Abstract
Immunoresponsive gene 1 (Irg1) is highly expressed in mammalian macrophages during inflammation, but its biological function has not yet been elucidated. Here, we identify Irg1 as the gene coding for an enzyme producing itaconic acid (also known as methylenesuccinic acid) through the decarboxylation of cis-aconitate, a tricarboxylic acid cycle intermediate. Using a gain-and-loss-of-function approach in both mouse and human immune cells, we found Irg1 expression levels correlating with the amounts of itaconic acid, a metabolite previously proposed to have an antimicrobial effect. We purified IRG1 protein and identified its cis-aconitate decarboxylating activity in an enzymatic assay. Itaconic acid is an organic compound that inhibits isocitrate lyase, the key enzyme of the glyoxylate shunt, a pathway essential for bacterial growth under specific conditions. Here we show that itaconic acid inhibits the growth of bacteria expressing isocitrate lyase, such as Salmonella enterica and Mycobacterium tuberculosis. Furthermore, Irg1 gene silencing in macrophages resulted in significantly decreased intracellular itaconic acid levels as well as significantly reduced antimicrobial activity during bacterial infections. Taken together, our results demonstrate that IRG1 links cellular metabolism with immune defense by catalyzing itaconic acid production.
During inflammatory responses, activated peripheral macrophages and microglial cells, the resident immune cells in the central nervous system, produce an array of inflammatory cytokines and free radicals that are responsible for their antimicrobial activity (1). Depending on the pathogens and the cytokine environment, macrophages are able to acquire a large spectrum of different phenotypes having specific pro- or anti-inflammatory properties (2, 3). Similar observations have been made in microglial cells, where the pro- or anti-inflammatory states have been elicited by different cytokines or bacterial components (4). In this context, gene expression profiling studies of murine macrophages and microglial cells have identified immunoresponsive gene 1 (Irg1) as one of the most highly up-regulated genes under proinflammatory conditions, such as bacterial infections (5–9). Increased Irg1 expression levels have also been detected in chicken spleen macrophages after infection with Salmonella enterica (10). Furthermore, it has been shown that IRG1 could play an important role in the susceptibility toward Marek disease virus (11). Irg1 was originally identified as a 2.3-kb cDNA from a library synthesized from mRNA isolated from a murine macrophage cell line after LPS stimulation (12). Irg1 is also highly expressed in the early events leading to implantation in the pregnant uterus (13, 14), the specific phase in which a high level of inflammatory cytokines are secreted by the endometrial cells as well as by cells of the immune system that are recruited to the site of implantation (15). Interestingly, Irg1 has been used to classify functional profiles of neurotoxic C-C chemokine receptor 9+Irg1+ and neurosupportive C-X-C chemokine receptor 3+Irg1− microglia in vivo (16). Although no mitochondria targeting signal sequence could be identified in the IRG1 amino acid sequence, IRG1 protein has been reported to associate with mitochondria (6).
Taken together, these data indicate an important role of Irg1 during immune response. Although expression levels of Irg1 have been extensively studied, its cellular function has not been addressed and is unknown. Based on sequence homology, IRG1 has been classified into the MmgE/PrpD family (17), which contains some proteins for which enzymatic activities have been identified in microorganisms (18). To elucidate if mammalian IRG1 exhibits an enzymatic function, we performed siRNA-mediated silencing of its expression in various cell models under inflammatory conditions and performed a nontargeted metabolomics analysis to detect significantly affected metabolites. Based on this analysis, we were able to elucidate the function of IRG1 as an enzyme catalyzing the production of the antimicrobial metabolite itaconic acid by decarboxylating cis-aconitate.
Results and Discussion
IRG1 Function.
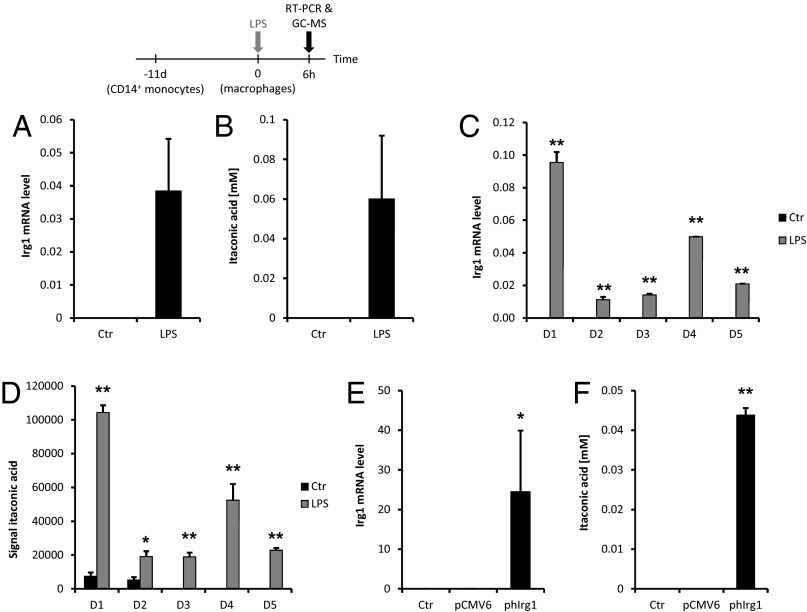
To investigate whether IRG1 has a potential enzymatic function, we analyzed the metabolomics profile of siRNA-mediated Irg1 silencing under LPS stimulation in RAW264.7 cells. We first confirmed that the silencing of Irg1 resulted in an 80% decrease of Irg1 mRNA level compared with nonspecific siRNA control (Fig. 1A). We detected very low levels of Irg1 mRNA (17-fold less compared with LPS-activated cells) in nonactivated macrophages. After silencing of Irg1 in RAW264.7 cells, we measured 260 intracellular metabolites and, of these, we found that 5 were significantly different compared with untreated RAW264.7 cells (Welch's t test, P < 0.05). Most strikingly, we observed that the metabolite most significantly affected by Irg1 silencing was itaconic acid (P = 2.5 × 10−8). This metabolite was very recently discovered by Strelko and colleagues in LPS-activated macrophages (19). However, until now, the enzyme catalyzing its production has not been described. Based on our experiments, the silencing of Irg1 elicited a 60% decrease of itaconic acid compared with control conditions (Fig. 1A). Only low levels of the metabolite (11.5-fold less compared with LPS-activated cells) were measured in resting macrophages. Having identified itaconic acid as the main affected metabolite by Irg1 silencing, we performed an intracellular quantification of this compound and found a concentration of 3 mM in BV2 retroviral-immortalized microglia (BV-2) cells and 8 mM in RAW264.7 mouse macrophages after LPS treatment (LPS 10 ng/mL) (Fig. 1B). We measured similar amounts in murine primary microglial cells induced by LPS treatment (Fig. S1). Such high intracellular itaconic acid concentrations after LPS treatment clearly point toward an immunological function of this metabolite.
Fig. 1.
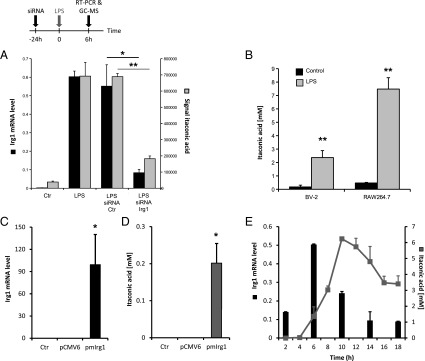
(A) Levels of mRNA (left y axis, black bars) or itaconic acid (right y axis, gray bars) in resting (Ctr) or LPS-activated RAW264.7 macrophages transfected with either siRNA specific for Irg1 or with siRNA Ctr. Metabolites and RNA extractions were performed after 6 h of stimulation. The levels of Irg1 mRNA were determined by real-time RT-PCR and normalized using L27 as a housekeeping gene. Each bar represents the average expression fold change (± SD). The levels of itaconic acid were determined by GC/MS measurements. Each bar represents itaconic acid levels (± SD). *P < 0.05, **P < 0.01. (B) Itaconic acid quantification (mM) in mouse microglial cells (BV-2 cell line) and mouse macrophages (RAW264.7 cell line) after 6 h of LPS stimulation at 10 ng/mL (gray bars). Untreated cells were used as a control (black bars). Bars represent the mean of itaconic acid concentration (± SEM). **P < 0.01. Levels of mRNA (C) or itaconic acid (D) in human A549 lung cancer cells transfected with the mouse Irg1 overexpressing plasmid (pmIrg1). Metabolites and RNA extractions were performed 24 h after transfection. Real-time RT-PCR results are normalized using L27 as a housekeeping gene and are shown as average expression fold change (± SEM). *P < 0.05, **P < 0.01. (E) RAW264.7 cells were treated with LPS (10 ng/mL) at different time points and analyzed for Irg1 expression and itaconic acid concentration (mM).
To further study the metabolic activity of IRG1, we overexpressed murine Irg1 in A549 human lung cancer cells. We found that these cells contained high amounts of both Irg1 gene transcript and itaconic acid metabolite (0.2 ± 0.05 mM) 24 h after transfection, but not in nontransfected cells or in cells transfected with an empty control plasmid, in which Irg1 mRNA and itaconic acid were below detection limit (Fig. 1 C and D).
Finally, to investigate the dynamics of Irg1 expression and itaconic acid production after a proinflammatory stimulus, we analyzed RAW264.7 cells activated with LPS (10 ng/mL) at different time points. Although Irg1 transcript was already produced after 2 h, significant amounts of itaconic acid could be measured starting 6 h after LPS treatment (Fig. 1E). The time-course of Irg1 expression is in line with observed expression profiles of other proinflammatory cytokines. The positive time-dependent correlation between Irg1 expression and itaconic acid levels further support the cellular role of IRG1 in itaconic acid production.
Itaconic Acid Metabolic Pathway.
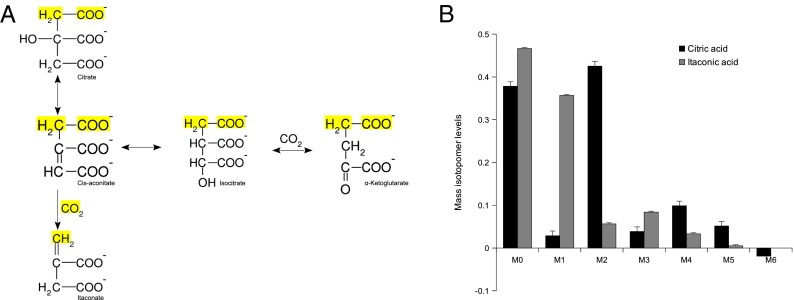
Intriguingly, murine IRG1 shows a 23% amino acid sequence identity to the enzyme cis-aconitate decarboxylase (CAD) expressed by the fungus Aspergillus terreus (Fig. S2). In fact, IRG1 is evolutionary conserved across a large set of species (Fig. S3). This fungus is commonly used for the biotechnological production of itaconic acid at an industrial scale (20). The biosynthesis of this dicarboxylic acid has been of interest because it can be used as a starting material for chemical synthesis of polymers (21). The fungal CAD enzyme catalyzes the formation of itaconic acid by decarboxylating cis-aconitate to itaconic acid (22). To determine if mammalian IRG1 has a similar function as CAD in A. terreus, we performed stable-isotope labeling experiments. We incubated LPS-activated RAW264.7 macrophages with uniformly 13C-labeled glucose (U-13C6). Citrate synthase catalyzes the transfer of two labeled carbon atoms from acetyl-CoA to oxaloacetate resulting in M2 cis-aconitate isotopologues (Fig. 2A). If the decarboxylation is performed by a CAD homolog, the first carbon atom of the molecule is expected to be removed during the decarboxylation, resulting in M1 isotopologues of itaconic acid. In our experimental setting, we determined 45% of the citric acid molecules as M2 isotopologues, whereas 38% of the itaconic acid molecules were M1 isotopologues (Fig. 2B). We also found a significant fraction of M2, M3, and M4 itaconic acid isotopologues. The M4 fraction of itaconic acid reflects pyruvate carboxylase or reverse malic enzyme activity. Because of the symmetry of succinic acid, subsequent turns of the tricarboxylic acid (TCA) cycle can result in M2 or M3 isotopologues of itaconic acid.
Fig. 2.
(A) Synthesis pathway of itaconic acid in the TCA cycle. Itaconic acid can only contain one labeled carbon if produced in the first round of the TCA cycle (yellow-marked atoms). (B) Labeling of citric acid (black bars) and itaconic acid (gray bars) using glucose as a tracer in RAW264.7 macrophages. The major fraction of labeled itaconic acid contains one isotope, whereas citric acid contains mainly two labeled atoms.
The observations described so far strongly suggest that Irg1 encodes a mammalian enzyme that catalyzes the decarboxylation of cis-aconitate to itaconic acid.
IRG1 Protein Purification and CAD Activity Assay.
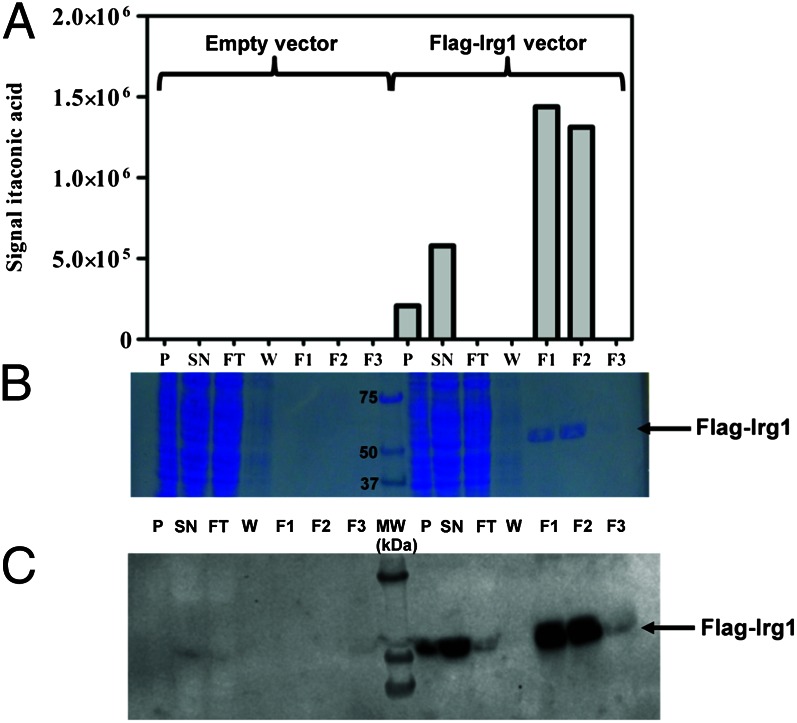
To directly demonstrate that the IRG1 protein catalyzes the decarboxylation of cis-aconitate, we purified FLAG-tagged IRG1 protein from human embryonic kidney 293T (HEK293T) cells transfected with a pCMV6-Entry-Irg1 expression plasmid. As depicted in Fig. 3A, protein extracts prepared from those cells catalyzed the conversion of cis-aconitate to itaconic acid. No itaconic acid formation was detected when extracts were prepared from cells transfected with an empty vector. Furthermore, affinity purification of the extract prepared from FLAG-Irg1 overexpressing cells clearly showed coelution of the cis-aconitate decarboxylase activity with a protein band identified as IRG1 by SDS/PAGE (expected molecular weight ∼55 kDa for Flag-Irg1; Fig. 3B) and Western blot analysis using anti-IRG1 antibody (Fig. 3C). SDS/PAGE analysis showed that this purification procedure yielded a homogenous preparation of the IRG1 protein (Fig. 3B), demonstrating that the cis-aconitate decarboxylase activity measured in the purified fractions was not due to another contaminating protein.
Fig. 3.
Purification of cis-aconitate decarboxylase from HEK293T cells transfected with the pCMV6-Entry-Irg1 expression plasmid. (A) Extracts from cells transfected with empty plasmid or Flag-Irg1 plasmid were loaded onto an affinity resin (Cell MM2, FlagM purification kit, Sigma Aldrich) and proteins were eluted with Flag peptide. Cis-aconitate decarboxylase activity was measured in cell extracts and purification fractions as described in Materials and Methods. (B) A total of 12 µL of each protein fraction was loaded onto an SDS/PAGE gel that was stained with Coomassie Blue. (C) Western blot analysis of the same protein fractions was performed using an IRG1-specific antibody. F1-F3, elution fractions; FT, flow through; P, pellet; SN, supernatant; W, wash.
Itaconic Acid Is Produced by Human Primary Macrophages but at Lower Levels Compared with Mouse Cells.
Because an Irg1 homologous gene is annotated in the human genome on chromosome 13, we were interested to further analyze IRG1 expression and itaconic acid amounts in human immune cells. To investigate this, we isolated CD14+ primary human monocytes from the blood of different donors, cultured them for differentiation into macrophages for 11 d, and stimulated an inflammatory response with LPS (10 µg/mL) for 6 h. In line with our previous observations in mouse macrophages, we observed that IRG1 expression in human peripheral blood mononuclear cell (PBMC)-derived macrophages was highly up-regulated after LPS activation compared with resting conditions in which IRG1 mRNA levels were almost undetectable (Fig. 4A). Our results are in accordance with those of Roach and colleagues (23), who analyzed LPS-activated PBMCs’ transcriptional profiles and observed IRG1 up-regulation compared with control conditions. At the metabolite level, itaconic acid amounts were highly increased under LPS-induced inflammatory conditions compared with resting cells in which the metabolite was measured in low amounts or below the detection limits (Fig. 4B). In line with the induction of itaconic acid production, we observed elevated IRG1 expression (Fig. 4 C and D). A similar trend was also mirrored by the expression of TNF-α mRNA, indicating that macrophages were activated (Fig. S4). Compared with the intracellular itaconic acid concentration in mouse immune cells, the concentration in human macrophages was two orders of magnitudes lower (8 mM vs. ∼60 µM).
Fig. 4.
Human IRG1 expression and itaconic acid production. Levels of mRNA and itaconic acid in resting (Ctr) and LPS-activated (10 µg/mL) PBMC-derived macrophages. RNA and metabolites extractions were performed after 6 h of stimulation. (A) The levels of IRG1 mRNA were determined by real-time RT-PCR and normalized using L27 as a housekeeping gene. Each bar represents the average expression fold change of five different donors (± SEM). (B) The corresponding levels of itaconic acid were determined by GC/MS measurements. Each bar represents itaconic acid levels (± SEM). *P < 0.05, **P < 0.01. (C and D) Differential IRG1 gene expression analysis and itaconic acid production between five different donors. (E and F) Levels of mRNA (E) or itaconic acid (F) in human A549 lung cancer cells transfected with the human IRG1 overexpressing plasmid (phIrg1). Metabolites and RNA extractions were performed 24 h after transfection. Real-time RT-PCR results are normalized using L27 as a housekeeping gene and are shown as average expression fold change (± SEM). *P < 0.05, **P < 0.01.
A major difference between mouse and human is the elevated production of NO in mouse macrophages under inflammatory conditions (24). It is well known that NO inhibits aconitase, the enzyme producing the itaconic acid precursor cis-aconitate (25–27). To test whether the NO-mediated inhibition of aconitase has an effect on itaconic acid production, we silenced the inducible nitric oxide synthetase (iNOS) gene to decrease NO levels in mouse macrophage (Fig. S5 A and B). On the other hand, we treated human PBMC-derived macrophages with the intracellular NO donor, diethylamine NONOate (28), to elevate the NO level in these cells (Fig. S6 A–C). We could not detect any effect on intracellular itaconic acid levels in either case. Based on these results, we assume that aconitase is not a rate-limiting step for itaconic acid synthesis.
Finally, we transfected A549 human lung cancer cells with a pCMV6 plasmid expressing human IRG1 cDNA (phIrg1) to show CAD activity of human IRG1. We observed high amounts of both IRG1 gene transcript and itaconic acid (0.044 ± 0.0018 mM) 24 h posttransfection (Fig. 4 E and F).
In Vivo Irg1 Expression and Itaconic Acid Production.
To confirm Irg1 expression and itaconic acid production in vivo, we intraperitoneally injected C57BL/6 mice with LPS (1 mg/kg) and harvested the peritoneal macrophages after 24 h. We were able to measure high Irg1 mRNA expression levels correlating with high amounts of intracellular itaconic acid compared with the saline-injected mice (Fig. 5 A and B).
Fig. 5.
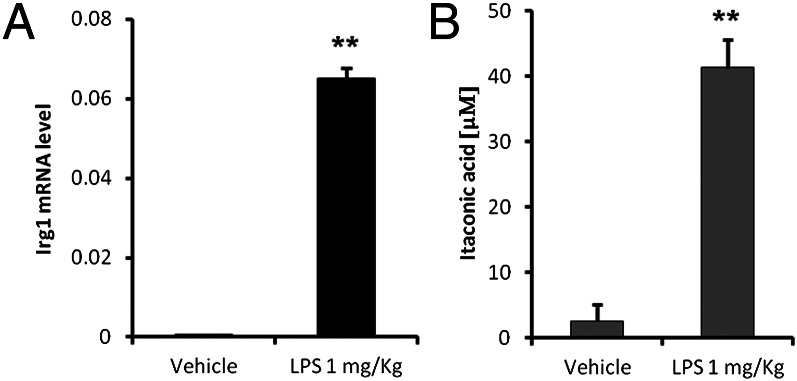
Mouse peritoneal macrophages from eight saline- and seven LPS-injected mice (1 mg/kg) were isolated and pooled 24 h after i.p. injection. (A) Irg1 expression levels and (B) itaconic acid production were analyzed compared with intraperitoneally saline-injected mice. Bars represent the mean of three technical replicates (± SEM).
Itaconic Acid Inhibits Bacterial Growth and Contributes to Antimicrobial Activity of Mouse Macrophages.
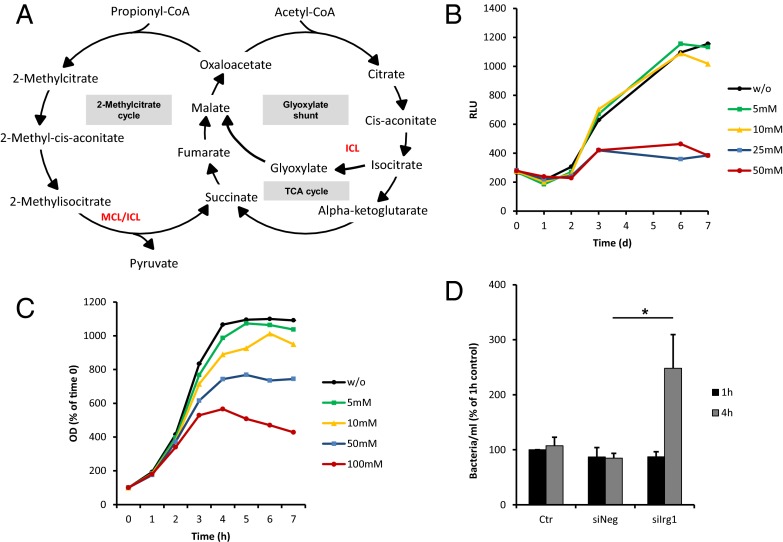
It has previously been shown that itaconic acid has an antimicrobial activity by inhibiting isocitrate lyase (ICL) (29, 30), an enzyme of the glyoxylate shunt (Fig. 6A). The glyoxylate shunt is not present in animals, but is essential for the survival of bacteria growing on fatty acids or acetate as the limiting carbon source (31). The strategy for survival during chronic stages of infection entails a metabolic shift in the bacteria’s carbon source to C2 substrates generated by β-oxidation of fatty acids (31). Under these conditions, glycolysis is decreased and the glyoxylate shunt is significantly up-regulated to allow anaplerotic maintenance of the TCA cycle and assimilation of carbon via gluconeogenesis (32). Highly elevated levels of ICL are observed in Mycobacterium tuberculosis grown on C2 sources (33) and shortly after uptake into human macrophages (34). Furthermore, it has been shown that persistence of M. tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme ICL (35). In fact, M. tuberculosis cannot persist in macrophages when both isoforms of icl are genetically knocked out (36). Because the glyoxylate shunt is exclusively found in prokaryotes, lower eukaryotes, and plants, it represents a unique target for drug development (37).
Fig. 6.
Effect of itaconic acid on the bacterial growth. (A) Schematic of the glyoxylate shunt and the 2-methylcitrate cycle. (B) GFP-expressing M. tuberculosis bacteria were cultured in 7H9 medium supplemented with acetate, without (w/o) or with various concentrations of itaconic acid (5, 10, 25, 50 mM). Growth was measured as relative light units (RLU) at indicated time points. Curves represent the mean of three technical replicates. (C) S. enterica was grown in liquid medium with acetate, without (w/o) or in the presence of different concentrations of itaconic acid (5, 10, 50, 100 mM). The OD was measured every hour. Curves are calculated in percent relative to time 0 and represent the mean of three independent experiments. (D) RAW264.7 cells were transfected with either siRNA specific for Irg1 (siIrg1) or with siRNA control (siNeg). After 24 h, the cells were infected with S. enterica at a multiplicity of infection of 1:10 and incubated for 1 h at 37 °C (Materials and Methods). Bars represent the mean of the numbers of bacteria per ml (± SEM) obtained from three independent experiments. *P < 0.05.
To confirm the antimicrobial effect of itaconic acid on bacterial replication (30), we cultured the pathogens M. tuberculosis and S. enterica (both known to synthesize ICL for biosynthesis through the glyoxylate shunt) in liquid minimal medium supplemented with acetate as the unique carbon source to force the bacterial metabolism to use the glyoxylate shunt.
We determined bacterial growth in this medium in the presence of increasing itaconic acid concentration and observed that the effective concentration of itaconic acid varies depending on the analyzed bacteria. The growth of M. tuberculosis in vitro was completely inhibited at 25–50 mM itaconic acid concentrations (Fig. 6B), whereas significant effects were already observed at 10 mM for S. enterica (Fig. 6C). To exclude secondary toxic effects of itaconic acid, we measured the growth of the bacteria on glycerol or glucose as a carbon source. In this case, bacterial metabolism does not rely on the glyoxylate shunt. Under these conditions, itaconic acid does not affect the bacterial growth (Figs. S7A and S8A). To further demonstrate the specificity of the antimicrobial activity of itaconic acid, we performed the same bacterial growth experiments, but by supplementing the medium with the itaconic acid precursor cis-aconitate. We observed that bacteria grown in the presence of cis-aconitate elicited an even more pronounced growth at different concentrations of this metabolite in both bacteria (Figs. S7 B and C and S8B), thus indicating that these bacteria started to use cis-aconitate as carbon source, in strong opposition with the effect we observed with itaconic acid.
It has been described that ICL exhibits additional methylisocitrate lyase (MCL) activity in M. tuberculosis. MCL is required for the detoxification of propionyl-CoA through the methylcitrate cycle (38, 39) (Fig. 6A). Propionyl-CoA accumulates during β-oxidation of odd-chain fatty acids and is produced from cholesterol of the host macrophages (40). As a result, inhibition of ICL in M. tuberculosis could have an additional toxic effect in the presence of propionate. To investigate the potential accentuated inhibition of the bacterial growth under these conditions, we incubated M. tuberculosis in glycerol with 0.1 µM propionate and increasing concentrations of itaconic acid. Indeed, the combination of these two effects inhibited M. tuberculosis growth at 5–10 mM of itaconic acid (Fig. S7D), thus confirming that MCL activity of ICL is affected by the metabolite.
To further investigate the involvement of itaconic acid in the antimicrobial activity of macrophages, we infected RAW264.7 cells with S. enterica and consequently observed an increased Irg1 expression associated with high intracellular amounts of itaconic acid (Fig. S9 A and B). We saw that silencing of Irg1 gene expression resulted in a decrease of intracellular itaconic acid concentration (Fig. S9B). We detected a significantly larger number of intracellularly viable bacteria in macrophages treated with siRNA targeting Irg1 compared with those treated with an unspecific control siRNA or with siRNA targeting murine aconitase 2 (Aco2) 4 h after infection (Fig. 6D and Fig. S10).
Taken together, our results clearly demonstrate the importance of Irg1 expression in macrophages during bacterial infection, thus contributing to their antimicrobial armature.
Conclusion
In this study, we report that the Irg1 gene encodes an enzyme synthesizing itaconic acid from the TCA cycle intermediate cis-aconitate. We showed a strong up-regulation of both Irg1 transcript and itaconic acid synthesis in macrophages in response to LPS activation. Furthermore, our data also show that IRG1-mediated itaconic acid production contributes to the antimicrobial activity of macrophages.
The discovery of an inducible enzyme linked to the TCA cycle and catalyzing the production of an antimicrobial metabolite is reminiscent of the mechanism found in the urea cycle responsible for the production of NO. iNOS is the enzyme that catalyzes the production of the antimicrobial compound NO from L-arginine (41) and, like IRG1, is induced in mouse macrophages and microglial cells under inflammatory conditions. In addition to other factors, NO and itaconic acid production by immune cells seem to represent an intrinsic property of these cells to self-react toward an inflammatory insult, and those metabolites seem to confer a form of innate metabolic immunity to macrophages and microglial cells.
Materials and Methods
Cell Culture.
Primary human monocytic CD14+ cells were isolated in two steps from blood samples provided by Red Cross Luxembourg. First, (PBMCs were separated in 50-mL Leucoseptubes (Greiner) through Ficoll-Paque Premium (GE Healthcare) density-gradient centrifugation at 1,000 × g for 10 min at room temperature with no brake. Second, CD14+ cells were purified with magnetic labeling. Therefore, 2 μL of CD14 Microbeads (Miltenyi Biotech) per 107 PBMCs were incubated for 30 min at 4 °C followed by a positive LS column (Miltenyi Biotech) magnetic selection. The purified CD14+ cells were differentiated in six-well plates for 11 d in RPMI1640 medium without L-glutamine and phenol red (Lonza) supplemented with 10% (vol/vol) human serum (A&E Scientific), 1% (vol/vol) penicillin/streptomycin (Invitrogen), and 0.05% (vol/vol) L-glutamine (Invitrogen). The medium was changed on days 4 and 7.
Four cell lines, specifically murine microglial BV-2 cells (42), murine macrophages RAW264.7 (43) (ATCC TIB-71), human epithelial A549 lung cancer cells (44) (ATCC CCL-185), and human HEK293T cells (45) were used for the experiments.
Mouse i.p. Injection of LPS and Peritoneal Macrophages Isolation.
All animal procedures, such as handling and euthanasia, have been performed according to the Federation of European Laboratory Animal Science Associations (FELASA) guidelines for the use of animals in research, and were institutionally approved by the Luxembourg Centre for System Biomedicine animal user committee and authorized by the local governmental agencies (Ministry of Health, Ministry of Agriculture, chief veterinarian of the Luxembourg government). Three-to-four-month-old SJL mice were injected i.p. with LPS (1 mg/kg) or with saline vehicle and were deeply anesthetized after 24 h by i.p. injection of 50 mg/kg of ketamine⋅HCl and 5 mg/kg xylazine⋅HCl. Mice were then euthanized by cervical dislocation. Eight saline- and seven LPS-injected mice were used for peritoneal macrophages isolation. A small incision was made in the upper abdomen, and peritoneal macrophages were washed out with 4–5 mL ice-cold sterile PBS/mouse and pooled into falcon tubes. The cell suspension was pelleted in a cooled centrifuge for 5 min at 250 × g and the resulting pellet was worked up for metabolites and RNA extractions.
M. tuberculosis Growth Analysis.
GFP-expressing M. tuberculosis H37Rv bacteria (47) were generated using the plasmid 32362:pMN437 (Addgene), kindly provided by M. Niederweis (University of Alabama, Birmingham, AL) (48). 1 × 106 bacteria were cultured in 7H9 medium supplemented with different carbon sources as indicated in a total volume of 100 μL in a black 96-well plate with a clear bottom (Corning Inc) sealed with an air-permeable membrane (Porvair Sciences). Growth was measured as relative light units at 528 nm after excitation at 485 nm in a fluorescence microplate reader (Synergy 2, Biotek) at indicated time points.
Statistical Analysis.
For comparison of means between two different treatments, the statistical analysis was done by the Student t test unless otherwise indicated. Error bars indicate SD or SEM as specified in the text.
For more information on protein purification and CAD activity assay, RNA isolation and RT-PCR, SDS/PAGE and Western blotting, GC/MS sample preparation and procedure as well as glucose labeling assay, please see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the laboratory of Prof. Paul Heuschling for mouse hosting, Prof. Stefan Ehlers for providing resources and facilitating interactions, and Lisa Niwinski for expert help and technical assistance. This study was supported by the Fonds National de la Recherche, Luxembourg (ATTRACT A10/03 and AFR 1328318).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218599110/-/DCSupplemental.
References
- 1.David S, Kroner A. Repertoire of microglial and macrophage responses after spinal cord injury. Nat Rev Neurosci. 2011;12(7):388–399. doi: 10.1038/nrn3053. [DOI] [PubMed] [Google Scholar]
- 2.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 3.Mantovani A, et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25(12):677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Michelucci A, Heurtaux T, Grandbarbe L, Morga E, Heuschling P. Characterization of the microglial phenotype under specific pro-inflammatory and anti-inflammatory conditions: Effects of oligomeric and fibrillar amyloid-beta. J Neuroimmunol. 2009;210(1-2):3–12. doi: 10.1016/j.jneuroim.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Tangsudjai S, et al. Involvement of the MyD88-independent pathway in controlling the intracellular fate of Burkholderia pseudomallei infection in the mouse macrophage cell line RAW 264.7. Microbiol Immunol. 2010;54(5):282–290. doi: 10.1111/j.1348-0421.2010.00205.x. [DOI] [PubMed] [Google Scholar]
- 6.Degrandi D, Hoffmann R, Beuter-Gunia C, Pfeffer K. The proinflammatory cytokine-induced IRG1 protein associates with mitochondria. J Interferon Cytokine Res. 2009;29(1):55–67. doi: 10.1089/jir.2008.0013. [DOI] [PubMed] [Google Scholar]
- 7.Thomas DM, Francescutti-Verbeem DM, Kuhn DM. Gene expression profile of activated microglia under conditions associated with dopamine neuronal damage. FASEB J. 2006;20(3):515–517. doi: 10.1096/fj.05-4873fje. [DOI] [PubMed] [Google Scholar]
- 8.Basler T, Jeckstadt S, Valentin-Weigand P, Goethe R. Mycobacterium paratuberculosis, Mycobacterium smegmatis, and lipopolysaccharide induce different transcriptional and post-transcriptional regulation of the IRG1 gene in murine macrophages. J Leukoc Biol. 2006;79(3):628–638. doi: 10.1189/jlb.0905520. [DOI] [PubMed] [Google Scholar]
- 9.Gautam A, et al. Interleukin-10 alters effector functions of multiple genes induced by Borrelia burgdorferi in macrophages to regulate Lyme disease inflammation. Infect Immun. 2011;79(12):4876–4892. doi: 10.1128/IAI.05451-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matulova M, et al. Characterization of chicken spleen transcriptome after infection with Salmonella enterica serovar Enteritidis. PLoS ONE. 2012;7(10):e48101. doi: 10.1371/journal.pone.0048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith J, et al. Systems analysis of immune responses in Marek’s disease virus-infected chickens identifies a gene involved in susceptibility and highlights a possible novel pathogenicity mechanism. J Virol. 2011;85(21):11146–11158. doi: 10.1128/JVI.05499-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee CG, Jenkins NA, Gilbert DJ, Copeland NG, O’Brien WE. Cloning and analysis of gene regulation of a novel LPS-inducible cDNA. Immunogenetics. 1995;41(5):263–270. doi: 10.1007/BF00172150. [DOI] [PubMed] [Google Scholar]
- 13.Cheon YP, Xu X, Bagchi MK, Bagchi IC. Immune-responsive gene 1 is a novel target of progesterone receptor and plays a critical role during implantation in the mouse. Endocrinology. 2003;144(12):5623–5630. doi: 10.1210/en.2003-0585. [DOI] [PubMed] [Google Scholar]
- 14.Chen B, Zhang D, Pollard JW. Progesterone regulation of the mammalian ortholog of methylcitrate dehydratase (immune response gene 1) in the uterine epithelium during implantation through the protein kinase C pathway. Mol Endocrinol. 2003;17(11):2340–2354. doi: 10.1210/me.2003-0207. [DOI] [PubMed] [Google Scholar]
- 15.Mor G, Cardenas I, Abrahams V, Guller S. Inflammation and pregnancy: The role of the immune system at the implantation site. Ann N Y Acad Sci. 2011;1221:80–87. doi: 10.1111/j.1749-6632.2010.05938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, et al. Different neurotropic pathogens elicit neurotoxic CCR9- or neurosupportive CXCR3-expressing microglia. J Immunol. 2006;177(6):3644–3656. doi: 10.4049/jimmunol.177.6.3644. [DOI] [PubMed] [Google Scholar]
- 17.Lohkamp B, Bäuerle B, Rieger PG, Schneider G. Three-dimensional structure of iminodisuccinate epimerase defines the fold of the MmgE/PrpD protein family. J Mol Biol. 2006;362(3):555–566. doi: 10.1016/j.jmb.2006.07.051. [DOI] [PubMed] [Google Scholar]
- 18.Kanamasa S, Dwiarti L, Okabe M, Park EY. Cloning and functional characterization of the cis-aconitic acid decarboxylase (CAD) gene from Aspergillus terreus. Appl Microbiol Biotechnol. 2008;80(2):223–229. doi: 10.1007/s00253-008-1523-1. [DOI] [PubMed] [Google Scholar]
- 19.Strelko CL, et al. Itaconic acid is a mammalian metabolite induced during macrophage activation. J Am Chem Soc. 2011;133(41):16386–16389. doi: 10.1021/ja2070889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bentley R, Thiessen CP. Biosynthesis of itaconic acid in Aspergillus terreus. I. Tracer studies with C14-labeled substrates. J Biol Chem. 1957;226(2):673–687. [PubMed] [Google Scholar]
- 21.Okabe M, Lies D, Kanamasa S, Park EY. Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Appl Microbiol Biotechnol. 2009;84(4):597–606. doi: 10.1007/s00253-009-2132-3. [DOI] [PubMed] [Google Scholar]
- 22.Bonnarme P, et al. Itaconate biosynthesis in Aspergillus terreus. J Bacteriol. 1995;177(12):3573–3578. doi: 10.1128/jb.177.12.3573-3578.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roach JC, et al. Transcription factor expression in lipopolysaccharide-activated peripheral-blood-derived mononuclear cells. Proc Natl Acad Sci USA. 2007;104(41):16245–16250. doi: 10.1073/pnas.0707757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroder K, et al. Conservation and divergence in Toll-like receptor 4-regulated gene expression in primary human versus mouse macrophages. Proc Natl Acad Sci USA. 2012;109(16):E944–E953. doi: 10.1073/pnas.1110156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardner PR, Costantino G, Szabó C, Salzman AL. Nitric oxide sensitivity of the aconitases. J Biol Chem. 1997;272(40):25071–25076. doi: 10.1074/jbc.272.40.25071. [DOI] [PubMed] [Google Scholar]
- 26.Drapier JC, Hibbs JB., Jr Aconitases: A class of metalloproteins highly sensitive to nitric oxide synthesis. Methods Enzymol. 1996;269:26–36. doi: 10.1016/s0076-6879(96)69006-5. [DOI] [PubMed] [Google Scholar]
- 27.Stadler J, et al. Effect of exogenous and endogenous nitric oxide on mitochondrial respiration of rat hepatocytes. Am J Physiol. 1991;260(5 Pt 1):C910–C916. doi: 10.1152/ajpcell.1991.260.5.C910. [DOI] [PubMed] [Google Scholar]
- 28.Kovaleva V, Berezhnaya E, Komandirov M, Rudkovskii M, Uzdensky A. Involvement of nitric oxide in photodynamic injury of neurons and glial cells. Nitric Oxide. 2013;29:46–52. doi: 10.1016/j.niox.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Patel TR, McFadden BA. Caenorhabditis elegans and Ascaris suum: Inhibition of isocitrate lyase by itaconate. Exp Parasitol. 1978;44(2):262–268. doi: 10.1016/0014-4894(78)90107-8. [DOI] [PubMed] [Google Scholar]
- 30.McFadden BA, Purohit S. Itaconate, an isocitrate lyase-directed inhibitor in Pseudomonas indigofera. J Bacteriol. 1977;131(1):136–144. doi: 10.1128/jb.131.1.136-144.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hillier S, Charnetzky WT. Glyoxylate bypass enzymes in Yersinia species and multiple forms of isocitrate lyase in Yersinia pestis. J Bacteriol. 1981;145(1):452–458. doi: 10.1128/jb.145.1.452-458.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma V, et al. Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis. Nat Struct Biol. 2000;7(8):663–668. doi: 10.1038/77964. [DOI] [PubMed] [Google Scholar]
- 33.Höner Zu Bentrup K, Miczak A, Swenson DL, Russell DG. Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J Bacteriol. 1999;181(23):7161–7167. doi: 10.1128/jb.181.23.7161-7167.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou JY, Graham JE, Clark-Curtiss JE. Mycobacterium avium genes expressed during growth in human macrophages detected by selective capture of transcribed sequences (SCOTS) Infect Immun. 2002;70(7):3714–3726. doi: 10.1128/IAI.70.7.3714-3726.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKinney JD, et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406(6797):735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 36.Muñoz-Elías EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11(6):638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell DG. Mycobacterium tuberculosis: Here today, and here tomorrow. Nat Rev Mol Cell Biol. 2001;2(8):569–577. doi: 10.1038/35085034. [DOI] [PubMed] [Google Scholar]
- 38.Muñoz-Elías EJ, Upton AM, Cherian J, McKinney JD. Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol Microbiol. 2006;60(5):1109–1122. doi: 10.1111/j.1365-2958.2006.05155.x. [DOI] [PubMed] [Google Scholar]
- 39.Savvi S, et al. Functional characterization of a vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis: Implications for propionate metabolism during growth on fatty acids. J Bacteriol. 2008;190(11):3886–3895. doi: 10.1128/JB.01767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell DG, et al. Mycobacterium tuberculosis wears what it eats. Cell Host Microbe. 2010;8(1):68–76. doi: 10.1016/j.chom.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knowles RG, Moncada S. Nitric oxide synthases in mammals. Biochem J. 1994;298(Pt 2):249–258. doi: 10.1042/bj2980249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bocchini V, et al. An immortalized cell line expresses properties of activated microglial cells. J Neurosci Res. 1992;31(4):616–621. doi: 10.1002/jnr.490310405. [DOI] [PubMed] [Google Scholar]
- 43.Raschke WC, Baird S, Ralph P, Nakoinz I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell. 1978;15(1):261–267. doi: 10.1016/0092-8674(78)90101-0. [DOI] [PubMed] [Google Scholar]
- 44.Giard DJ, et al. In vitro cultivation of human tumors: Establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51(5):1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 45.Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 46.Smal C, et al. Identification of in vivo phosphorylation sites on human deoxycytidine kinase. Role of Ser-74 in the control of enzyme activity. J Biol Chem. 2006;281(8):4887–4893. doi: 10.1074/jbc.M512129200. [DOI] [PubMed] [Google Scholar]
- 47.Steinhäuser C, et al. Lipid-labeling facilitates a novel magnetic isolation procedure to characterize pathogen-containing phagosomes. Traffic. 2013;14(3):321–336. doi: 10.1111/tra.12031. [DOI] [PubMed] [Google Scholar]
- 48.Song H, Sandie R, Wang Y, Andrade-Navarro MA, Niederweis M. Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2008;88(6):526–544. doi: 10.1016/j.tube.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.