Significance
Infection triggers the innate immune response in all metazoans, activating regulatory pathways that result in expression of effector proteins, including potent antimicrobial peptides. These pathways can also be activated in the brain by aging, stress, and injury. Although nominally protective, excessive neuroinflammatory responses may themselves contribute to neurodegenerative disease by mechanisms that remain unclear. We found that hyperactivation of innate immunity in the Drosophila brain as a result of mutation or bacterial injection causes neurodegeneration because of neurotoxic effects of antimicrobial peptides. These findings have important implications for the role of neuroinflammation in human neurodegenerative disease.
Keywords: neuroprotection, neuronal cell death, neurotoxic mechanism
Abstract
A growing body of evidence in humans implicates chronic activation of the innate immune response in the brain as a major cause of neuropathology in various neurodegenerative conditions, although the mechanisms remain unclear. In an unbiased genetic screen for mutants exhibiting neurodegeneration in Drosophila, we have recovered a mutation of dnr1 (defense repressor 1), a negative regulator of the Imd (immune deficiency) innate immune-response pathway. dnr1 mutants exhibit shortened lifespan and progressive, age-dependent neuropathology associated with activation of the Imd pathway and elevated expression of AMP (antimicrobial peptide) genes. To test the hypothesis that overactivation of innate immune-response pathways in the brain is responsible for neurodegeneration, we demonstrated that direct bacterial infection in the brain of wild-type flies also triggers neurodegeneration. In both cases, neurodegeneration is dependent on the NF-κB transcription factor, Relish. Moreover, we found that neural overexpression of individual AMP genes is sufficient to cause neurodegeneration. These results provide a mechanistic link between innate immune responses and neurodegeneration and may have important implications for the role of neuroinflammation in human neurodegenerative diseases as well.
Like all metazoans, Drosophila mounts a potent innate immune response as a defense mechanism to protect against microbial infection. Characteristic features of this response include receptor-based recognition of invading microorganisms, triggering downstream signaling pathways, and activation of NF-κB family transcription factors that elicit effector mechanisms, including synthesis of powerful antimicrobial peptides (AMPs). Two distinct pathways mediate synthesis of AMPs in Drosophila: the Toll pathway, which is activated primarily by fungi and Gram-positive bacteria, and the Imd (immune deficiency) pathway, which is activated primarily by Gram-negative bacteria (1). Activation of the Toll pathway ultimately results in degradation of Cactus, an Iκκ (Inhibitor of κB kinase), thereby enabling NF-κB Dorsal and Dif (Dorsal-related immunity factor), to initiate transcription of effector genes, including those encoding a subset of AMPs (2). The Imd pathway is triggered by pathogen stimulation of membrane-bound and intracellular receptors (3) that signal through Imd to activate Relish (Rel), another NF-κB transcription factor, which then transcribes an overlapping subset of AMPs as well as other genes. Activation of Rel is mediated by cleavage of its N-terminal autoinhibitory domain following activation of the Dredd (death-related ced-3/Nedd2-like protein) caspase (4). Both the Toll and Imd pathways are evolutionarily conserved in mammals, including humans, with homologous genes encoding most of the key components in the mammalian TLR (Toll-like receptor) and TNF-receptor proinflammatory pathways, respectively (5).
In addition to the core components of the innate immune-response pathways, there are a number of regulatory proteins in both flies and mammals that modulate these pathways to prevent an uncontrolled immune response. One recently identified regulator for the Imd pathway is Dnr1 (defense repressor 1) (Fig. S1), a protein that contains a RING (Really Interesting New Gene) domain (an E3 ubiquitin ligase domain) that shares strong similarity with the RING domain found in inhibitor of apoptosis proteins that inhibit caspase activity by promoting their proteolytic degradation (6, 7). Dnr1 acts in a similar manner to down-regulate Dredd. RNAi-mediated depletion of Dnr1 results in induction of Rel-dependent transcripts both in cultured S2 cells and in vivo, whereas induction of the Imd pathway via bacterial infection is blocked by heat-shock mediated expression of Dnr1 (7).
Regulation of innate immune pathways is particularly important in the nervous system, where a large body of evidence demonstrates that maintenance of neuronal viability in response to aging, disease, and environmental factors requires controlled activity of immune responses that may have either beneficial or deleterious consequences on neuronal survival (8). Reflecting its likely evolutionary origin, the innate immune response is triggered not only by pathogen-derived molecules but also by endogenous molecules generated by stressed and injured cells (9). The aberrant protein aggregates associated with most human degenerative diseases can be a strong trigger of neuroinflammatory pathways that limit accumulation of these aggregates and promote their clearance. For example, evidence indicates that activation of innate immune responses in Alzheimer’s Disease (AD) contributes to the removal of β-amyloid fibrils (10, 11). Conversely, other results, including the observed hyperactivation of inflammatory pathways in aging brains and the finding that long-term use of anti-inflammatory drugs substantially reduces the risk for AD (12) and Parkinson Disease (13), indicate that excessive or uncontrolled inflammatory responses may themselves be key contributors to neurodegenerative disease.
Recent studies in Drosophila demonstrate that activation of both the Toll and Imd pathways in the nervous system can contribute to neurodegeneration. In a fly model of AD, cell death and tissue degeneration caused by expression of human amyloid-β peptide (Aβ)42 in the compound eye was suppressed by mutations of Toll and its downstream effectors, including Dorsal and Dif (14). Retinal degeneration in Drosophila caused by a mutation in the eye-specific phospholipase C encoded by norpA (no receptor potential) is suppressed by mutations of Dredd and Relish, implicating involvement of Imd pathway components (15). Flies mutant for ATM (A-T mutated), the gene responsible for Ataxia-telangiectasia in humans, exhibit shortened lifespan, impaired locomotor activity, and neurodegeneration in the central brain (16). These phenotypes were associated with elevated brain expression of many innate immune-response genes. Together, these examples indicate that activation of immune pathways in Drosophila, as in mammals, can factor importantly in neurodegeneration, although the precise mechanisms are not understood.
Here we describe the identification of a mutation in dnr1 in an unbiased forward genetic screen for mutants exhibiting neurodegeneration in Drosophila. Dnr1 mutants exhibit shortened lifespan and progressive, age-dependent neuropathology associated with activation of the Imd pathway and elevated expression of AMP genes. To examine the hypothesis that overactivation of the Imd pathway in the brain is directly responsible for neurodegeneration, we tested whether direct bacterial infection in the brain of wild-type flies also triggers neurodegeneration. In both dnr1 mutants and brain-infected flies, neurodegeneration is dependent on Relish. Moreover, we show that neural overexpression of individual AMP genes is sufficient to cause neurodegeneration. These results provide direct evidence that regulatory proteins such as dnr1 that down-regulate the innate immune response serve a neuroprotective role and may have important implications for involvement of neuroinflammatory pathways in human neurodegenerative disease as well.
Results
Loss of dnr1 Causes Neurodegeneration.
To identify new genes required for maintaining neuronal viability, we performed histological analysis on a collection of Drosophila P-element insertion lines to screen for any lines exhibiting neurodegeneration as manifested by the appearance of vacuolar lesions in the central brain at 15 and 25 d of age. One of the lines we identified in this screen, 2-133, exhibited occasional neuropathological lesions in the neuropil of 15-d-old flies that were absent in age-matched controls and became more pronounced in size and frequency in 25-d-old flies, indicating that this mutation results in progressive, age-dependent neurodegeneration.
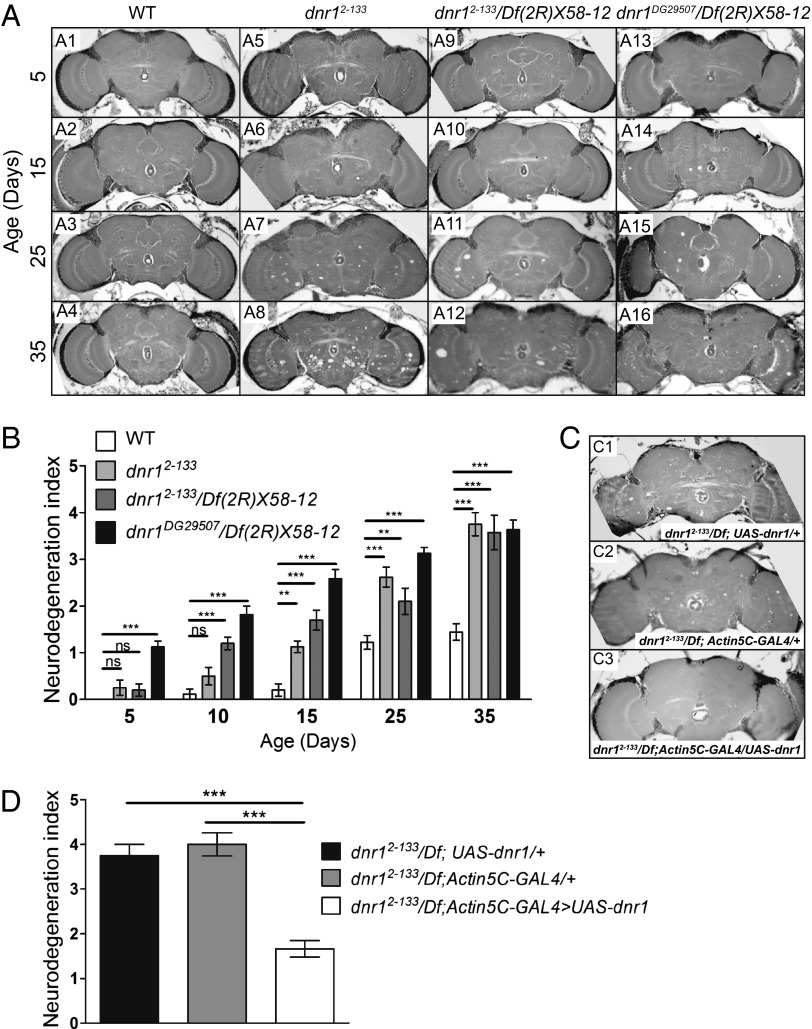
We mapped the P-element insertion in 2-133 by thermal asymmetric interlaced (TAIL) PCR (17) to the 5′ UTR of the gene defense repressor 1 (dnr1) (Fig. S2A), identifying it as the likely candidate gene whose disruption is responsible for the observed phenotype. Several lines of evidence confirm this identity. First, we measured dnr1 mRNA expression levels in the heads of 10-d-old 2-133 flies by quantitative real-time PCR (qPCR). In comparison with wild-type flies, dnr1 expression is reduced by about 90% in 2-133 (Fig. S2B). Second, we examined 2-133/Df(2R)X58-12 flies and found that deficiencies that uncovered dnr1 failed to complement the neurodegeneration phenotype of 2-133, demonstrating that this phenotype and the P-element insertion mapped to the same small chromosome interval (Fig. 1 A and B). Third, we examined a second independent P-element insertion in dnr1 (dnr1DG29507) and found that it was also associated with age-dependent neurodegeneration (Fig. 1 A and B). Finally, we used a ubiquitous Actin5c-GAL4 driver to express wild-type UAS-dnr1 and were able to rescue neurodegeneration (Fig. 1 C and D). Taken together, these results demonstrate that dnr1 is the gene that is disrupted in the 2-133 insertion strain and that loss of dnr1 function results in neurodegeneration. We will subsequently refer to 2-133 as dnr12-133.
Fig. 1.
Mutations in dnr1 cause neurodegeneration. (A) Representative 5-μm paraffin sections at approximately midbrain of wild-type (Canton-S) (A1- A4), dnr12-133 (A5–A8), dnr12-133/Df(2R)X58-12 (A9-A12), and dnr1DG29507/Df(2R)X58-12 (A11-A16) flies at the indicated ages. (B) Quantification of neurodegeneration in the brains of Canton-S, dnr12-133, dnr12-133/Df(2R)X58-12 and dnr1DG29507/Df(2R)X58-12 flies based on the neurodegeneration index scale shown in Fig. S3. Values shown are mean ± SEM. Values are listed in Table S1. (C) The 5-μm paraffin sections at approximately midbrain of 25-d-old dnr12-133;UAS-dnr1/+ (C1), dnr12-133;Actin5C-GAL4/+ (C2), and dnr12-133;Actin5C-GAL4/UAS-dnr1 (C3) flies. Quantification of neurodegeneration in these genotypes is shown in D. The neurodegeneration index of the rescued flies is restored back to normal (see, for example, 25-d-old wild-type in B). Values shown are mean ± SEM. **P < 0.01, ***P < 0.005 based on one-way ANOVA with Dunnett's posttest. ns, not significant.
To obtain a more complete picture of the age dependence of neurodegeneration in dnr1 mutants, we first examined the lifespan of dnr12-133 homozygotes at 29 °C and found that the mutants had a substantially reduced lifespan compared with wild-type controls or dnr1/+ heterozygotes, with only 50% survival at 30 d and almost no survivors by 40 d (Fig. S2C). We thus chose 35 d as our final time point and performed histological analysis on 5- to 35-d-old flies. As evident in Fig. 1A, neurodegeneration in the mutant progresses as a function of age. To quantify the phenotype we developed a neurodegeneration index (Materials and Methods and Fig. S3) based on the severity of the appearance of the vacuolar lesions according to their size and abundance. This index provided a useful and reliable metric for comparing neurodegeneration among different genotypes and at different ages (Fig. 1B and Table S1). As for lifespan, the neurodegeneration phenotype of dnr1 is recessive (Fig. S2 D and E).
Neurodegeneration in dnr1 Mutants Requires Relish Activity.
Dnr1 was originally identified as a negative regulator of the Dredd caspase in the Imd pathway (6, 18). Dredd is required for activation of Relish by associating with Fadd (Fas-associated death domain protein) to cleave off the autoinhibitory domain from Relish, an NF-κB family transcription factor to generate its active form (4, 19, 20). In Drosophila S2 cells, reduction of dnr1 expression by RNAi results in transcriptional activation of the Dipt-lacZ (Diptericin) reporter, which is a widely accepted signature of Relish activation via the Imd pathway (7). These results suggest the possibility that the Imd pathway also mediates the neurodegeneration phenotype we observe in dnr1 mutants. Alternatively, dnr1 mutations could result in neurodegeneration via some alternative pathway that has not yet been characterized.
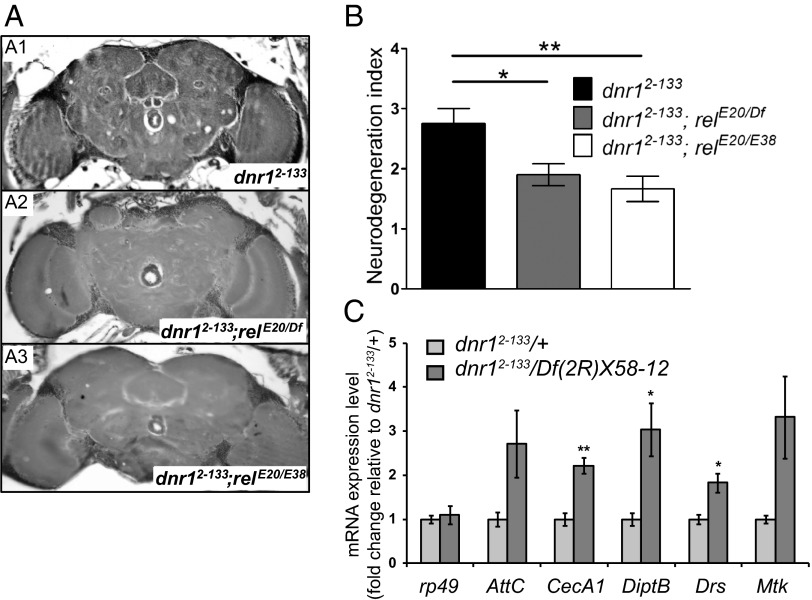
To distinguish these possibilities we tested whether neurodegeneration in dnr1 mutants requires Relish activity. Compared with dnr12-133 alone, neurodegeneration in dnr12-133;relE20/Df(3R)ED05331 and dnr12-133;relE20/relE38 flies aged at 29 °C for 25 d was reduced nearly back to levels seen in wild-type flies (Fig. 2 A and B, and Table S1). These data demonstrate that dnr1-associated neurodegeneration requires Relish activity and is thus likely to be mediated via the innate immune-response pathway.
Fig. 2.
Neurodegeneration in dnr1 requires Relish activity and is associated with increased transcription of AMP genes. (A) Representative 5-μm paraffin sections at approximately midbrain of 25-d-old dnr12-133 (A1), dnr12-133;relE20/Df(3R)ED05331 (A2), and dnr12-133;relE20/relE38 (A3) flies. (B) Quantification of neurodegeneration in the brains of 25-d-old dnr12-133, dnr12-133;relE20/Df(3R)ED05331, and dnr12-133;relE20/relE38 flies. Values shown are mean ± SEM. *P < 0.05 and **P < 0.01 based on one-way ANOVA with Dunnett's posttest. (C). qPCR analysis of mRNA levels of rp49, AttacinC (AttC), CecropinA1 (CecA1), DiptericinB (DiptB), Drosomycin (Drs), and Metchnikowin (Mtk) in the heads of dnr12-133/Df(2R)X58-12 and dnr12-133/+ at 10 d of age at 29 °C. mRNA fold changes are normalized to that in dnr12-133/+. Values shown are mean ± SEM. *P < 0.05 and **P < 0.01 based on Student's t test.
Neuronal Apoptosis Is Not Perturbed in dnr1 Mutants.
Because Dnr1 has also been implicated in regulating apoptosis in cultured cells, another possibility is that the neurodegeneration in dnr1 mutants results from activation of apoptosis. To test whether Dnr1 regulates apoptosis in the nervous system, we performed an immunohistochemical assay for cleaved (activated) caspase 3 in dnr1 mutant brains. As a positive control we used ATM8 flies (16). As previously reported, clusters of caspase 3-positive cells were readily detected in the brains of ATM8 flies (Fig. S4 A and F). In contrast, dnr1 mutants at 20 and 25 d of age did not differ significantly from age-matched wild-type controls with very few caspase 3-positive cells detected in either background (Fig. S4 B–F). Thus, loss of Dnr1 does not appear to play a significant role in regulating apoptosis in the nervous system, and neurodegeneration in dnr1 mutants is unlikely to involve activation of the canonical apoptosis pathway.
AMP Gene Expression Is Elevated in dnr1 Mutants.
Because Dnr1 is a negative regulator of Imd signaling, it is expected that loss of dnr1 should lead to constitutive activation of Relish and increased expression of immune-response genes, including those encoding AMPs, such as diptericin, cecropin, and attacin (7). To test this prediction, we performed qPCR analysis of diptericin, cecropin, and attacin mRNA levels in heads of dnr12-133/Df(2R)X58-12 flies and found that they are elevated compared with dnr12-133/+ control flies (Fig. 2C). Unexpectedly, we also found elevated RNA levels in dnr1 mutants for some AMPs, such as drosomycin and metchnikowin, the expression of which is normally regulated by the Toll pathway, a separate branch of the innate immune response dependent on the NF-κB transcription factors Dorsal and Dif rather than Relish (Fig. 2C). Thus, dnr1 may also negatively regulate the Toll pathway by a mechanism that has not yet been defined or by cross-regulation between the two pathways (21). In either case, loss of dnr1 leads to increased expression of AMPs downstream of both the Imd and the Toll innate immune-response pathways.
Neurodegeneration Can Be Triggered by Bacterial Infection in the Brain.
To further test the hypothesis that activation of the immune response in the nervous system is responsible for neurodegeneration, we developed a method to trigger activation of the immune response in the nervous system that did not depend on loss of dnr1. Instead, we used an injection method to introduce bacteria into the CNS. For the experimental group of flies, we poked a thin metal needle coated with a mixture of Gram-negative (Escherichia coli) and Gram-positive (Micrococcus luteus) bacteria through the eye into the central brain. Control flies were similarly poked with a sterile metal needle, to control for the effects of injury alone. As an additional control, we also pricked flies in the thorax, as previously described (22), to induce the innate immune response and AMP expression in fat body. Because of the blood–brain barrier, we did not expect bacterial injection in the thorax to elicit an immune response in the brain, nor did we expect AMPs expressed in cells outside the CNS to enter the brain.
To monitor expected activation of the immune response following bacterial infection, we used the attacin-GFP reporter line. In flies injected with bacteria in the head or thorax, we detected GFP signal in whole flies, as well as dissected fat bodies at 48 h after injection. The GFP signal was detectable in dissected brains only when the injection was done in the head. We did not detect expression of the reporter in control flies poked with a sterile needle (Fig. S5), confirming preferential activation of the immune response in bacterially injected flies.
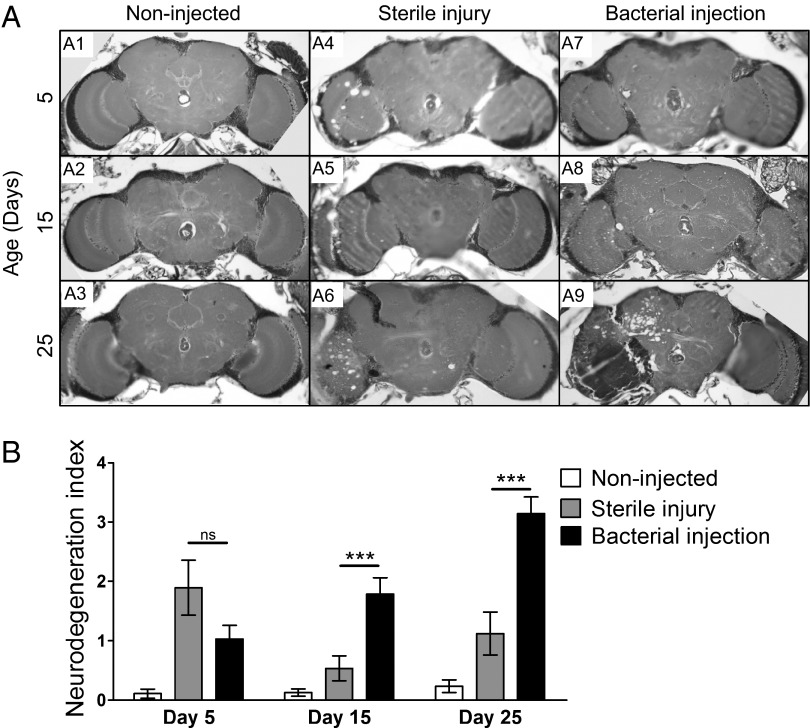
To assess whether activation of the immune response via bacterial injection could trigger neurodegeneration, 1- to 3-d-old adults were injected and aged for 5–25 d before histological analysis. As expected, we did not observe any neurodegeneration in flies that were injected in the thorax either with bacteria or a sterile needle (Fig. S6). Flies that were injected in the brain with a sterile needle did display some vacuolar neuropathology on the left side of the brain at the injury site (Fig. 3A), which presumably reflects physical damage from the needle itself. However, in comparison with the experimental group, the neuropathology in these flies was less severe and it did not progress with age (Fig. 3 A and B). In a single experiment, 5-d-old flies injected with the sterile needle showed elevated levels of neurodegeneration (Fig. 3B), which was likely because of excessive mechanical damage from the needle used in that experiment. In contrast, flies injected with bacteria exhibited clear evidence of neuropathology localized to the left side of the brain at the injection site. Overall, the severity of the lesions and the percentage of flies manifesting neuropathology were substantially greater in the flies injected in the head with bacteria versus those that received a sterile injury (Fig. 3). Moreover, the neuropathology observed in the experimental group clearly progressed as a function of age (Fig. 3). These results provide further support for the idea that strong activation of the immune response in the brain either via loss of dnr1 or by bacterial infection can trigger neurodegeneration.
Fig. 3.
Bacterial injection in the Drosophila CNS triggers neurodegeneration. (A) Wild-type (Canton-S) flies were subjected to the following experimental manipulations before aging and histological analysis: untreated (noninjected) (A1–A3); injected in the head with a sterile needle (sterile injury) (A4–A6); injected in the head with bacteria (bacterial injection) (A7–A9). In all cases, the injection site was the left optic lobe. Representative 5-μm paraffin sections at approximately midbrain of flies aged at 25 °C for 5, 15, and 25 d before sectioning. (B) Quantification of neurodegeneration in brains of noninjected, sterile-injected, and bacterially injected flies represented in A. ***P < .005 based on Student's t test. ns, not significant.
Bacterial Injection-Triggered Neurodegeneration Requires Relish Activity.
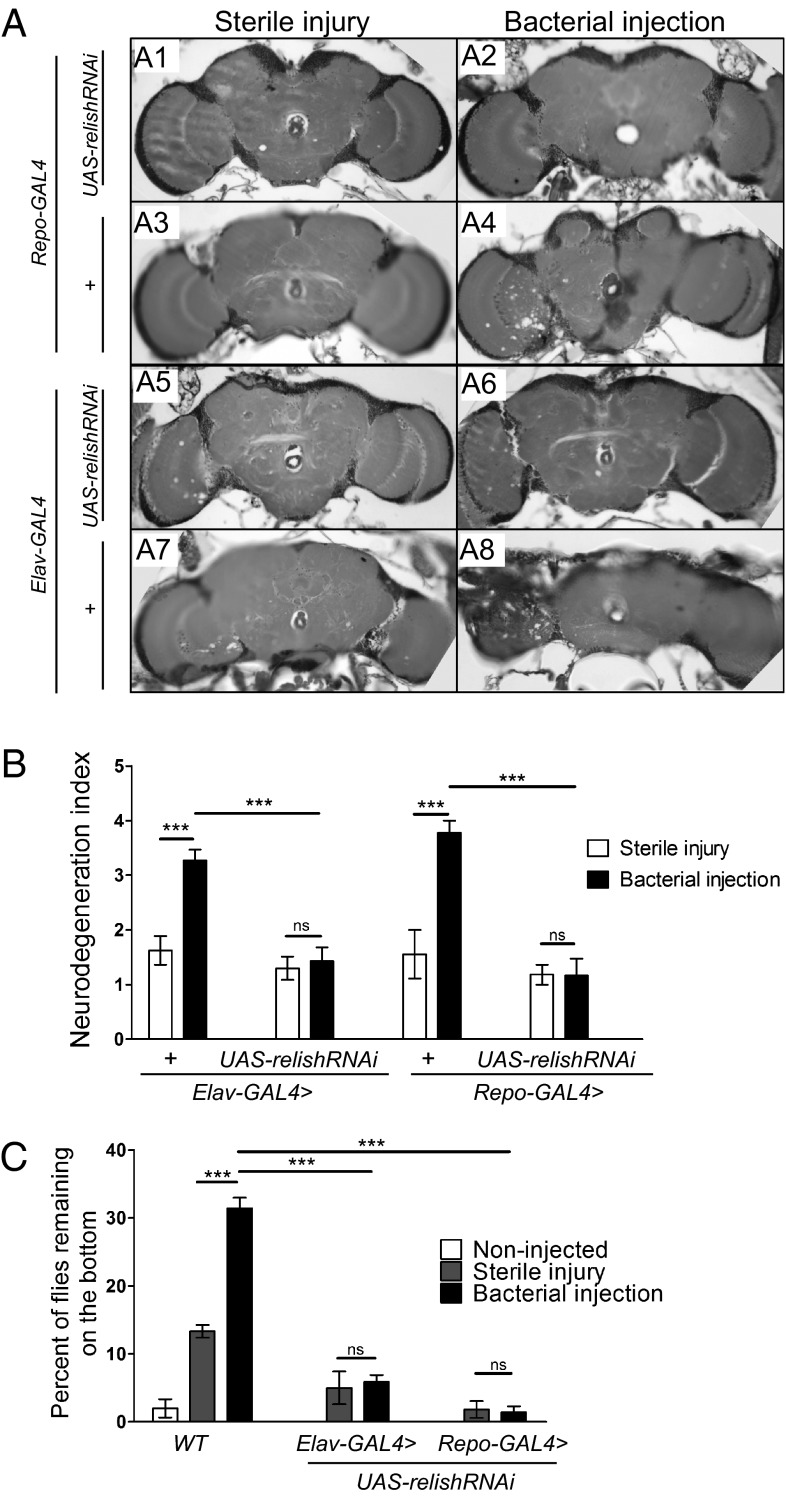
If the neuropathology induced by bacterial injection in the head is dependent on activation of the immune response pathway, as we have suggested, then as for dnr1 mutants, this phenotype should be suppressed if Relish activity is reduced. We used a UAS-relishRNAi construct, under the control of Elav-GAL4 or Repo-GAL4 drivers to knock down relish expression specifically in neurons or glia, respectively (23, 24), and examined whether this knockdown affected manifestation of neuropathology in bacterially injected flies. As in previous observations, brains of Repo-GALl4 and Elav-GAL4 control flies given a sterile injection appeared relatively normal when examined histologically at 25 d of age, whereas prominent neuropathology was observed at the injection site of bacterially injected flies (Fig. 4 A and B). In contrast, the neuropathological phenotype was strikingly suppressed when bacterial injection was performed on heads of Repo-GAL4 > UAS-relishRNAi or Elav-GAL4 > UAS-relishRNAi flies (Fig. 4 A and B).
Fig. 4.
Neurodegeneration triggered by bacterial injection is suppressed by loss of relish in either glia or neurons. (A) Flies of the indicated genotypes were injected in the left optic lobe, either with a sterile needle (sterile injury) (Left) or with bacteria (bacterial injection) (Right) and aged for 25 d at 25 °C before histological analysis. Representative 5-μm paraffin sections at approximately midbrain of Repo-GAL4 > UAS-relishRNAi (A1, A2), Repo-GAL4/+ (A3, A4), Elav-GAL4 > UAS-relishRNAi (A5, A6), and Elav-GAL4/+ (A7, A8). (B) Quantification of neurodegeneration in the brains of flies represented in A based on the neurodegeneration index scale. Values shown are mean ± SEM. (C) Bacterially injected flies exhibit defective locomotion that is suppressed by loss of relish. The histogram shows the percentage of flies (aged for 25 d at 25 °C) of the indicated genotype and treatment that remain on the bottom of the vial 10 s after being tapped down. Values shown are mean ± SEM. ***P < 0.005 based on Student's t test. ns, not significant.
In parallel with the progressive, age-dependent neuropathology observed in bacterially injected flies, we noticed that these flies also displayed a locomotor defect and spent more time on the bottom of culture vials than control flies. To quantify this phenotype, flies subjected to the various injection protocols were tapped to the bottom of their vials, and the percent of flies remaining on the bottom of the vial after 10 s was determined (Fig. 4C). At 25 d of age, about 30% of the infected flies remained on the bottom of the vial, which is at least twofold greater than control flies injected with a sterile needle (Fig. 4C). This locomotor defect was also suppressed by loss of Relish (Fig. 4C). In contrast with injection of wild-type flies, only about 5% of injected Elav-GAL4 > UAS-relishRNAi flies and 3% of injected Repo-GAL4 > UAS-relishRNAi were similarly impaired (Fig. 4C). Taken together, these results demonstrate that both the neuropathological consequences of bacterial injection in the brain and the accompanying locomotor impairment are suppressed by loss of Relish, supporting the idea that these phenotypes are because of activation of the innate immune response in neurons and glial cells in the CNS.
Overexpression of AMPs in the Nervous System Causes Neurodegeneration.
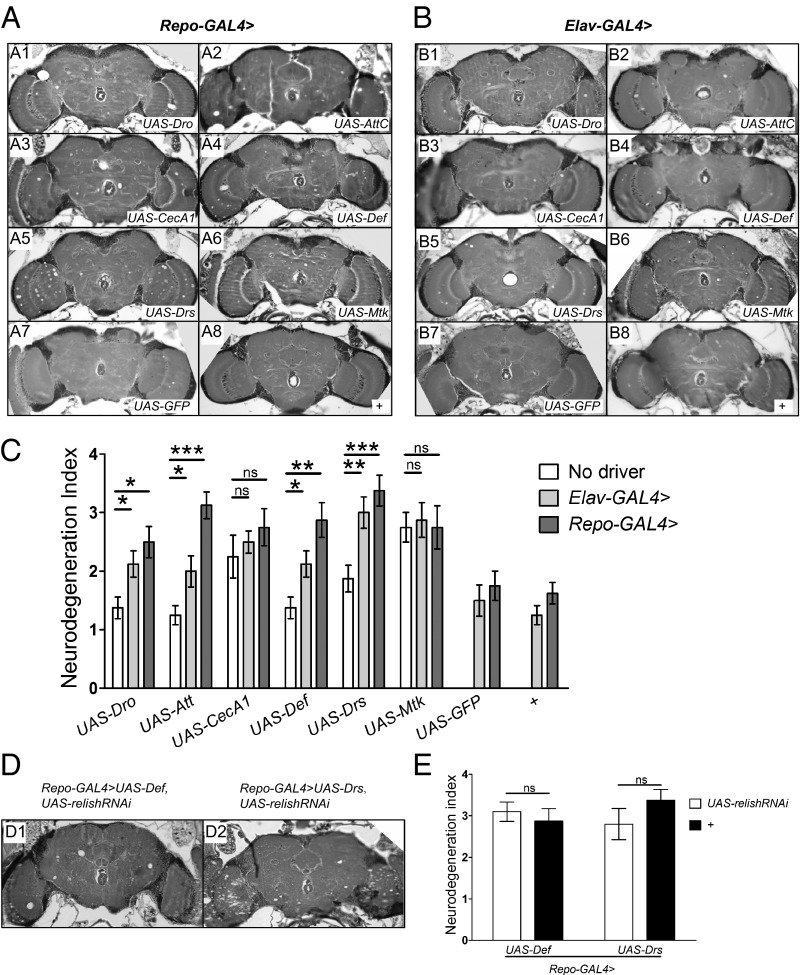
In general, AMPs exert their toxic effect on invading bacteria by disrupting their cell membranes (25). Thus, one possibility is that elevated expression of AMPs in the CNS is directly responsible for neurodegeneration because of toxic effects on neurons and glia as well. To test this hypothesis, we drove expression of various AMPs in neurons or glial cells, respectively, and examined the effects on neuronal integrity as a function of age. At 2 d of age, brains of Elav-GAL4 > UAS-AMP and Repo-GAL4 > UAS-AMP flies looked normal and were indistinguishable from wild-type or from Elav-GAL4 and Repo-GAL4 controls (Fig. S7). However, by 25 d of age, overexpression of AMPs either in neurons or in glia causes obvious vacuolar lesions in the brain (Fig. 5 A and B). Quantification of the neurodegeneration index revealed a significant increase compared with controls for overexpression of all AMPs tested except CecropinA1 and Metchnickowin (Fig. 5C). In these two cases, there was an unexplained elevation of neurodegeneration even in the controls, with transgenic cDNA construct alone confounding the analysis. Nonetheless, the results with the other four AMPs tested clearly indicate that overexpression of AMPs either in glial cells or in neurons can cause vacuolar pathology indicative of neurodegeneration. Indeed, the degree of neuropathology manifested when even a single AMP is overexpressed is remarkable. Taken together, these results support the idea that elevated expression of multiple AMPs in a dnr1 mutant background is sufficient to cause the observed neurodegeneration phenotype.
Fig. 5.
Overexpression of AMPs in the Drosophila central nervous system causes neurodegeneration. (A and B) Representative 5-μm paraffin sections at approximately midbrain of 25-d-old flies expressing individual AMPs in glia (Repo-GAL4 > UAS-AMP) (A) or in neurons (Elav-GAL4 > UAS-AMP) (B). Panels A8 and B8 are driver-only controls, where “+” refers to a wild-type (Canton-S) third chromosome and X chromosome, respectively. (C) Quantification of neurodegeneration in brains of Repo-GAL4 > UAS-AMP and Elav-GAL4 > UAS-AMP flies at 25 d of age. Values shown are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005 based on one-way ANOVA with Dunnett's posttest. (D) Representative 5-μm paraffin sections at approximately midbrain of 25-d-old Repo-GAL4 > UAS-Defensin, UAS-relishRNAi flies (D1) and Repo-GAL4 > UAS-Drosomycin, UAS-relishRNAi flies (D2). (E) Quantification of neurodegeneration in the brains of 25-d-old flies of the genotypes indicated. Values shown are mean ± SEM. ns, not significant based on Student's t test. Att, Attacin; CecA1, Cecropin A1;Def, Defensin; Dro, Drosocin; Drs, Drosomycin; Mtk, Metchnikowin.
In contrast with the neurodegeneration observed in dnr1 mutants or in bacterially injected flies, which was suppressed when Relish is knocked down, we would not expect this to be the case for GAL4-driven overexpression of AMPs if this elevated expression is directly responsible for neurodegeneration. To test this theory, we drove UAS-relishRNAi with Repo-GAL4 to knock down Relish expression in glia and asked whether this affected the neuropathology caused by overexpresson of Defensin or Drosomycin. We found that neurodegeneration in Repo-GAL4 > UAS-Defensin, UAS-relishRNAi and Repo-GAL4 > UAS-Drosomycin, UAS-relishRNAi flies at 25 d of age was not suppressed compared with Repo-GAL4 > UAS-Defensin and Repo-GAL4 > UAS-Drosomycin, respectively (Fig. 5E). Thus, neurodegeneration caused by direct overexpression of AMPs in glia cannot be suppressed by knocking down Relish, supporting our model that AMP up-regulation as a consequence of constant activation of the Imd pathway in the nervous system is necessary and sufficient to cause neurodegeneration.
Discussion
There is considerable evidence linking immune-response pathways with neurodegenerative disease in humans. The role of the immune response in neurodegeneration is complex and has been referred to as a double-edged sword (26). On the one hand, the immune response appears to serve a protective role by promoting the clearance of potentially toxic protein aggregates and the phagocytosis of dead or damaged cells (27–29). On the other hand, a prolonged elevated inflammatory response can exacerbate neuronal cell damage and cause additional deleterious effects. Although the precise mechanisms are not yet fully understood, the release of neurotoxic factors and the generation of reactive oxygen species by stimulated microglia have been implicated in several studies (30). The harmful consequences of an exaggerated or sustained immune response in the nervous system emphasize the importance of regulatory mechanisms that modulate the response to limit its duration and amplitude. This study, together with other recent reports (16, 31) linking elevated immune responses with neurodegeneration in Drosophila, make this organism an excellent model for investigating the underlying cellular and molecular mechanisms.
Here we demonstrate that loss of dnr1, which encodes a negative regulator of the Imd immune response pathway in Drosophila, is associated with progressive, age-dependent neurodegeneration. Our investigation leads us to suggest that the elevated immune response in dnr1 mutants is directly responsible for the resultant neurodegeneration. In particular, increased expression of AMPs that normally protect against invading pathogens also appear to be toxic to neurons when expressed at high levels over an extended time. This mechanism can account for a significant portion of the neurodegeneration observed in dnr1 mutants.
The Dnr1 protein contains a RING ubiquitin ligase domain closely related to those found in inhibitor of apoptosis proteins in flies and mammals that are known to inhibit initiator caspases in the apoptotic pathway. Previous biochemical and genetic evidence demonstrate that Dnr1 negatively regulates Dredd by physically associating with it to promote its proteolytic degradation. Because Dredd activates the NF-κB transcription factor Relish by cleavage of a C-terminal autoinhibitory domain, loss of Dnr1 should cause persistent activation of the Imd pathway in the absence of any other inducing trigger, such as bacterial infection or mutation in another pathway such as ATM. In support of this conclusion, we observe elevated expression of AMP genes in dnr1 mutants. We also examined the possibility that loss of Dnr1 might lead to neurodegeneration because of inappropriate activation of apoptosis because Dnr1 appears to negatively regulate apoptosis in S2 cells (6). However, we found little evidence for caspase 3-positive cells in brains of 20- to 25-d-old dnr1 mutants, suggesting that Dnr1 does not have a critical role in regulating apoptosis in the CNS. On the other hand, suppression of neurodegeneration in dnr1 mutants by loss of Relish supports the idea that excessive activation of the Imd innate immune-response pathway in the nervous system is necessary and sufficient to cause neurodegeneration.
Consistent with this model, we found that direct activation of the innate immune response in the brain by piercing it with a fine bacteria-coated needle resulted in localized neurodegeneration at the injection site. Moreover, as with dnr1 mutants, neurodegeneration caused by bacterial injection in the brain is dependent on Relish, as expected if it is mediated by an elevated immune response. Because we injected a mixture of Gram-positive and Gram-negative bacteria into the brain, which activate the Toll and Imd pathways, respectively, one might not have expected that knocking down Relish alone would suppress neurodegeneration as fully as it did. However, comparable observation have been made in previous studies where injection of a bacterial mixture of E. coli and M. luteus in the Drosophila thorax induces expression of all AMP genes, including those regulated by the Toll pathway. Interestingly, expression of Toll-dependent AMPs Metchnikowin and Drosomycin was reduced to 50% and 80%, respectively, of normal levels in relE20 and dredd mutants, suggesting that the Imd pathway can affect expression of these AMPs directly or indirectly (19). Moreover, in surface epithelia it has been found that expression of Drosomycin and Metchnikowin is dependent on Imd and not the Toll pathway (31, 32). Thus, by analogy with these previous studies, we suggest that knocking down Relish in the nervous system not only leads to down-regulation of Imd-dependent AMPs, but also to a decrease in Drosomycin and Metchnikowin traditionally thought of as being Toll-dependent.
Although glia are major contributors to the innate immune response in the nervous system of both Drosophila and mammals, it was of interest that neurodegeneration associated with bacterial infection in the brain could be suppressed by knocking down Relish activity with RNAi either in glia or neurons. This result is consistent with studies in mammals demonstrating that in addition to the contributions from activated glia, cell-autonomous immune responses in neurons can play a critical role in neurodegeneration (8).
Importantly, we found that bacterial infection only evoked neurodegeneration when the injection was directly in the brain; systemic activation of the immune response by injection in the thorax did not cause neurodegeneration. In contrast, in mouse and humans various studies have shown that changes in peripheral immune responses, such as systemic inflammation, can contribute to neurodegeneration through release of factors, such as TNF, and by infiltration of the brain by circulating immune cells (33). The difference between flies and mammals in the effect of systemic immune responses on neurodegeneration is likely to be associated with differences in the blood–brain barrier, as well as differences in the nature of their circulatory systems.
Activation of the immune response by loss of dnr1 or by bacterial injection in the brain results in increased expression of many genes that are part of the defense mechanisms used to contain and attack microbial invaders. Consequently, there are many potential candidate genes whose elevated expression could contribute to neurodegeneration by a severe or prolonged activation of the immune response. Because of their prominent role in the innate immune response and because they can exert direct toxic effects on bacterial membranes (5), we focused on the possible role of AMPs. We found that driving the overexpression of various individual AMPs, either in neurons or glia, could cause the appearance of vacuolar neuropathology in brains reminiscent of that seen in dnr1 mutants or bacterially injected flies. Although the neurodegeneration caused by overexpression of single AMPs is relatively modest, it is notable that we see any neuropathology at all in these flies. The combined effect of expressing many AMPs simultaneously, as would occur in dnr1 mutants or bacterially injected flies, is expected to be much more severe. Thus, although we cannot rule out the possibility that other effector mechanisms of the innate immune response can exert toxic effects on neurons under some conditions, it seems that AMP-associated toxicity can account for a significant portion of the phenotypes observed in dnr1 mutants. Loss of dnr1 and constitutive activation of the innate immune response does not have any obvious cytotoxic effects in other organs, such as the fat body, suggesting the possibility that neurons might be particularly sensitive to toxic effects of AMPs. The reason for such a differential toxicity remains unknown and will be of interest to investigate further. Recent studies demonstrating that Drosomycin can interact with voltage-activated sodium channels in Drosophila and modify their activity (34) suggests one possible mechanism by which Drosomycin, and perhaps other AMPs, could exert specific effects on neurons.
The proposed mechanism of AMP-induced neurodegeneration could also account, at least partially, for the neurodegeneration observed in ATM mutants and retinal degeneration in norpA mutants, both of which result in activation of immune-response pathways with increased expression of AMP genes (15, 16). Interestingly, although retinal degeneration in norpA depends on Dredd and Relish, it is apparently independent of Imd/dFadd signaling, suggesting that Dredd may be activated via an alternative pathway. In mammals, there are cytosolic as well as membrane-associated innate immune receptors that can activate common downstream pathways. Possibly, such a mechanism is responsible for Dredd activation in norpA mutants.
There is a growing body of evidence that misregulation of the innate immune response in the human CNS, including elevated expression of AMPs, plays a key role in the neuropathology associated with various neurodegenerative disorders, aging, diabetes, and traumatic brain injury, although the mechanisms remain to be elucidated. A number of AMPs, including counterparts of some invertebrate AMPs, are expressed in mammalian brains (35) and are part of the innate immune-response mechanism, although the pathways regulating their expression may differ from Drosophila. In addition to providing defense against infection, many of these AMPs also function as signaling molecules that additionally regulate components of the immune response. Most of the conditions that result in neuropathology, such as neurodegenerative disease, traumatic brain injury, and aging are associated with a proinflammatory state with aberrant expression of AMPs. It has been proposed that these changes exacerbate the proinflammatory state in the brain and potentiate the neurodegenerative process (36). Among the various means that have been considered by which altered AMP expression in the brain could cause deleterious effects, one possibility is a direct cytotoxic effect on brain cells. The results we have presented here strongly support this possibility and provide a direct link between mechanisms of neurodegeneration in flies and humans.
In this regard, the most remarkable link between AMPs and human neurodegenerative disorders is the recent evidence that the Amyloid-β peptide, the normal in vivo function of which has been enigmatic, is in fact an AMP with antimicrobial and immune regulatory activities (37). This finding provides an explanation for the large body of evidence linking neuroinflammation with AD neuropathology. Together with our demonstration that AMPs can be neurotoxic, the Aβ-AMP link also suggests new ways of thinking about the pathophysiology underlying neurodegeneration in AD and related disorders, as well as opportunities for therapeutic intervention.
In summary, our identification of a mutation in a negative regulator of the immune response pathway in an unbiased screen for mutants exhibiting neurodegeneration has provided compelling evidence that constitutive activation of the innate immune response can cause neurodegeneration in the absence of any other triggers. On the basis of our results, we suggest that toxic effects of AMPs expressed persistently or at high levels in neurons or glia can largely account for this neurodegeneration. These results highlight the important neuroprotective role of negative regulators, such as Dnr1, that normally act to constrain the immune response and may be particularly critical for preserving neuronal viability. Recent important findings linking misexpression of AMPs with neurodegeneration in humans strongly suggest that our results will be of general significance.
Materials and Methods
Drosophila Genetics.
Flies were maintained on cornmeal-molasses medium at 25 °C unless otherwise stated. Canton-S was used as the wild-type control. dnr12-133 is from a laboratory collection of P-element insertion lines. UAS-dnr1 was obtained from Edan Foley (University of Alberta, Edmonton, AB, Canada); Repo-GAL4, UAS-Drosocin, UAS-Attacin, UAS-CecropinA1, UAS-Defensin, UAS-Drosomycin, and UAS-Metchnikowin (38) were obtained from David A. Wassarman (University of Wisconsin, Madison, WI); dnr1DG29507, Df(2R)X58-12, relishE20, relishE38, and Df(3R)ED05331 were obtained from the Bloomington Stock Center; UAS-relishRNAi (15) were obtained from Vienna Drosophila RNAi Center.
Histology.
Flies were collected upon eclosion and aged at 29 °C, except for the bacterial injection experiments for which the flies were raised and aged at 25 °C. Fly heads were severed and placed in fresh Carnoy’s fixative (ethanol:chloroform:acetic acid at 6:3:1) overnight at 4 °C. Heads were then washed and placed in 70% (vol/vol) ethanol and processed into paraffin using standard histological procedures. Embedded heads were sectioned at 5 µm, and stained with H&E. Images were taken under a Nikon light microscope (Nikon), equipped with a QImaging camera and images were generated using QImaging software (QImaging).
Lifespan Analysis.
One-hundred flies (five vials of 20 flies each) for each genotype were collected and transferred to 29 °C at 1 d posteclosion. Flies were transferred to fresh vials every other day. The number of surviving flies was recorded every 3 d during the first 21 d, and daily thereafter.
Scoring of Neurodegeneration.
Neurodegeneration is indicated by the appearance of vacuolar lesions in the brain neuropil. Six levels of neurodegeneration (0, 1, 2, 3, 4, and 5) are defined for quantification (Fig. S3). The higher number indicates more severe neurodegeneration. Scoring of brains was done blind with respect to genotype. For most genotypes, sample sizes of 6–12 brains were examined and quantified. Values shown are mean ± SEM. All values are listed in Table S1.
Molecular Analysis.
TAIL PCR was used to identify the site of P-element insertion in dnr2-133. Genomic DNA was isolated from dnr2-133 adults. Primers 2223, pry4, and TAIL1 were used in a primary PCR. Primers p5out2, 2231, and TAIL2 were used in a secondary PCR. Primers p5out1, 2229, and TAIL3 were used in the tertiary PCR. All primer sequences are provided in Table S2. qPCR was used to measure mRNA expression levels. Flies were collected at indicated time points and RNA was isolated using the RNeasy Plus Mini Kit (Qiagen). cDNA was generated using an iScript cDNA Synthesis Kit (Bio-Rad). Real-time PCR was carried out by using iQ SYBR Green Supermix (Bio-Rad). Primer sequences are provided in Table S3. PCR was performed as follows: 35 cycles: step 1: 95 °C for 10 s, step 2: 60 °C for 30 s, step 3: 72 °C for 40 s each cycle.
Bacterial Injection.
Bacteria used in this study, E. coli (Gram-negative) and M. luteus (Gram-positive) (kindly provided by Heidi Goodrich-Blair, Department of Bacteriology, University of Wisconsin, Madison, WI) were cultured overnight in LB medium at 37 °C. Infection of brains with bacteria was done manually with a 0.1-mm minuten pin (Fine Science Tools) mounted on a small holder. The minuten pin was dipped into 1:1 mixture of concentrated E. coli and M. luteus bacterial pellets (OD = 200) obtained after centrifugation of the liquid cultures in their exponential phase of growth. The pin was inserted into the brain through the left eye of CO2-anesthetized flies. The reproducibility of injection was monitored by the appearance of a melanization spot at the injection site and by subsequent histological analysis, where the injection track could be observed. The same procedure was followed for control experiments except with a sterile pin. For thorax injection experiments, flies were pricked with the pin, either sterile or coated with bacteria, through the cuticle on one side of the thorax. All infection experiments were carried out at 25 °C.
Behavior Test.
A climbing assay was used to quantify mobility. Groups of 10–15 adult flies of each indicated genotype were aged for 25 d at 25 °C. Flies were tapped to the bottom of the vial and videotaped for 10 s. The number of flies remaining on bottom was scored. For each genotype, five independent replicates were averaged.
Statistical Analysis.
Data were analyzed using GraphPad Prism software (GraphPad Software). The mean values were compared by nonparametric unpaired Student's t test between two groups or by one-way ANOVA with Dunnett’s posttest between more than two groups. In all tests, P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Edan Folley and Heidi Goodrich-Blair for fly stocks and bacterial strains; Aki Ikeda for use of his microtome facility; and Grace Boekhoff-Falk, Arash Bashirullah, David A. Wassarman, Richard Daniels, Daniel Babcock, Daniel Miller, and all members of the B.G. laboratory for fly stocks, advice, technical help, and discussions on the project. This work was supported by research Grant R01 AG033620 from the National Institutes of Health (to B.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306220110/-/DCSupplemental.
References
- 1.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21(11):2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86(6):973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 3.Kurata S. Extracellular and intracellular pathogen recognition by Drosophila PGRP-LE and PGRP-LC. Int Immunol. 2010;22(3):143–148. doi: 10.1093/intimm/dxp128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stöven S, Ando I, Kadalayil L, Engström Y, Hultmark D. Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000;1(4):347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 6.Primrose DA, et al. Interactions of DNR1 with the apoptotic machinery of Drosophila melanogaster. J Cell Sci. 2007;120(Pt 7):1189–1199. doi: 10.1242/jcs.03417. [DOI] [PubMed] [Google Scholar]
- 7.Guntermann S, Primrose DA, Foley E. Dnr1-dependent regulation of the Drosophila immune deficiency signaling pathway. Dev Comp Immunol. 2009;33(1):127–134. doi: 10.1016/j.dci.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 8.Czirr E, Wyss-Coray T. The immunology of neurodegeneration. J Clin Invest. 2012;122(4):1156–1163. doi: 10.1172/JCI58656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matzinger P. The danger model: A renewed sense of self. Science. 2002;296(5566):301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 10.Geylis V, Steinitz M. Immunotherapy of Alzheimer’s disease (AD): From murine models to anti-amyloid beta (Abeta) human monoclonal antibodies. Autoimmun Rev. 2006;5(1):33–39. doi: 10.1016/j.autrev.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Wilcock DM, et al. Microglial activation facilitates Abeta plaque removal following intracranial anti-Abeta antibody administration. Neurobiol Dis. 2004;15(1):11–20. doi: 10.1016/j.nbd.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 12.in ’t Veld BA, et al. NSAIDs and incident Alzheimer’s disease. The Rotterdam Study. Neurobiol Aging. 1998;19(6):607–611. doi: 10.1016/s0197-4580(98)00096-7. [DOI] [PubMed] [Google Scholar]
- 13.Gao XA, Chen HL, Schwarzschild MA, Ascherio A. Use of ibuprofen and risk of Parkinson disease. Neurology. 2011;76(10):863–869. doi: 10.1212/WNL.0b013e31820f2d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan L, Schedl P, Song HJ, Garza D, Konsolaki M. The Toll—>NFkappaB signaling pathway mediates the neuropathological effects of the human Alzheimer’s Abeta42 polypeptide in Drosophila. PLoS ONE. 2008;3(12):e3966. doi: 10.1371/journal.pone.0003966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinchore Y, Gerber GF, Dolph PJ. Alternative pathway of cell death in Drosophila mediated by NF-kappaB transcription factor Relish. Proc Natl Acad Sci USA. 2012;109(10):E605–E612. doi: 10.1073/pnas.1110666109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen AJ, Rimkus SA, Wassarman DA. ATM kinase inhibition in glial cells activates the innate immune response and causes neurodegeneration in Drosophila. Proc Natl Acad Sci USA. 2012;109(11):E656–E664. doi: 10.1073/pnas.1110470109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25(3):674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- 18.Foley E, O’Farrell PH. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2004;2(8):E203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 2000;1(4):353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naitza S, et al. The Drosophila immune defense against Gram-negative infection requires the death protein dFADD. Immunity. 2002;17(5):575–581. doi: 10.1016/s1074-7613(02)00454-5. [DOI] [PubMed] [Google Scholar]
- 21.Tanji T, Yun EY, Ip YT. Heterodimers of NF-kappaB transcription factors DIF and Relish regulate antimicrobial peptide genes in Drosophila. Proc Natl Acad Sci USA. 2010;107(33):14715–14720. doi: 10.1073/pnas.1009473107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzou P, Meister M, Lemaitre B. Methods for studying infection and immunity in Drosophila. Method Microbiol. 2002;31:507–529. [Google Scholar]
- 23.Yao KM, White K. Neural specificity of elav expression: Defining a Drosophila promoter for directing expression to the nervous system. J Neurochem. 1994;63(1):41–51. doi: 10.1046/j.1471-4159.1994.63010041.x. [DOI] [PubMed] [Google Scholar]
- 24.Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8(8):981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- 25.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415(6870):389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 26.Wyss-Coray T, Mucke L. Inflammation in neurodegenerative disease—A double-edged sword. Neuron. 2002;35(3):419–432. doi: 10.1016/s0896-6273(02)00794-8. [DOI] [PubMed] [Google Scholar]
- 27.Mattson MP. NF-kappaB in the survival and plasticity of neurons. Neurochem Res. 2005;30(6-7):883–893. doi: 10.1007/s11064-005-6961-x. [DOI] [PubMed] [Google Scholar]
- 28.Mattson MP, Meffert MK. Roles for NF-kappaB in nerve cell survival, plasticity, and disease. Cell Death Differ. 2006;13(5):852–860. doi: 10.1038/sj.cdd.4401837. [DOI] [PubMed] [Google Scholar]
- 29.Sarnico I, et al. NF-kappaB dimers in the regulation of neuronal survival. Int Rev Neurobiol. 2009;85:351–362. doi: 10.1016/S0074-7742(09)85024-1. [DOI] [PubMed] [Google Scholar]
- 30.Block ML, Zecca L, Hong JS. Microglia-mediated neurotoxicity: Uncovering the molecular mechanisms. Nat Rev Neurosci. 2007;8(1):57–69. doi: 10.1038/nrn2038. [DOI] [PubMed] [Google Scholar]
- 31.Tzou P, et al. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 2000;13(5):737–748. doi: 10.1016/s1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- 32.Ferrandon D, et al. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 1998;17(5):1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmes C, et al. Systemic inflammation and disease progression in Alzheimer disease. Neurology. 2009;73(10):768–774. doi: 10.1212/WNL.0b013e3181b6bb95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen L, et al. Drosomycin, an innate immunity peptide of Drosophila melanogaster, interacts with the fly voltage-gated sodium channel. J Biol Chem. 2009;284(35):23558–23563. doi: 10.1074/jbc.M109.023358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schluesener HJ, Su Y, Ebrahimi A, Pouladsaz D. Antimicrobial peptides in the brain: Neuropeptides and amyloid. Front Biosci (Schol Ed) 2012;4:1375–1380. doi: 10.2741/s339. [DOI] [PubMed] [Google Scholar]
- 36.Williams WM, Castellani RJ, Weinberg A, Perry G, Smith MA. Do β-defensins and other antimicrobial peptides play a role in neuroimmune function and neurodegeneration? ScientificWorldJournal. 2012;2012:905785. doi: 10.1100/2012/905785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soscia SJ, et al. The Alzheimer’s disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS ONE. 2010;5(3):e9505. doi: 10.1371/journal.pone.0009505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonthius DJ, et al. Deficiency of neuronal nitric oxide synthase (nNOS) worsens alcohol-induced microencephaly and neuronal loss in developing mice. Brain Res Dev Brain Res. 2002;138(1):45–59. doi: 10.1016/s0165-3806(02)00458-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.