therefore, for data analysis, we included reversible steps for the formation of both productive and nonproductive complexes at each step during elongation (Fig. 3B). Comparative analysis of three different models describing the rates of transcription through the nucleosome (Fig. S9 and SI Discussion) suggests that only this model adequately describes the process of transcription through the nucleosome. The data show an excellent fit of the entire set to the sequential model (Fig. S10). In all cases, FACT strongly (more than sevenfold) and positively affects only the rates of conversion from nonproductive to productive complexes (Fig. 3C). Thus, FACT facilitates formation of productive ECs, and thereby transcription, through the nucleosome. Our previous analysis of the structures of the intermediates formed during transcription through the nucleosome by Pol II (15) allows approximate assignment of the kinetic steps to corresponding structural changes (Fig. 3C).
hFACT Prevents Nucleosome Displacement During Transcription.
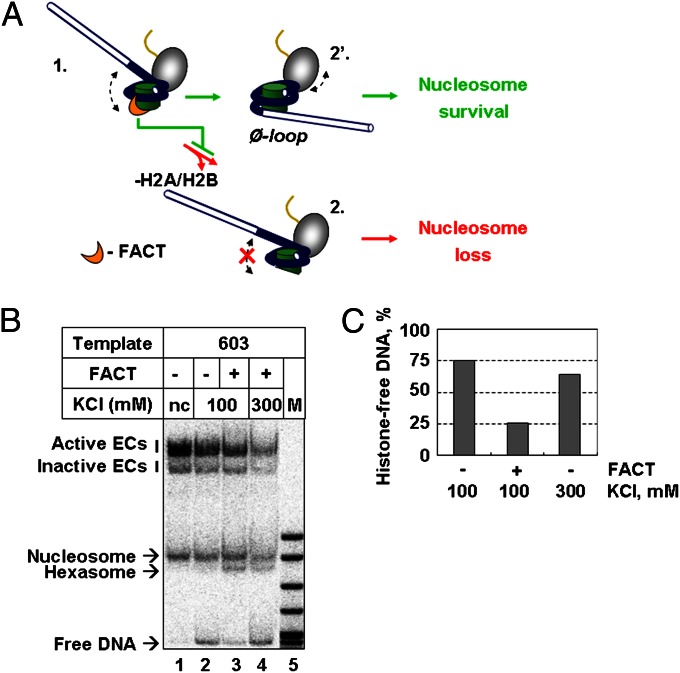
The model predicts that FACT substitutes displaced DNA in the nucleosome and thus helps retain H2A/H2B dimers within the nucleosome during transcription (Fig. 1E). Because the d-dimer is most likely constitutively lost during both FACT-dependent and FACT-independent transcription (6, 15, 28), only proximal P-dimer could be stabilized by FACT. We have proposed that the loss of proximal dimer during transcription could cause displacement of the entire octamer from DNA (Fig. 4A) (15). Given that binding of dimers to the H3/H4 tetramer depends on the presence of nucleosomal DNA (29), FACT could increase the efficiency of nucleosome (hexasome) survival by increasing retention of the P-dimer.
Fig. 4.
FACT prevents histone octamer displacement during Pol II transcription. (A) During single-round transcription without FACT, ∼50% of templates lose histone octamer (pathway 1 → 2), likely owing to displacement of promoter-proximal H2A/H2B dimer (P-dimer) by Pol II (15). FACT–P-dimer interaction likely stabilizes the dimer–tetramer interaction and prevents loss of the octamer (pathway 1 → 2′). (B) The presence of FACT reduces the amount of histone-free DNA produced during transcription by Pol II. DNA-labeled 603 nucleosomes were transcribed at different KCl concentrations in the absence or presence of FACT, followed by sample analysis by native PAGE. (C) The amounts of histone-free DNA (presented as a fraction of transcribed templates) produced after transcription.
For analysis of the fate of nucleosomes after transcription, 603 nucleosomes were DNA-labeled and transcribed in the absence or presence of hFACT, and the resulting products were analyzed by native PAGE (Fig. 4B). Transcription in the presence of hFACT results in a strong decrease in the amount of released DNA and a corresponding increase in the amount of the hexasomes after transcription by Pol II (Fig. 4 B and C). This observation is consistent with a role of hFACT as an assembly factor. Transcription of the same template by E. coli RNAP in the presence of hFACT produces a similar outcome (Fig. S11).
In summary, our in vitro system recapitulates the positive effect of FACT on nucleosome survival observed in vivo (9–11). Survival is likely mediated by the interactions of hFACT with the P-dimer, leading to better H2A/H2B dimer retention during transcription and more efficient formation of the intermediate containing a small intranucleosomal DNA loop (Ø-loop) required for nucleosome survival (Fig. 4A).
Mechanism of FACT Action During Transcription Through the Nucleosome.
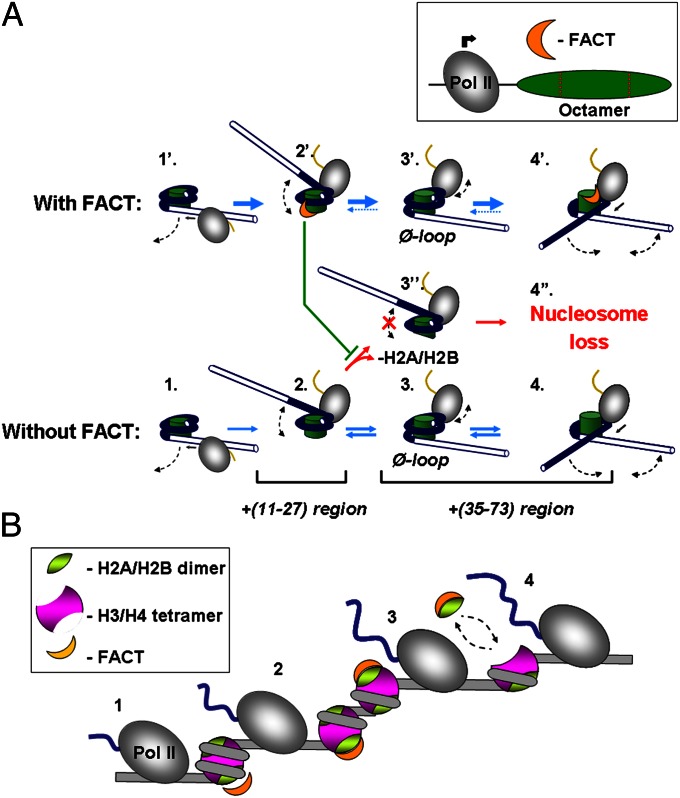
Transcription through a nucleosome in the absence of FACT (Fig. 5A, pathways 1–4) involves sequential, partial, and reversible uncoiling of nucleosomal DNA from the octamer. Biochemical analysis of several key intermediates formed during transcription through the nucleosome in the presence of hFACT using DNase I footprinting (EC+39, EC+41, and EC+49) suggests that hFACT does not detectably affect their steady-state structures. Instead, hFACT likely forms transient complexes with H2A/H2B dimers and thus facilitates DNA uncoiling from the octamer (Fig. 5A, pathways 1′–4′). As Pol II enters the nucleosome and transcribes through the +(15–36) region, it initially uncoils promoter-proximal nucleosomal DNA (complexes 1′ and 2′). FACT interacts with the exposed DNA-binding surface of the P-dimer and thus prevents the loss of promoter-proximal dimer (complex 3′′) that otherwise would result in nucleosome loss (complex 4′′). Instead, during transcription through the +(36–73) region DNA behind Pol II is recoiled on the surface of the octamer, the Ø-loop is formed, and the DNA in front of the complex becomes uncoiled from the octamer (complexes 3′ → 4′), allowing further transcription and nucleosome recovery. DNA recoiling displaces FACT from its binding site on the proximal dimer, and facilitates its interaction with the distal dimer, promoting uncoiling of the downstream DNA. Given that the distal dimer is eventually displaced from the nucleosome, FACT is likely to be displaced as well, leaving DNA-bound histone hexamer behind Pol II.
Fig. 5.
Proposed mechanism of FACT action during transcription by Pol II. (A) Pol II transcription through the nucleosome (complexes 1–4) (15) is accompanied by transient partial uncoiling of nucleosomal DNA from the octamer. (B) Proposed mechanism of FACT action during moderate-level transcription of genes; see the text for details.
Discussion
Our data show that hFACT strongly facilitates transcription through chromatin, and that hFACT activity during transcription is mediated by H2A/H2B dimers (Fig. 1). The activity of hFACT is not polymerase-specific (Fig. S5) and can be strongly affected by the sequence of nucleosomal DNA (Fig. S6). Mutations in histones destabilizing the dimer–tetramer interface and cross-linking of all core histones in the octamer minimally affect hFACT action during transcription by Pol II (Fig. 2). Analysis of the time courses of transcription suggests that hFACT decreases the lifetime of nonproductive Pol II–nucleosome complexes by facilitating their conversion into productive ECs (Fig. 3). Finally, in agreement with in vivo studies (8–11), the presence of hFACT results in more efficient nucleosome survival (Fig. 4).
Previously proposed models of FACT action during Pol II transcription suggest that during moderate-level transcription of genes by Pol II, a histone H2A/H2B dimer is displaced from its intranucleosomal location [displacement could be coupled with FACT binding to the nucleosome (30)], and either remains tethered to the nucleosome via FACT (31, 32) or is displaced into solution (6). Then FACT-bound dimer could be reloaded on chromatin in the wake of the progressing Pol II. Some studies have suggested that FACT interacts with the H3/H4 tetramer and thus could stabilize nucleosomal structure during transcription (33, 34). Our data suggest that hFACT action does not require displacement of dimer or reloading of dimer onto the nucleosome after transcription. In fact, according to our model, FACT action is mediated by H2A/H2B dimers bound in their intranucleosomal locations (Fig. 5A). In turn, hFACT likely facilitates retention of the P-dimer in the nucleosome during transcription. Our data are consistent with the results of recent studies showing that hFACT competes with DNA for a shared interaction interface on H2A/H2B dimer, with this high-affinity binding largely mediated by the acidic C-terminal domain of the Spt16 subunit (35).
Our data, considered along with the previously reported results, suggest the following scenario during moderate-level transcription through chromatin by Pol II (Fig. 5B). As the enzyme approaches a nucleosome, FACT may destabilize the nucleosome in front of the enzyme (complex 1). As Pol II enters the nucleosome and partially uncoils nucleosomal DNA, FACT sequentially binds to promoter-proximal (complex 2) and promoter-distal H2A/H2B dimers (complex 3). The FACT–dimer interactions facilitate nucleosome survival (likely mediated by the proximal dimer–FACT interactions) and transcription through chromatin. During further transcription through the nucleosome in vitro, the H2A/H2B dimer is eventually displaced from DNA (28). In vivo, FACT could remain associated with the displaced dimer and may help deliver it to chromatin behind the transcribing Pol II (complex 4). The proposed mechanism is likely relevant for other processive enzymes working in chromatin. Indeed, as these DNA-targeted enzymes (e.g., DNA polymerases, ATP-dependent chromatin remodelers) encounter nucleosomes and attempt to displace histones from DNA, their progression could be facilitated by the FACT–dimer interactions. Although our data do not provide direct support for the models involving an interaction of FACT with H3/H4 tetramer and reloading of FACT-bound H2A/H2B dimer on the nucleosome behind transcribing Pol II, our model of FACT action is not mutually exclusive with any of the previously proposed models (6, 12, 13).
Recent studies have shown that inactivation of FACT leads to phosphorylation of p53 by casein kinase 2 and inhibition of NF-κB–dependent genes; the level of FACT expression is higher in nondifferentiated and cancer cells (3, 4). It has been proposed that the requirement for FACT during transcript elongation dictates higher levels of FACT in cells in which gene expression occurs at a higher level, such as cancer or undifferentiated cells (36). Our present results further suggest that FACT activity in undifferentiated and cancer cells is likely activated by the alternating interactions with the DNA-binding surface of the H2A/H2B dimers within the nucleosome.
Materials and Methods
Preparation of Proteins, DNA Templates, and Nucleosomes.
Nucleosomes were assembled on DNA fragments carrying various high-affinity positioning sequence using salt gradient method as described in SI Materials and Methods. Yeast Pol II (28), FACT complex (6), and histones (37) were purified as described previously.
Transcription and Kinetics Assay.
Transcription by yeast Pol II and E. coli RNAP was conducted as described previously (15).The kinetics of Pol II transcription through nucleosome was measured by performing transcription for indicated short time intervals at 22 °C. The yields of RNA products at each time point were quantified and fit to the model (Fig. 5B) using the KinTek Explorer program (38, 39). Details of these procedures are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (Grants GM58650 to V.M.S., GM51966 to S.S.P, GM55310 to S.S.P., GM064844 to D.R., R37GM037120 to D.R., and GM088409 to K.L.) and the Ministry of Education and Science of the Russian Federation (Contract 14.512.11.0028 to V.M.S.). K.L. and D.R. are also supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222198110/-/DCSupplemental.
References
- 1.Wittmeyer J, Formosa T. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol Cell Biol. 1997;17(7):4178–4190. doi: 10.1128/mcb.17.7.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92(1):105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Ramirez M, Dong F, Ausio J. Role of the histone “tails” in the folding of oligonucleosomes depleted of histone H1. J Biol Chem. 1992;267(27):19587–19595. [PubMed] [Google Scholar]
- 4.Gasparian AV, et al. Curaxins: Anticancer compounds that simultaneously suppress NF-κB and activate p53 by targeting FACT. Sci Transl Med. 2011;3(95):95ra74. doi: 10.1126/scitranslmed.3002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400(6741):284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 6.Belotserkovskaya R, et al. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301(5636):1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 7.Saunders A, et al. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301(5636):1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- 8.Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23(22):8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Formosa T, et al. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: Polymerase passage may degrade chromatin structure. Genetics. 2002;162(4):1557–1571. doi: 10.1093/genetics/162.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung V, et al. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6(11):e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301(5636):1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 12.McCullough L, et al. Insight into the mechanism of nucleosome reorganization from histone mutants that suppress defects in the FACT histone chaperone. Genetics. 2011;188(4):835–846. doi: 10.1534/genetics.111.128769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xin H, et al. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol Cell. 2009;35(3):365–376. doi: 10.1016/j.molcel.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bondarenko VA, et al. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24(3):469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Kulaeva OI, et al. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat Struct Mol Biol. 2009;16(12):1272–1278. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulaeva OI, Gaykalova DA, Studitsky VM. Transcription through chromatin by RNA polymerase II: Histone displacement and exchange. Mutat Res. 2007;618(1-2):116–129. doi: 10.1016/j.mrfmmm.2006.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulaeva OI, Hsieh FK, Studitsky VM. RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc Natl Acad Sci USA. 2010;107(25):11325–11330. doi: 10.1073/pnas.1001148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulaeva OI, Studitsky VM. Mechanism of histone survival during transcription by RNA polymerase II. Transcription. 2010;1(2):85–88. doi: 10.4161/trns.1.2.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone–DNA interactions. Mol Cell. 2010;37(6):834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall MA, et al. High-resolution dynamic mapping of histone–DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16(2):124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh FK, Fisher M, Ujvári A, Studitsky VM, Luse DS. Histone Sin mutations promote nucleosome traversal and histone displacement by RNA polymerase II. EMBO Rep. 2010;11(9):705–710. doi: 10.1038/embor.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luse DS, Studitsky VM. The mechanism of nucleosome traversal by RNA polymerase II: Roles for template uncoiling and transcript elongation factors. RNA Biol. 2011;8(4):581–585. doi: 10.4161/rna.8.4.15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walter W, Kireeva ML, Studitsky VM, Kashlev M. Bacterial polymerase and yeast polymerase II use similar mechanisms for transcription through nucleosomes. J Biol Chem. 2003;278(38):36148–36156. doi: 10.1074/jbc.M305647200. [DOI] [PubMed] [Google Scholar]
- 24.Gaykalova DA, et al. A polar barrier to transcription can be circumvented by remodeler-induced nucleosome translocation. Nucleic Acids Res. 2011;39(9):3520–3528. doi: 10.1093/nar/gkq1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandey M, Levin MK, Patel SS. Experimental and computational analysis of DNA unwinding and polymerization kinetics. Methods Mol Biol. 2010;587:57–83. doi: 10.1007/978-1-60327-355-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: Inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5(4):683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 27.Kireeva ML, et al. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18(1):97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 28.Kireeva ML, et al. Nucleosome remodeling induced by RNA polymerase II: Loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9(3):541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 29.Feng HP, Scherl DS, Widom J. Lifetime of the histone octamer studied by continuous-flow quasielastic light scattering: Test of a model for nucleosome transcription. Biochemistry. 1993;32(30):7824–7831. doi: 10.1021/bi00081a030. [DOI] [PubMed] [Google Scholar]
- 30.Ruone S, Rhoades AR, Formosa T. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes. J Biol Chem. 2003;278(46):45288–45295. doi: 10.1074/jbc.M307291200. [DOI] [PubMed] [Google Scholar]
- 31.Reinberg D, Sims RJ., 3rd de FACTo nucleosome dynamics. J Biol Chem. 2006;281(33):23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- 32.Winkler DD, Luger K. The histone chaperone FACT: Structural insights and mechanisms for nucleosome reorganization. J Biol Chem. 2011;286(21):18369–18374. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers CN, et al. Mutant versions of the S. cerevisiae transcription elongation factor Spt16 define regions of Spt16 that functionally interact with histone H3. PLoS ONE. 2011;6(6):e20847. doi: 10.1371/journal.pone.0020847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamai A, Puglisi A, Strubin M. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol Cell. 2009;35(3):377–383. doi: 10.1016/j.molcel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Winkler DD, Muthurajan UM, Hieb AR, Luger K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem. 2011;286(48):41883–41892. doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsieh FK, Kulaeva OI, Orlovsky IV, Studitsky VM. FACT in cell differentiation and carcinogenesis. Oncotarget. 2011;2(11):830–832. doi: 10.18632/oncotarget.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 38.Johnson KA, Simpson ZB, Blom T. Global Kinetic Explorer: A new computer program for dynamic simulation and fitting of kinetic data. Anal Biochem. 2009;387(1):20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 39.Johnson KA, Simpson ZB, Blom T. FitSpace Explorer: An algorithm to evaluate multidimensional parameter space in fitting kinetic data. Anal Biochem. 2009;387(1):30–41. doi: 10.1016/j.ab.2008.12.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.