Significance
It has been proposed that differential physical interactions of apolipoprotein E (apoE) isoforms with soluble amyloid-β (Aβ) in brain fluids influence the metabolism of Aβ, providing a major mechanism to account for how APOE influences Alzheimer’s disease risk. The current study challenges this proposal and clearly shows that lipoproteins containing apoE isoforms are unlikely to play a significant role in Aβ metabolism by binding directly to Aβ in physiological fluids such as cerebrospinal fluid or interstitial fluid. Our in vitro and in vivo results suggest that apoE isoforms influence Aβ metabolism by competing for the same clearance pathways within the brain.
Keywords: neurodegeneration, cholesterol efflux
Abstract
Apolipoprotein E gene (APOE) alleles may shift the onset of Alzheimer’s disease (AD) through apoE protein isoforms changing the probability of amyloid-β (Aβ) accumulation. It has been proposed that differential physical interactions of apoE isoforms with soluble Aβ (sAβ) in brain fluids influence the metabolism of Aβ, providing a mechanism to account for how APOE influences AD risk. In contrast, we provide clear evidence that apoE and sAβ interactions occur minimally in solution and in the cerebrospinal fluid of human subjects, producing apoE3 and apoE4 isoforms as assessed by multiple biochemical and analytical techniques. Despite minimal extracellular interactions with sAβ in fluid, we find that apoE isoforms regulate the metabolism of sAβ by astrocytes and in the interstitial fluid of mice that received apoE infusions during brain Aβ microdialysis. We find that a significant portion of apoE and sAβ compete for the low-density lipoprotein receptor-related protein 1 (LRP1)–dependent cellular uptake pathway in astrocytes, providing a mechanism to account for apoE’s regulation of sAβ metabolism despite minimal evidence of direct interactions in extracellular fluids. We propose that apoE influences sAβ metabolism not through direct binding to sAβ in solution but through its actions with other interacting receptors/transporters and cell surfaces. These results provide an alternative frame work for the mechanistic explanations on how apoE isoforms influence the risk of AD pathogenesis.
Alzheimer’s disease (AD), the most common cause of dementia, is a neurodegenerative disease pathologically characterized by extracellular accumulation of amyloid-β (Aβ), intracellular accumulation of tau, neuronal and synaptic loss, brain atrophy, and inflammation (1–3). There is significant evidence that the accumulation of Aβ peptides in the brain generates a cascade of events that initiates neurodegeneration and clinical dementia of the Alzheimer’s type (4–7). Thus it is important to understand factors that govern Aβ metabolism and how they lead to Aβ accumulation. The human apolipoprotein E gene (APOE) encodes the apoE protein, a key protein involved in lipid metabolism (8). It has been established that APOE strongly influences the risk for sporadic, late-onset AD (9–11). The presence of one copy of the APOE ε4 allele increases the risk of late-onset AD by about 3.7 times, and the presence of two copies increases the risk by about 12 times (9) (www.alzgene.org/meta.asp?geneID=83), relative to the ε3 isoform. ApoE is an exchangeable apolipoprotein with three major polymorphic forms: apoE2 (Cys112, Cys158), apoE3 (Cys112, Arg158), and apoE4 (Arg112, Arg158) (8, 12). The differences in amino acids at these positions are important because they change the charge and structural properties of the protein that influence the functional properties of apoE isoforms. Several mechanisms have been proposed to explain the mechanism by which apoE isoforms affect AD risk. An abundance of evidence from in vitro, animal, and human studies suggests that a major reason that apoE isoforms affect risk for AD is that they differentially modulate Aβ clearance and accumulation in the brain. ApoE4 increases Aβ aggregation and impairs clearance relative to other apoE isoforms (12–21). A recent in vivo study shows that the clearance of soluble Aβ in the brain interstitial fluid (ISF) depends on the isoform of human apoE expressed (apoE4 < apoE3 ≤ apoE2) (21).
ApoE is a component of many cerebral amyloid deposits, including Aβ deposits in AD and prion protein deposits in Creutzfeldt–Jakob disease (22, 23). The identified role of apoE isoforms in influencing AD onset, the association of apoE with extracellular amyloid plaques, and the ability of apoE to affect Aβ aggregation and clearance in vivo led us to hypothesize, as have others, that apoE isoforms interact directly with Aβ to influence Aβ metabolism. In the past two decades, numerous in vitro studies have tested this hypothesis, reporting that that apoE interacts with monomeric, fibrillar, and/or oligomeric Aβ with isoform specificity depending upon the type of apoE (lipidated or nonlipidated) and Aβ used in these studies (10, 24–34; reviewed in ref. 13). It has been suggested that the differential direct interaction of apoE isoforms with Aβ influences Aβ clearance and/or aggregation in the CNS, thus providing a mechanistic explanation for how apoE isoforms influence the risk of AD pathogenesis.
ApoE, along with apolipoprotein A1, plays an important role as a cholesterol and phospholipid acceptor in reverse cholesterol transport and subsequently in the distribution of cholesterol among different cells (35, 36). Lipoproteins and their transporters/receptors form an efficient transport system to distribute lipids, peptides, and other water-soluble and insoluble molecules. Given the role of apoE in modulating the metabolism of Aβ, apoE-containing lipoproteins in the brain also could act as efficient acceptors by binding directly to Aβ secreted from neurons, thus playing a major role in the clearance or aggregation of Aβ in an isoform-dependent manner in the CNS. To date, most apoE/Aβ-binding studies in solution have been performed with synthetic preparations of Aβ using supraphysiological (high micromolar to millimolar) concentrations and have not assessed the formation of apoE/Aβ complexes quantitatively. Importantly, the precise amount of direct binding between physiological preparations of apoE and Aβ occurring in solution and biological fluids, the molecular nature of these interactions, and how the direct interaction of apoE/Aβ influences Aβ metabolism have not been detailed. Moreover, most apoE–Aβ interaction studies were not done in the presence of primary brain-derived cells to test whether apoE could act as an extracellular acceptor for Aβ to alter its metabolism.
In this study, we hypothesized that apoE lipoproteins function as extracellular acceptors/carriers for Aβ with isoform specificity to regulate subsequent Aβ clearance or aggregation. Our current findings reject this hypothesis, because we show that the association of human apoE particles (reconstituted, cell-secreted, and human CSF) with Aβ (synthetic, cell-secreted, and CSF) in the presence and absence of cells is minimal in extracellular solutions. Despite minimal interaction in extracellular solution, apoE isoforms influence the clearance of cell-secreted and synthetic sAβ in astrocytes and the clearance of extracellular Aβ in a mouse model of β-amyloidosis during in vivo microdialysis. ApoE isoforms block the uptake and subsequent degradation of Aβ in astrocytes by competing for the same cellular clearance pathways. Our results suggest that the ability of apoE to influence Aβ metabolism probably is mediated not through direct binding to Aβ in extracellular solutions but rather through its actions with other interacting cellular receptors, transporters, and/or cellular/membrane surfaces.
Results
Association of apoE with Cell-Secreted Soluble Aβ and Synthetic Soluble Aβ in the Presence and Absence of Cells Is Minimal.
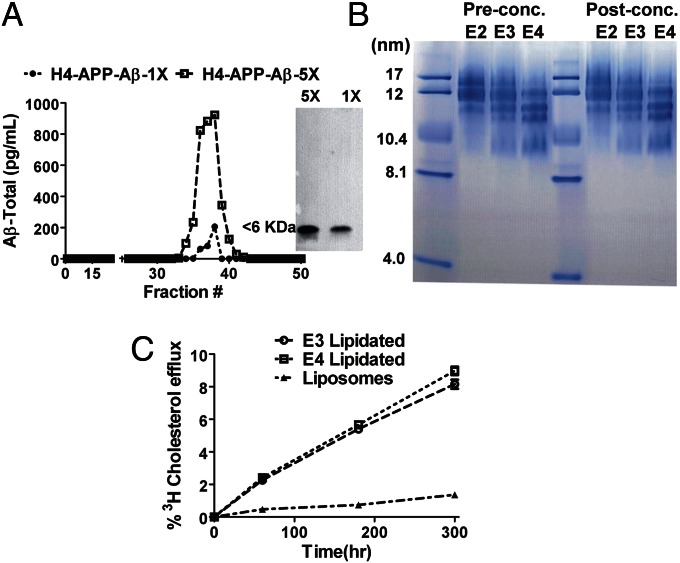
For apoE- and Aβ-binding studies, we primarily used cell-derived soluble Aβ (sAβ) and a range of concentrations of sAβ and apoE that are of physiological relevance in biological fluid. H4 neuroglioma cells expressing human amyloid precursor protein (APP) with the Swedish mutation (APP695∆NL) were used to collect cell-derived Aβ secreted into the culture medium. We characterized the H4 APP695∆NL cell-derived Aβ by size-exclusion chromatography (SEC); the elution profiles show that Aβ exists as a monomeric species, as was further confirmed by Western blot analysis showing that the Aβ collected was less than 6 kDa (Fig. 1A). The H4 APP695∆NL cell-secreted Aβ consists of ∼90–95% Aβ40 and 5–10% Aβ42 (Fig. S1), a ratio similar to the Aβ40:42 ratio found in human CSF in normal subjects and in patients with late-onset AD (37, 38). In this study, we used apoE from multiple sources: (i) reconstituted lipidated apoE isoform particles (rapoE2, rapoE3, and rapoE4); (ii) astrocyte-secreted human apoE particles and astrocyte-secreted mouse apoE containing HDL, purified as previously described (39, 40); and (iii) human apoE present in human cerebrospinal fluid (CSF) and isolated by SEC (41). The rapoE isoform particles were prepared using specified ratios of cholesterol and phospholipids and were characterized by nondenaturing gradient gel electrophoresis (Fig. 1B and SI Materials and Methods). The rapoE isoforms were heterogeneous in size with particle diameters ranging from 10–17 nm, similar to the range of HDL-like particles isolated from human CSF. Although the particle sizes differed slightly by rapoE isoform in the preparation shown in Fig. 1B, there were no obvious differences across preparations, and most preparations contained rapoE isoforms of the same size (21). To test the lipid mobilization efficiency and thus the functionality of rapoE isoform particles, the isoforms were tested for their cholesterol efflux efficiency relative to liposomes not containing rapoE. The apoE isoform particles were efficient acceptors, promoting cholesterol efflux in a dose-dependent manner (Fig. 1C), whereas liposomes effluxed relatively little cholesterol over the time course. There were no significant isoform-specific effects on the efflux properties as observed in previous studies (Fig. 1C) (42).
Fig. 1.
Characterization of rapoE isoforms and cell-secreted sAβ. (A) H4 APP695∆NL cells were incubated with serum-free Opti-MEM medium for 24 h. Then the medium was collected and concentrated fivefold with a 3-kDa cutoff concentrator and subjected to SEC. The SEC-purified Aβ was separated in a 16.5% tricine gel with nondenaturing PAGE and immunoblotted with 82E1 (anti–Aβ1–5) antibody (Right). (B) rapoE2, rapoE3, and rapoE4 particles (8 µg) were loaded on a 4–20% Tris⋅glycine gel for native PAGE to assess particle size using a standard containing proteins of specified hydrodynamic radii; particles that were concentrated for in vivo microdialysis experiments in Fig. 6 also were analyzed by native PAGE. (C) [3H]-cholesterol–labeled H4 APP695∆NL cells were incubated with 20 µg rapoE3 (1:50:10) and rapoE4 (1:50:10) particles at different time points to assess cholesterol efflux relative to liposomes, expressed as the percentage of radiolabeled cholesterol released into the medium. Differences were assessed using Student's t tests (n = 4; P < 0.05).
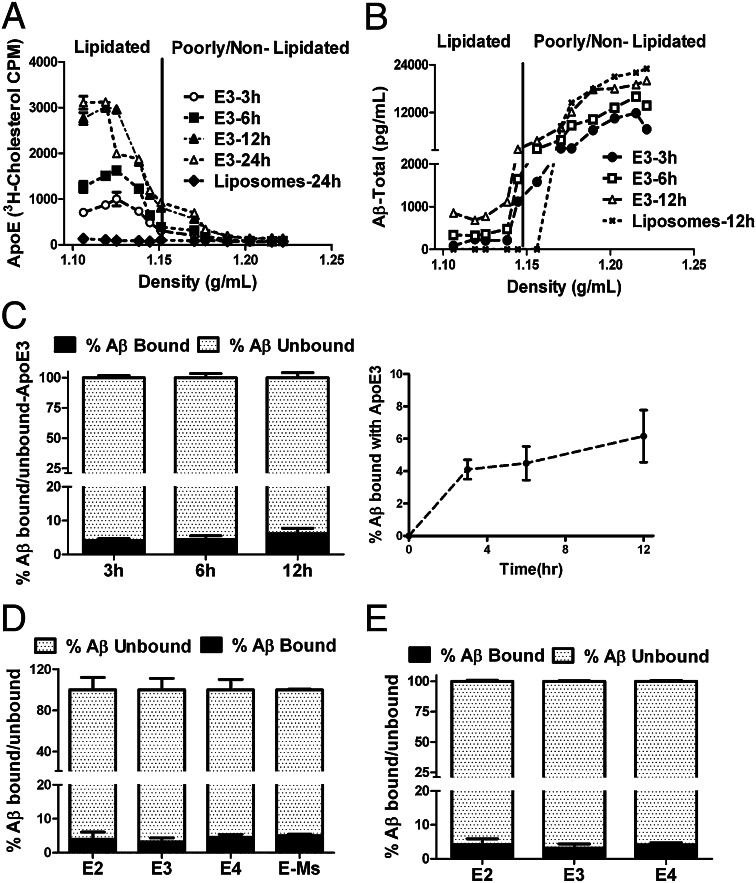
First, we wanted to test whether apoE could act as an efficient external Aβ acceptor by binding directly to Aβ. The molar ratio of Aβ:apoE present in human CSF is 1:50–75 (38, 43), so we performed Aβ and apoE-binding studies in this physiological range of molar ratios of Aβ:apoE. To study the direct binding of apoE and Aβ in the presence of cells that secrete Aβ, H4 APP695∆NL cells were labeled with 3H-cholesterol and were incubated with rapoE3 for 3, 6, or 12 h with molar ratios of Aβ:apoE (as observed at the end of the incubation time) of 1:50, 1:35, and 1:25. Samples collected at different time points were subjected to KBr gradient ultracentrifugation. We examined the density distribution of rapoE3 and Aβ in the KBr gradient. More than 95% of lipidated rapoE3, as determined by 3H cholesterol, was located in the region of the KBr gradient with density less than 1.15 g/cm3 (Fig. 2A). Using Aβ ELISA, the KBr gradient-separated samples were analyzed over the entire range of lipidated and poorly lipidated particles to determine the extent to which Aβ was associated with rapoE3 particles and whether the association changes with incubation time. We found that ∼95% of Aβ is unbound at 12 h and is not associated with lipidated rapoE3 (Fig. 2 B and C). There were small and time-dependent increases in the amount of Aβ present in the lipoprotein fractions containing rapoE at 3 and 12 h (4.5 vs. 6.1%), but the increase observed was not significant (Fig. 2C). Surprisingly, quantification of both Aβ and apoE in the lipidated apoE fractions (density less than 1.15) indicates a ratio of one molecule of Aβ(1–40) for every 430 molecules of apoE3 at 12 h (Fig. S2). Further, the association between cell-secreted Aβ and human apoE isoform particles did not differ among isoforms, nor did these particles differ from mouse apoE particles in their association with cell-secreted Aβ (Fig. 2D). These results suggest that lipidated apoE is a poor external acceptor of Aβ from CNS-derived cells and has an extremely low ability to associate with cell-secreted sAβ.
Fig. 2.
KBr density gradient studies showing that the association of apoE with cell-secreted sAβ in the presence and absence of cells is minimal. (A) H4 APP695∆NL cells were incubated with 20 μg of lipidated rapoE3 particles and liposome particles. Medium was collected at indicated time points and subjected to ultracentrifugation in a KBr density gradient. Aliquots of the density gradient were used to determine radioactivity and density. rapoE3 (1:50:10) particles were distributed primarily below the density of 1.15 g/cm3. (B) H4 APP695∆NL cells were incubated with 20 μg of lipidated rapoE3 particles (1:50:10) and liposomes (phospholipid:cholesterol ratio, 50:10) for 3, 6, and 12 h [Aβ:apoE molar ratios of 1:50 (3 h), 1:35 (6 h), and 1:25 (12 h)]. Medium was collected at indicated time points and subjected to ultracentrifugation in a KBr density gradient. Aliquots of the density gradient were used for Aβ1-x ELlSA. (C) (Left) The bar graph represents the percentage of sAβ present in the region with a KBr density less than 1.15 g/cm3 (percent Aβ bound) and in the region with a KBr density greater than 1.15 g/cm3 (percent Aβ unbound). (Right) Time-dependent binding of Aβ and rapoE3. Differences were assessed using one-way ANOVA followed by a Dunnet post test (n = 5). (D) The bar graph represents the percentage of sAβ present in the region with a KBr density less than 1.15 g/cm3 (percent Aβ bound) and in the region with a KBr density greater than 1.15 g/cm3 (percent Aβ unbound) when H4 APP695∆NL cells were incubated with 20 μg of lipidated rapoE2, rapoE3, rapoE4 (1:50:10) particles and mouse astrocyte-derived apoE particles for 12 h (Aβ:apoE molar ratio, 1:25). Differences were assessed using Student's t tests (n = 5). (E) The bar graph represents the percentage of sAβ present in the region with a KBr density less than 1.15 g/cm3 (percent Aβ bound) and in the region with a KBr density greater than 1.15 g/cm3 (percent Aβ unbound) when H4 APP695∆NL CM-derived Aβ was incubated with 20 μg of lipidated rapoE2, rapoE3, rapoE4 particles (in a ratio of 1:50:10) for 12 h (Aβ:apoE molar ratio, 1:25 at 12 h) without cells. Differences were assessed using one-way ANOVA followed by a Dunnet posttest (n = 4).
To investigate further the binding ability of apoE with cell-secreted sAβ in the absence of cells, the size-exclusion–purified Aβ from H4 APP695∆NL cells was incubated with rapoE3 and rapoE4 particles for 12 h (molar ratio of Aβ:apoE, 1:25). The collected samples were subjected to KBr gradient ultracentrifugation. We then examined the density distribution of Aβ in the KBr gradient. Similar to the findings shown in Fig. 2D, ∼95–97% of Aβ was not associated with rapoE3 or rapoE4 particles (Fig. 2E). Further, we used synthetic sAβ (molar ratio of Aβ:apoE, 1:25) in similar studies and found that more than 95% of Aβ is not associated with rapoE3 or rapoE4 (Fig. S3). These results further suggest that in the presence or absence of cells, in solution, and regardless of the source, Aβ is an extremely poor binding partner of lipidated apoE isoforms.
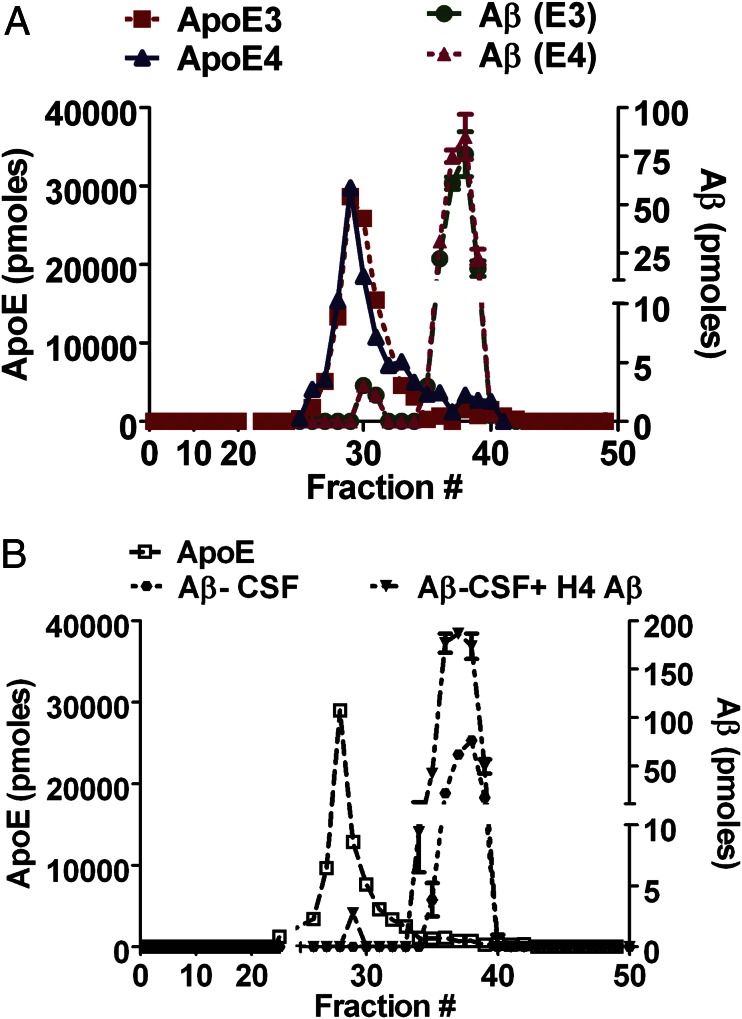
When a KBr-gradient ultracentrifugation technique is used, the presence of high salt and the high centrifugal force possibly could dissociate Aβ from apoE. To circumvent this potential limitation, we used SEC in physiological salt concentrations to isolate and quantify the Aβ associated with rapoE isoforms and astrocyte-secreted apoE isoforms. The APP695∆NL conditioned media (CM) (Aβ,100 ng/mL) was incubated with rapoE3 (apoE3:phospholipids:cholesterol ratio, 1:50:10), rapoE4 (apoE4:phospholipids:cholesterol ratio, 1:50:10) or astrocyte-secreted, immunopurified apoE3 or apoE4 for 3 or 6 h (molar ratio of Aβ:apoE, 1:25). The collected samples were separated on a size-exclusion column, and the distribution of apoE and Aβ in the fractions was analyzed further. Using sensitive sandwich ELISAs for apoE and Aβ, we found that apoE and Aβ were found in separate peaks, and more than 95% of Aβ was not associated with rapoE3, rapoE4, astrocyte-secreted apoE3, or astrocyte-secreted apoE4 (Fig. 3 A–C). The H4 APP neuroglioma cells secrete mostly monomeric sAβ (Fig. 1). Recent studies have suggested that apoE associates with higher-order Aβ more efficiently than with monomeric Aβ. To address this question, we used 7PA2 cells that previously have been shown to secrete low- and high-order Aβ species (44). The 7PA2 CM (Aβ,100 ng/mL) was incubated with astrocyte-secreted apoE3 and apoE4 for 6 h (molar ratio of Aβ:apoE, 1:20). The collected samples were separated on a size-exclusion column, and the distribution of apoE and Aβ in the fractions was analyzed. Aβ was eluted in multiple peaks corresponding to high and lower molecular weights, suggesting that both high- and low-order species of Aβ are present in the medium (Fig. S4A). However, in the presence of astrocyte-secreted apoE3 or apoE4, the SEC elution profile of Aβ remained the same, and there was no change in peak height or pattern, thus suggesting minimal association of apoE3 and apoE4 with higher- or lower-order Aβ species secreted by 7PA2 cells (Fig. S4B).
Fig. 3.
SEC studies showing that the association of apoE with cell-secreted sAβ is minimal. (A and B) H4 APP695∆NL CM-derived sAβ (100 ng/mL) was incubated with 10 μg of astrocyte-derived apoE3 and apoE4 particles for 6 h (A) or 20 μg of rapoE3 and rapoE4 particles (1:50:10) for 3 or 6 h (B). Medium was subjected to SEC with a Superose 6/10 column. Fractions were analyzed for apoE and Aβ by ELISAs. (C) The bar graph represents the percentage of sAβ present in the region of apoE elution, detected by ELISA (fraction number, 25–32) (percent Aβ bound) and in the region in which apoE is not detectable with ELISA (fraction number, 34–40) (percent Aβ unbound). Differences were assessed using t tests (n = 5). (D) [3H]-cholesterol–labeled H4 cells were incubated with 20 µg rapoE3 particles (1:50:10) and the indicated concentrations of Aβ purified from H4 APP695∆NL CM at different time points to assess cholesterol efflux, expressed as the percentage of radiolabeled cholesterol released into the medium. The efflux properties of apoE3 did not change significantly in the presence or absence of Aβ. Significance was assessed using one-way ANOVA followed by a Dunnet posttest (n = 4).
It has been suggested that the ability of apoE to efflux cholesterol is compromised by its putative interaction with Aβ (45). To investigate this possibility, we performed cholesterol efflux of rapoE isoform particles in the presence of cell-secreted sAβ with Aβ concentrations up to eightfold higher than the Aβ levels found in CSF (2–15 nM) (38). The rapoE isoforms increased cholesterol efflux in a time-dependent fashion (Fig. 3D). However, there was no compromise in the cholesterol efflux properties of apoE isoforms in the presence of cell-secreted sAβ at any concentration tested (Fig. 3D). These results further suggest that apoE isoform functions are not compromised in the presence of physiological or supraphysiological concentrations of Aβ.
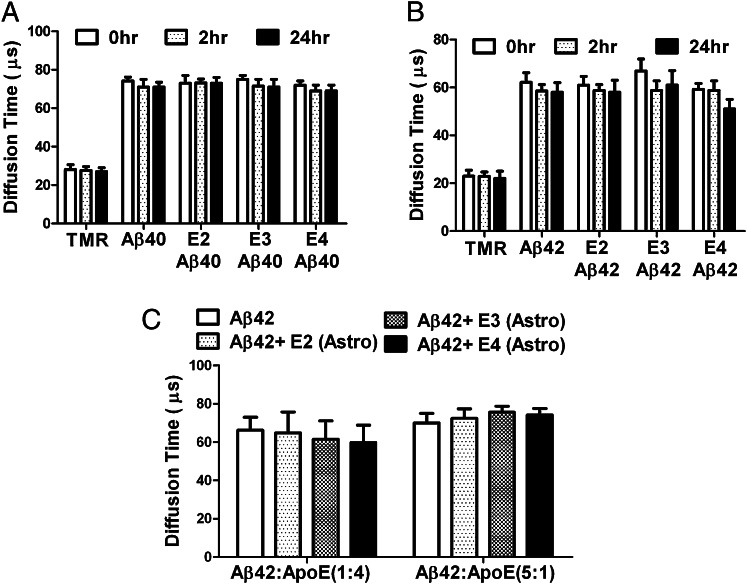
Physical separation processes such as chromatographic or high-speed ultracentrifugation techniques possibly could dissociate protein complexes if the interactions between proteins are very weak. To circumvent this issue, we used fluorescence correlation spectroscopy (FCS), a highly sensitive single-molecule technique that measures the diffusion time of molecules and does not involve a physical separation process (46). If Aβ binds directly to apoE in solution, the diffusion time of Aβ should increase significantly because of the change in the size of the Aβ/apoE complex. Synthetic Aβ40 and Aβ42 were labeled with tetramethylrhodamine (TMR) and incubated with rapoE2, rapoE3, and rapoE4 particles for 2 or 24 h. The diffusion time for TMR alone was 24 µs and increased to 62 µs when fluorescein was tagged with Aβ40 and Aβ42 (Fig. 4A). However, no increase in the diffusion time was observed when TMR-labeled Aβ40 and Aβ42 were incubated with rapoE2, rapoE3, and rapoE4 particles even at Aβ:apoE molar ratios of 1:1 (Fig. 4 A and B). Further, we performed binding studies with astrocyte-secreted apoE2, apoE3, and apoE4 and TMR-labeled Aβ42 (at Aβ42:apoE molar ratios of 1:1 and 5:1) for 24 h. The diffusion time of Aβ was not increased in the presence of astrocyte-secreted apoE even with high Aβ:apoE molar ratios (Fig. 4C). These results further confirm that lipid-associated apoE interacts negligibly or extremely poorly with sAβ derived from synthetic or cellular sources in solution.
Fig. 4.
FCS study showing that the association of apoE with synthetic Aβ is negligible. (A and B) Diffusion time was obtained from incubation of 100 nM TMR, 100 nM TMR-Aβ40 and TMR-Aβ42 and 100 nM rapoE2, rapoE3, and rapoE4 particles (molar ratio 1:1) for 2 h or 24 h (n = 4). (C) Diffusion time was obtained from incubation of 100 nM and 500 nM TMR-Aβ42 and 100 nM astrocyte-derived apoE2, apoE3, and apoE4 for 24 h (n = 4).
Association of apoE with sAβ in Human apoE3 and apoE4 CSF Is Minimal.
Previous studies have reported that Aβ is associated with apoE in human CSF (26, 41). However, these studies were conducted with concentrated CSF (with a 10-kDa cutoff membrane) to 10–30 times their original volumes, and the amount of apoE/Aβ complex formation or association was not determined. To investigate further the putative associations in human CSF, we separated apoE and Aβ from nonconcentrated CSF (0.5 mL) collected from age-matched APOE ε3 homozygous and APOE ε4 homozygous human subjects by SEC and analyzed the distribution of apoE and Aβ in the fractions. ApoE and Aβ eluted in separate peaks, and more than 95% of Aβ was not coeluted with apoE3 and apoE4 peaks (Fig. 5A), similar to our observations with cell-secreted Aβ incubated with rapoE particles. Further, there were no significant differences between apoE isoforms in the coelution profile (Fig. 5A). Quantification of Aβ and apoE in the peak fractions containing both apoE and Aβ indicates an Aβ:apoE ratio of 1:3,000–5,000 (Fig. 5A). The recovery of apoE and Aβ using 0.5 mL of CSF by SEC columns was ∼40%. To improve the recovery, we mildly concentrated pooled CSF samples from a 4.0-mL sample to four times the original volume with a 3-kDa cutoff concentrator and performed SEC. The recovery of apoE and Aβ increased significantly (50–60%), as did the presence of Aβ in the apoE peak (Fig. S5). However, the stoichiometry of apoE:Aβ remained the same in concentrated and nonconcentrated samples (ratio of Aβ:apoE, 1:3,000–5,000) (Fig. S5). To understand whether apoE in CSF binds to the cell-secreted exogenous Aβ, the nonconcentrated pooled human CSF (1.0 mL) was incubated with cell-secreted sAβ for 6 h. ApoE and Aβ were separated by size exclusion, and the distribution of apoE and sAβ in the fractions was analyzed. ApoE and sAβ eluted in distinct peaks, and more than 95% of Aβ (exogenous as well as endogenous sAβ) was not present in the same fractions as apoE3 and apoE4 (Fig. 5B). The exogenously added cell-secreted sAβ eluted along with the endogenous CSF sAβ peak. These results strongly suggest that the majority of apoE in physiological solutions (i.e., CSF) is not associated with Aβ.
Fig. 5.
Association of apoE with sAβ in human APOE ε3/ε3 and APOE ε4/ε4 CSF is minimal. (A) CSF from APOE ε3/ε3 and APOE ε4/ε4 human subjects (0.5 mL) was subjected to SEC. Fractions were analyzed for apoE and Aβ1-x by ELISAs as described in SI Materials and Methods. Differences were assessed using Student's t tests (n = 4). (B) Pooled CSF from human subjects (0.8 mL) was incubated with Aβ purified from CM of H4 APP695∆NL cells for 6 h and subjected to SEC. Fractions were analyzed for apoE and Aβ1-x by ELISAs as described in SI Materials and Methods. Differences were assessed using Student's t tests (n = 4).
Mechanism of Aβ Clearance Through apoE.
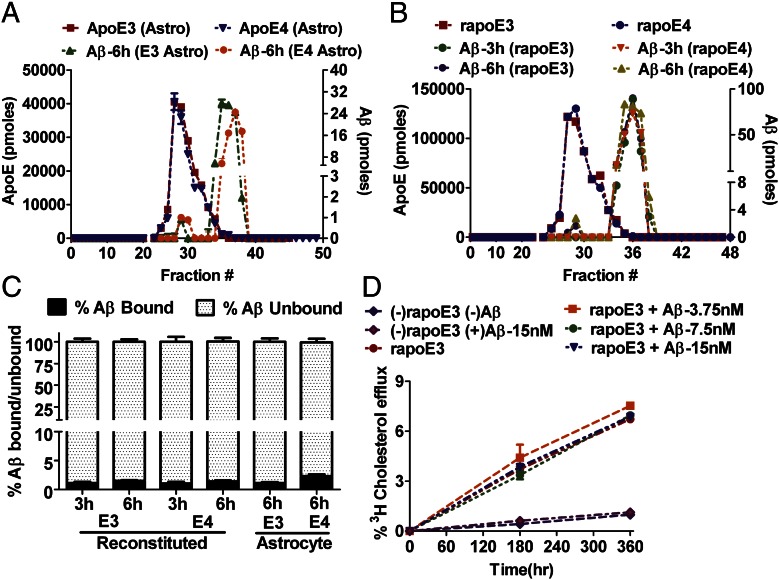
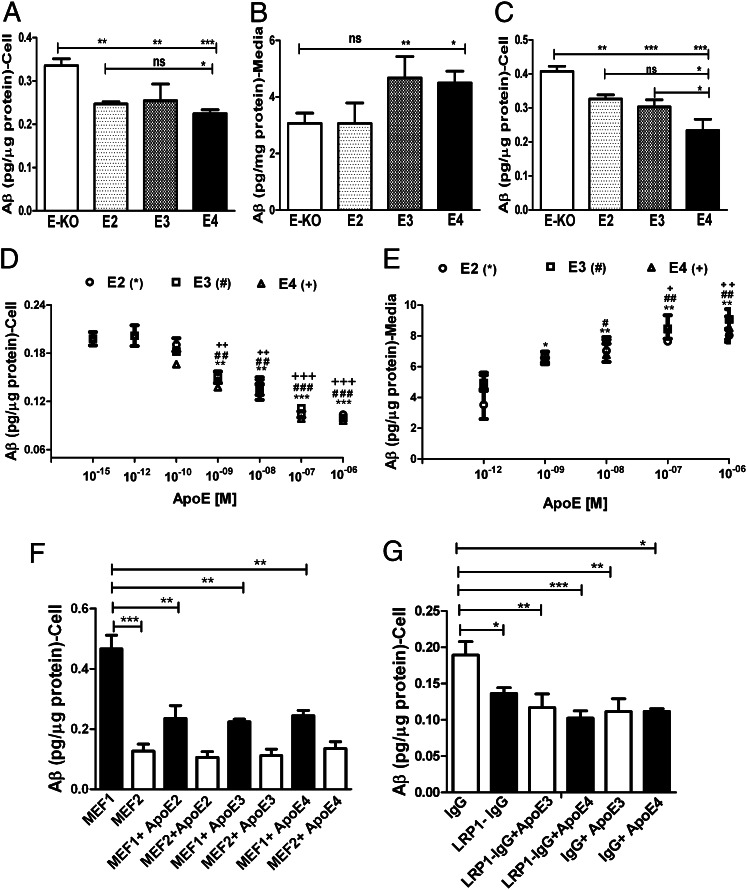
Our current results suggest that sAβ binds very poorly to apoE in lipoproteins, and therefore the ability of apoE-containing lipoproteins to influence Aβ clearance may not be mediated through its direct binding to Aβ in the extracellular fluid of the CNS where Aβ plaques accumulate. Astrocytes produce the majority of apoE in the CNS, and previous evidence suggests that they are one of the main cell types in the brain that play a central role in the cellular clearance of Aβ (47–49). To test whether particles containing the apoE isoform influence Aβ clearance by astrocytes, we cultured immortalized astrocytes derived from apoE2, apoE3, and apoE4 knock-in or apoE-KO mice (apoE2, apoE3, and apoE4 astrocytes secreted 4–6 ng/mL of apoE into culture medium/h; Fig. S6) and incubated them with sAβ (cell-secreted H4APP CM, 100 ng/mL sAβ) to assess cellular Aβ clearance. After 12-h incubation, the medium was collected, and cells were homogenized in lysis buffer after trypsin treatment to remove extracellular membrane-bound Aβ. Aβ levels in the medium and cell homogenates were determined by ELISA to examine cellular uptake and clearance. The apoE-KO cells had significantly higher cellular clearance of Aβ than cells producing apoE2, apoE3, or apoE4 (Fig. 6A). There was a significant increase in the clearance of Aβ in the presence of apoE2 compared with apoE4; however, there was no significant difference in cellular clearance between apoE2 vs. apoE3, nor was there a difference between apoE3 and apoE4 (Fig. 6A). The Aβ present in the medium was significantly lower in the presence of apoE-KO astrocytes than in the presence of cells producing apoE3 or apoE4. This result is consistent with the observation of increased cellular levels of Aβ in apoE-KO cells and decreased cellular Aβ in the cells producing apoE3 and apoE4 (Fig. 6B). To confirm our results and assess another cellular source of Aβ, we used Aβ CM from 7PA2 cells and incubated the Aβ with apoE2, apoE3, apoE4, and apoE-KO astrocytes to evaluate Aβ uptake. We observed similar results, in that the apoE-KO cells had significantly higher cellular clearance of Aβ than did cells secreting apoE2, apoE3, or apoE4 (EKO > E2 > E3 > E4) (Fig. 6C). In other words, we observed that cells expressing apoE4 had lower clearance of 7PA2 CM-derived Aβ than cells expressing apoE2 or apoE3 (Fig. 6C), similar to our observations shown in Fig. 6A. Overall, these results suggest that apoE is not required for the cellular clearance of sAβ in astrocytes, and apoE isoforms significantly inhibit the uptake of sAβ, either by competing for the same pathways or through other effects on Aβ clearance pathways in astrocytes.
Fig. 6.
Aβ and apoE share clearance pathways without direct binding. (A and B) To assess the effect of apoE isoforms on Aβ uptake, apoE2, apoE3, apoE4, and apoE-KO astrocytes were incubated with CM from H4 APP695∆NL cells secreting sAβ (sAβ, 100 ng/mL) for 12 h. The amount of Aβ in the cell lysate (A) and medium (B) then was assessed by ELISA as described in Materials and Methods. (C) apoE2, apoE3, apoE4, and apoE-KO–expressing astrocytes were incubated with CM from 7PA2 cells (sAβ,100 ng/mL) for 12 h. The amount of Aβ in the cell lysate then was assessed by ELISA as described in SI Materials and Methods. (D–F) To determine whether apoE and Aβ compete for the same cellular uptake pathways, apoE-KO astrocytes were incubated with CM from H4 APP695∆NL cells (sAβ,100 ng/mL) and indicated concentrations of rapoE2, rapoE3, and rapoE4 (apoE:phospholipid:cholesterol ratio, 1:50:10) for 12 h. The amount of Aβ in the cell lysate (D) and in the medium from D (E) then was assessed by ELISA as described in SI Materials and Methods. To determine the role of LRP1 in apoE-dependent Aβ competition, MEF1 (LRP1-expressing cells), and MEF2 (LRP1-KO cells) were incubated with CM from H4 APP695∆NL cells (sAβ,100 ng/mL) and 10−7 M rapoE2, rapoE3, and rapoE4 (apoE:phospholipid:cholesterol ratio, 1:50:10) for 12 h. (F) The amount of Aβ in the cell lysate then was assessed by ELISA as described in SI Materials and Methods. (G) ApoE-KO astrocytes were incubated with CM from H4 APP695∆NL cells (sAβ,100 ng/mL) in the presence or absence of nonimmune IgG or anti-LRP1 IgG (75 µg/mL) and 10−7 M rapoE3 and rapoE4 (apoE:phospholipid:cholesterol ratio, 1:50:10) for 12 h. The amount of Aβ in the cell lysate then was assessed by ELISA as described in SI Materials and Methods. Significance was assessed using one-way ANOVA followed by a Dunnett posttest (n = 4–6). Data are shown as mean ± SEM (n ≥ 4) *, #, +P < 0.05; **, ##, ++P < 0.005; ***, ###, +++P < 0.0005; n.s., not significant.
To determine whether apoE and Aβ compete for the same cellular clearance pathways, we performed a competition assay with lipidated apoE isoforms and sAβ in apoE-KO astrocytes. Cell-secreted sAβ (H4APP CM; Aβ, 100 ng/mL) was incubated with apoE-KO astrocytes in the presence of increasing concentrations of rapoE2, rapoE3, and rapoE4 particles (apoE:phospholipid:cholesterol ratio, 1:50:10), and the cellular clearance of Aβ was determined. ApoE isoform particles significantly inhibited the cellular clearance of Aβ by astrocytes (Fig. 6D). The inhibition of Aβ cellular clearance by apoE exhibited saturation at ∼100 nM of apoE (Fig. 6D), suggesting that apoE blocked receptor-mediated uptake of Aβ. The significant increase in Aβ levels in media with increasing concentrations of apoE isoforms (Fig. 6E) is consistent with the decreased Aβ clearance by the cells in the presence of apoE. ApoE isoform particles were able to block ∼50% of the total Aβ cellular clearance in this setting, suggesting the presence of other pathways of Aβ uptake independent of externally applied apoE. Further, we tested the role of lipidation of apoE isoforms in inhibiting Aβ clearance in astrocytes. Cell-secreted sAβ (H4APP CM; Aβ, 100 ng/mL) was incubated with apoE-KO astrocytes in the presence of increasing concentrations of poorly lipidated rapoE2, rapoE3, and rapoE4 particles (apoE:phospholipid:cholesterol ratio, 1:5:1) and the cellular clearance of Aβ was determined. rapoE isoform particles significantly inhibited the uptake of Aβ by astrocytes as observed with highly lipidated apoE isoforms (Fig. S7). We did not observe a difference among apoE isoforms in the competition for blocking Aβ uptake by astrocytes.
Recent studies have suggested that low-density lipoprotein receptor-related protein 1 (LRP1) and LDL receptor (LDLR) play a major role in the metabolism of Aβ in brain (47, 50-53). The equal efficiency of apoE2, apoE3, and apoE4 in blocking Aβ uptake/clearance in astrocytes ruled out the possibility that LDLR plays a role in the competitive inhibition of Aβ uptake. It also has been suggested that LDLR has a role in Aβ metabolism that is independent of apoE (47, 54). To test whether LRP1 has any role in the apoE-sensitive clearance of sAβ in cells, we used mouse embryonic fibroblasts, MEF1 (LRP1-expressing cells) and MEF2 (LRP1-deficient cells) cells to test the clearance of cell-secreted sAβ in the presence of extracellular apoE2, apoE3, and apoE4. The levels of sAβ clearance in MEF1 cells were similar to the levels in astrocytes, and 50% of the sAβ clearance was inhibited in the presence of apoE3 and apoE4 (apoE, 100 nM) (Fig. 6F). The LRP1-deficient cells had significantly lower uptake of sAβ, as observed in previous studies (50). However, the effect of apoE on the inhibition of Aβ uptake was completely absent in LRP1-deficient cells, suggesting that LRP1 plays a role in apoE-dependent competition of sAβ uptake in cells (Fig. 6F). To determine further the role of LRP1 in apoE-dependent competition in sAβ clearance by astrocytes, we assessed the effects of an LRP1-blocking antibody in apoE-KO astrocytes in the presence and absence of apoE3 and apoE4. The LRP1 antibody significantly decreased the uptake of sAβ (∼30%) in the absence of apoE (Fig. 6F). However, the effect of apoE (apoE, 100 nM) on Aβ uptake in the presence of the LRP1-blocking antibody did not exceed the maximum inhibition (i.e., ∼50% inhibition as observed in Fig. 6D), further suggesting that LRP1 may play a significant role in the apoE-dependent inhibition of cellular uptake of sAβ by astrocytes (Fig. 6F).
Activation of endoplasmic reticulum (ER) stress response has been reported in Arginine 61 apoE mice and in macrophages isolated from apoE4 mice (55, 56). We wanted to test whether ER stress is activated in astrocytes expressing the apoE isoform and whether this activation could account for the observed difference in sAβ clearance in the apoE isoform (Fig. 6 A and C). To test whether apoE isoforms are associated with differential activation of ER stress in our experimental conditions, we cultured immortalized astrocytes derived from apoE2, apoE3, and apoE4 knock-in or apoE-KO mice and incubated them with sAβ (cell-secreted H4APP CM,100 ng/mL sAβ) for 12 h. After 12-h incubation, cells were homogenized, and Western blot analysis was performed for ER stress markers. ApoE4 cells had significantly higher levels of the ER stress-marker proteins binding immunoglobulin protein (BiP), inositol-requiring enzyme 1 α (IRE1α), and C/EBP homology protein (CHOP), whereas programmed death-1 (PD-1) levels were similar in all genotypes (Fig. S8). ApoE2, ApoE3, and ApoE-KO cells had similar levels of all ER stress markers tested (Fig. S8). These results suggest that cells expressing apoE4 have higher ER stress, which may affect the general metabolism and specifically the clearance of Aβ. However, the similar levels of ER stress markers in apoE2, apoE3, and apoE-KO cells does not fit the pattern of sAβ clearance seen in the presence vs. absence of apoE. This result suggests that ER stress and the inhibition of sAβ clearance by apoE in these cells probably are not linked phenomena.
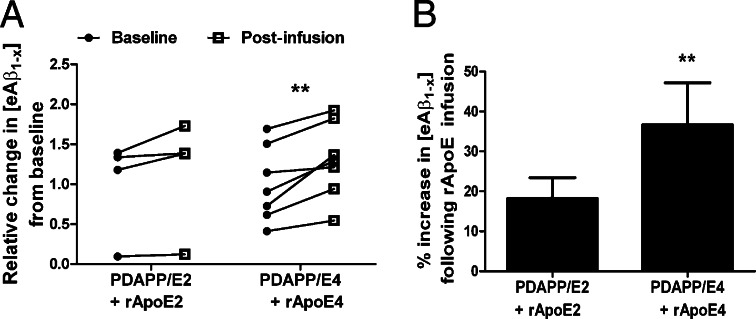
To determine further the effects of increasing apoE concentration on Aβ in vivo, we assessed Aβ present in the ISF of human-APP transgenic mice (PDAPP-V717F) by in vivo microdialysis (21, 57) while simultaneously infusing rapoE particles containing apoE2 or apoE4. We characterized the particles before in vivo microdialysis by nondenaturing PAGE (Fig. 1B) before and after the concentration procedure needed for infusion in vivo and found that these particles were essentially the same as those used in our in vitro studies. An equimolar amount of rapoE2 or rapoE4 particles was infused directly around an implanted 38-kDa microdialysis probe during brain ISF sampling of freely behaving PDAPP/E2 or PDAPP/E4 mice to examine the acute effects of rapoE treatment on extracellular Aβ levels. Treatment with rapoE2 particles increased steady-state levels of ISF Aβ by ∼18.2% compared with a baseline period of steady-state ISF Aβ obtained in PDAPP/E2 mice, although this effect did not reach statistical significance (Fig. 7). In contrast, infusion of the same amount of rapoE4 particles in PDAPP/E4 mice significantly increased ISF Aβ levels by 36.6% above baseline levels. These results further suggest that apoE and Aβ may compete for similar clearance pathways in vivo, as suggested by our studies in astrocytes (Fig. 6), and that the extent of this competition may depend on the isoform.
Fig. 7.
Infusing CNS fluid with rapoE particles increases ISF Aβ levels. (A) A 38-kDa cutoff microdialysis probe with a specialized side infusion port was implanted in hippocampi of young PDAPP/TRE mice. After a stable 6-h baseline, 1 μg of freshly prepared rapoE2 or rapoE4 (in artificial CSF solution) was infused at a flow rate of 0.07 μL/min directly at the site of microdialysis in PDAPP/E2 or PDAPP/E4 mice, respectively, and ISF [eAβ1-x] was monitored for an additional 6 h. Shown is the relative change in [eAβ1-x] within each mouse after rapoE2 or rapoE4 infusion compared with its mean baseline period. (B) The mean percent change in [eAβ1-x] following rapoE infusion relative to the baseline period was calculated for each mouse in each group. Differences between baseline and treatment periods were assessed for each group using paired t tests. n = 4–7 mice per group; **P < 0.01.
Discussion
The experiments in this study provide multiple lines of evidence that sAβ is a very poor binding partner of apoE-containing lipoproteins and that the influence of apoE on sAβ metabolism may not require direct binding of apoE with sAβ in solution. We used native, cell-derived sAβ and lipidated apoE from multiple sources to study the direct association of sAβ and apoE isoforms. Using density-gradient centrifugation and SEC as separation techniques and a nonseparation technique such as FCS, we show both in vitro and in vivo that more than 95% of sAβ is not associated with apoE-containing lipoproteins and binds to apoE isoforms in extracellular solution only to a very small extent. Our surprising findings strongly suggest that lipoproteins containing apoE isoforms likely do not play a significant role in sAβ metabolism by binding directly to Aβ in physiological fluids such as CSF or ISF. Our in vitro and in vivo results also suggest that apoE isoforms influence sAβ metabolism through a different mechanism: by competing for the same clearance pathways within the brain.
Several intriguing questions regarding apoE and sAβ binding remain and should motivate further study. What is the physiological relevance of the observed small percentage (3–6%) of sAβ associated with apoE lipoproteins? The half-life of sAβ in the brain ISF of mouse models overexpressing h-APP is ∼0.5–1.5 h (E4 > E2 = E3) and in the CSF of humans is 6 h (21, 58). The turnover rate of Aβ40/42 in human CSF is around 8%/h (58, 59). Considering the slow binding kinetics of apoE and sAβ (∼5% of binding in 12 h) (Fig. 2C) and the faster clearance rate of Aβ in extracellular fluid, a significant role of apoE/Aβ complexes in mediating clearance differences of sAβ in solution is exceedingly improbable. The low percentage of binding could contribute to Aβ aggregation pathways in the long term. However, no significant apoE isoform-dependent association of Aβ was observed in apoE-containing lipoproteins in plasma or CSF in our studies or in others (41, 60).
Human CSF contains 2–3 nM of Aβ and 150–300 nM of apoE-containing lipoproteins (molar ratio: 1–2 molecules of Aβ for every 75–150 molecules of apoE). This extracellular location is an excellent space to study apoE and Aβ interactions and the subsequent metabolic fates of both molecules. Previous studies from our laboratory and by others have shown that some fraction of Aβ associates with apoE containing lipoproteins (41, 60). However, several studies suggest that the majority of the endogenous Aβ is present in the CSF lipoproteins (26, 61). In one study, the authors used a 10-kDa cutoff concentrator to concentrate 30 mL of CSF to 1 mL for SEC separation, but the amount of Aβ filtered through the 10-kDa cutoff concentrator and the molar ratios of apoE and Aβ present in the coeluted apoE-Aβ fractions were unclear from this study (26). A separate study used gradient ultracentrifugation to examine apoE–Aβ interactions, finding that 70% of endogenous Aβ is present in the lipoproteins of CSF and plasma (61). However, by using a similar gradient ultracentrifugation technique, it was reported that 90% of endogenous Aβ was present along with free protein (26). Our study convincingly shows that a very small percent of Aβ actually is associated with or coelutes in fractions that contain lipoproteins.
What is the relevance of apoE and sAβ surface-binding studies? The binding affinities of lipid-free and lipidated apoE isoforms and Aβ by surface plasmon resonance (SPR) spectroscopy or plate-based assays have been reported to be in the low nanomolar range; however, the maximal molar apoE:Aβ ratios on the chip surface have been estimated as 130:1 (62). Apolipoproteins such as apoAI and apoAII also are reported to bind to Aβ with high affinity and in a range similar to that found for apoE (34, 62). We have repeated the surface affinity binding studies using SPR, finding that the binding of lipidated apoE and Aβ is in the nanomolar range (Table S1). However, the observed Kd values were higher than previously reported, and lowering the lipidation of apoE isoforms significantly increased the binding affinity to Aβ40 and Aβ42 (Table S1). These results suggests that a small percentage of apoE or Aβ protein conformations may change during the process of binding to a surface, facilitating the high-affinity binding of apoE and Aβ on surfaces such as cells or the extracellular matrix. Whether apoE or sAβ binding to the surface changes the structure of apoE/sAβ to assist their binding and whether these surface-binding properties of apoE and Aβ complex formation are physiologically relevant requires further investigation. It will be important to understand whether parenchymal Aβ interacts with apoE on surfaces to influence Aβ clearance or aggregation in an isoform-dependent manner.
Our data raise the question of whether apoE must bind to sAβ to influence its metabolism. Multiple pathways exist in brain for Aβ clearance, including blood–brain barrier-mediated clearance, cellular uptake, and passive elimination (47, 52, 63–65). Astrocytes represent a major cell type mediating clearance of Aβ from the extracellular space in the brain. Recently, it was shown that LDLR plays an important role in the clearance of sAβ in astrocytes and modifies plaque pathology in an APP mouse model independent of apoE, suggesting that sAβ can be metabolized by cells by binding directly to lipoprotein receptors such as LDLR without requiring apoE to facilitate this clearance (47, 54). Human and mouse apoE dose-dependently increase Aβ levels and plaque burden in APP transgenic mouse models (16, 66, 67). The levels of apoE and Aβ pathology are inversely correlated in mice, suggesting that decreasing the apoE level augments Aβ clearance and/or reduce Aβ aggregation. Indeed, we found that acutely increasing apoE2 or apoE4 levels in the ISF of PDAPP/TRE mice resulted in greater retention of Aβ in the ISF. Furthermore, particles containing apoE isoforms decrease the uptake of Aβ by astrocytes. ApoE and sAβ are transported through similar pathways, probably through lipoprotein receptors/transporters, resulting in competition with each other for cellular uptake that is not dependent on a direct interaction.
The isoform specificity for Aβ uptake is not completely consistent among different sources of Aβ, including CM of H4APP and 7PA2 cells Aβ (Fig. 6). Aβ present in the CM of H4 APP and 7PA2 cells resulted in differences among apoE isoforms in Aβ clearance, differences that were more robust using CM obtained from 7PA2 cells. This result probably reflects differences in the Aβ species present in the CM; i.e., H4APP cells produced mostly monomeric Aβ, whereas the CM of 7PA2 had both monomeric and higher-order species. The role of apoE isoforms in mediating the uptake and degradation of higher-order species of Aβ (in physiologically relevant concentrations) remains to be determined.
In skin fibroblasts, it was shown that Aβ competes with apoE containing very low-density lipoprotein (VLDL) for uptake into cells in a manner that does not depend on the apoE isoform (68). In the present study, we show that apoE-containing lipoproteins efficiently blocked 50% of sAβ uptake into astrocytes with no isoform-specific differences. The saturation of Aβ uptake in apoE competition studies suggests that the uptake mechanism functions through a receptor(s). The absence of apoE isoform-dependent differences observed in our apoE competition assay (apoE-KO cells incubated exogenously with increasing concentrations of apoE2/E3/E4 and a constant amount of sAβ) compared with results obtained with human apoE isoform-expressing astrocytes (only endogenously produced apoE and exogenous Aβ incubation) likely can be explained by differences in exogenous vs. endogenous apoE uptake/recycling pathways. In peripheral cells, it was observed that apoE intracellular transport pathways are different for apoE that is endogenously produced (and recycled) and for exogenously incubated (and recycled) apoE (69). In brief, our results suggest that apoE isoforms block the clearance of Aβ in astrocytes by competing for the same clearance pathways and/or through defective apoE4 and/or apoE-receptor recycling, as observed in human hepatoma cells and neuronal cells, respectively (70, 71). Consistent with our in vitro studies, we sampled the hippocampal ISF of a mouse model of β-amyloidosis expressing human apoE isoforms (PDAPP/TRE2 and PDAPP/TRE4) after acute infusion of lipidated apoE2 and apoE4. Infusion of either apoE2 or apoE4 particles elevated the concentration of sAβ in the brain ISF, suggesting that apoE and Aβ may compete for the same clearance pathways in vivo, as was suggested by our studies in astrocytes (Fig. 6). However, the extent of this competition is isoform dependent in vivo, whereas we do not observe this dependence under in vitro conditions. The precise reasons for this difference between in vitro and in vivo results are unknown. The difference may result from the contribution of multiple cell types, the blood–brain barrier, and the extracellular matrix present in the brain that are involved in the clearance of Aβ in an apoE-dependent fashion.
Elucidation of receptors that are responsible for the uptake of endogenous and exogenous apoE and sAβ will be important to understand further how apoE influences sAβ metabolism in the CNS. We have identified LRP1 as a potential candidate in apoE-dependent sAβ uptake in astrocytes and in MEF cells (Fig. 6 E and F). LRP1 is an endocytic receptor involved in the metabolism of various extracellular ligands (72). Studies have shown that LRP1 is involved in the metabolism of Aβ in the brain by directly binding to Aβ for cellular uptake/clearance or indirectly through other pathways (73-75). ApoE isoforms interact directly with LRP1 and hence have the potential to compete with sAβ in the cellular clearance process. The association and dissociation rates of the ligands of LRP1 determine the competition efficiency and subsequent uptake and metabolism of ligands. Hence, apoE isoforms with differential binding and recycling kinetics with LRP1 could differentially influence Aβ clearance in brain. It is important to understand further how apoE isoforms and levels modulate LRP1 function on the cell surface to affect sAβ clearance in vivo.
In conclusion, we present several lines of evidence that do not support the existence of significant direct interactions of apoE isoforms with sAβ in CNS fluids. Instead, we propose that the ability of apoE to influence Aβ clearance or aggregation probably is mediated not through direct binding to sAβ in solution but rather through its actions with LRP1 and other interacting receptors/transporters. Evidence indicates that decreasing apoE levels in the CNS and modulating the level and activity of apoE receptors and transporters may provide opportunities for developing AD therapeutics.
Materials and Methods
Aβ Clearance Assays.
Human apoE isoform-expressing (apoE2, apoE3, and apoE4) and apoE-KO immortalized astrocytes, MEF1 (LRP1-expressing cells, and MEF2 (LRP1-KO) cells were plated in six- or 12-well plates and were grown to confluence. To measure Aβ uptake, cells first were washed three times with serum-free medium (SFM) followed by the addition of fresh SFM. CM containing H4 APP695∆NL cell Aβ or CM containing 7PA2 cell-secreted Aβ then was added to the medium at an approximate concentration of 100 ng/mL, and the cells were incubated at 37 °C for 12 h. Then the medium was collected, and the cells were washed twice with PBS. To remove cell surface-bound Aβ, the cells were incubated with 0.05% trypsin/EDTA for 2 min and then were washed twice with PBS. Cells were removed and homogenized by adding 50 mM Tris⋅Hcl, 150 mM NaCl, 1% Triton X-100 (pH 7.6), and complete protease inhibitor mixture (Roche), and the cell lysates were cleared by centrifugation at 14,000 × g (Beckman Coulter, Rotor number 301.5) for 12 min. Protein content was measured in all cell lysates using a bicinchoninic acid (BCA) protein assay (Thermo Scientific). The Aβ and apoE in the media and cell lysates were measured by sandwich ELISAs for Aβ using m266 (anti–Aβ13–28) and biotinylated 3D6 (anti–Aβ1–5) antibodies and HJ6.2 and biotinylated HJ6.1 to detect apoE (21, 47, 76). In LRP1-blocking experiments, nonimmune IgG or anti-LRP1 IgG (75 µg/mL) was added 1–1.5 h before apoE and Aβ treatment.
For apoE/Aβ competition assays, apoE-KO cells were plated into 12-well plates, and CM containing either H4 APP695∆NL cell-derived Aβ or 7PA2 cell-derived Aβ was added to the medium at a concentration of 100 ng/mL. Where indicated, cells were incubated with 10−15 M, 10−12 M, 10−10 M, 10−9 M, 10−8 M, 10−7 M, and 10−6 M rapoE2, rApoE3, rApoE4 (1:50:10 or 1:5:1) at 37 °C for 12 h. Then the medium was collected, and the cells were washed twice with PBS. To remove cell surface-bound Aβ, the cells were incubated with 0.05% trypsin/EDTA for 2 min and then were washed twice with PBS. The cells were removed and were homogenized by 50 mM Tris⋅Hcl, 150 mM NaCl, 1% Triton X-100 (pH 7.6), and complete protease inhibitor mixture (Roche), and the cell lysates were cleared by centrifugation at 14,000 × g (Beckman Coulter, Rotor number 301.5). Protein content was measured in all cell lysates using a BCA protein assay (Thermo Scientific). The Aβ and apoE in the media and cell lysates were measured by sandwich ELISAs m266 (anti–Aβ13–28) and 3D6b (anti–Aβ1–5) and HJ 6.2 and HJ6.1b, respectively (21, 47, 76).
Statistics.
All data are presented as mean ±SEM, and different conditions were compared using one-way ANOVA followed by Dunnett’s and Bonferroni post hoc test to compare control with treatment groups. The Student's t test was used for comparing conditions with only two groups. Statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001) was determined using GraphPad Prism Software.
Additional materials and methods are available in SI Methods and Materials.
Supplementary Material
Acknowledgments
We thank Anne Fagan and the investigators and staff of the Charles F. and Joanne Knight Alzheimer’s Disease Research Center (Washington University, St. Louis) for human CSF samples (supported by National Institutes of Health Grants AG03991, AG026276, and AG056810). The m266, 2G3, and 21F12 antibodies were a generous gift from Eli Lilly and Co. We also thank Ronald B. DeMattos (Eli Lilly and Co., Indianapolis) for contributing PDAPP/TRE mice and Dennis J. Selkoe (Harvard Medical School, Boston) for providing 7PA2 cells. This work was supported by a Fellowship Grant from the American Health Assistance Foundation (to P.B.V.) and by National Institutes of Health Grants AG034004 (to J.M.C.), AG13956 (to D.M.H.), NS074969 (to D.M.H. and G.B.), and AG027924 (to G.B.).
Footnotes
Conflict of interest statement: D.M.H. is a scientific advisor to C2N Diagnostics and is a co-inventor on US patent 7,892,845 “Methods for measuring the metabolism of neurally derived biomolecules in vivo.” Washington University with D.M.H. as co-inventor also has submitted the US non-provisional patent application “Methods for measuring the metabolism of CNS derived biomolecules in vivo,” serial #12/267,974. D.M.H. has consulted for Bristol-Myers Squibb, Pfizer, Satori, Innogenetics, and AstraZeneca.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220484110/-/DCSupplemental.
References
- 1.Morris JC. Clinical assessment of Alzheimer’s disease. Neurology. 1997;49(3, Suppl 3):S7–S10. doi: 10.1212/wnl.49.3_suppl_3.s7. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe DJ. Alzheimer's disease is a synaptic failure. Science. 2002;298(5594):789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 3.Holtzman DM, Morris JC, Goate AM. Alzheimer’s disease: The challenge of the second century. Sci Transl Med. 2011;3(77):sr1. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 5.Karran E, Mercken M, De Strooper B. The amyloid cascade hypothesis for Alzheimer’s disease: An appraisal for the development of therapeutics. Nat Rev Drug Discov. 2011;10(9):698–712. doi: 10.1038/nrd3505. [DOI] [PubMed] [Google Scholar]
- 6.Golde TE, Dickson D, Hutton M. Filling the gaps in the abeta cascade hypothesis of Alzheimer’s disease. Curr Alzheimer Res. 2006;3(5):421–430. doi: 10.2174/156720506779025189. [DOI] [PubMed] [Google Scholar]
- 7.Tanzi RE, Bertram L. Twenty years of the Alzheimer’s disease amyloid hypothesis: A genetic perspective. Cell. 2005;120(4):545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Mahley RW, Rall SC., Jr Apolipoprotein E: Far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 9.Corder EH, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 10.Strittmatter WJ, et al. Apolipoprotein E: High-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saunders AM, et al. Apolipoprotein E epsilon 4 allele distributions in late-onset Alzheimer’s disease and in other amyloid-forming diseases. Lancet. 1993;342(8873):710–711. doi: 10.1016/0140-6736(93)91709-u. [DOI] [PubMed] [Google Scholar]
- 12.Verghese PB, Castellano JM, Holtzman DM. Apolipoprotein E in Alzheimer’s disease and other neurological disorders. Lancet Neurol. 2011;10(3):241–252. doi: 10.1016/S1474-4422(10)70325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer’s disease. Neuron. 2009;63(3):287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma J, Yee A, Brewer HB, Jr, Das S, Potter H. Amyloid-associated proteins alpha 1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer beta-protein into filaments. Nature. 1994;372(6501):92–94. doi: 10.1038/372092a0. [DOI] [PubMed] [Google Scholar]
- 15.Castano EM, et al. Fibrillogenesis in Alzheimer’s disease of amyloid beta peptides and apolipoprotein E. Biochem J. 1995;306(Pt 2):599–604. doi: 10.1042/bj3060599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bales KR, et al. Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nat Genet. 1997;17(3):263–264. doi: 10.1038/ng1197-263. [DOI] [PubMed] [Google Scholar]
- 17.Bales KR, et al. Apolipoprotein E is essential for amyloid deposition in the APP(V717F) transgenic mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 1999;96(26):15233–15238. doi: 10.1073/pnas.96.26.15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holtzman DM, et al. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 2000;97(6):2892–2897. doi: 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmechel DE, et al. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc Natl Acad Sci USA. 1993;90(20):9649–9653. doi: 10.1073/pnas.90.20.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rebeck GW, Reiter JS, Strickland DK, Hyman BT. Apolipoprotein E in sporadic Alzheimer’s disease: Allelic variation and receptor interactions. Neuron. 1993;11(4):575–580. doi: 10.1016/0896-6273(93)90070-8. [DOI] [PubMed] [Google Scholar]
- 21.Castellano JM, et al. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci Transl Med. 2011;3(89):89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Namba Y, Tomonaga M, Kawasaki H, Otomo E, Ikeda K. Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer’s disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res. 1991;541(1):163–166. doi: 10.1016/0006-8993(91)91092-f. [DOI] [PubMed] [Google Scholar]
- 23.Wisniewski T, Frangione B. Apolipoprotein E: A pathological chaperone protein in patients with cerebral and systemic amyloid. Neurosci Lett. 1992;135(2):235–238. doi: 10.1016/0304-3940(92)90444-c. [DOI] [PubMed] [Google Scholar]
- 24.Strittmatter WJ, et al. Binding of human apolipoprotein E to synthetic amyloid beta peptide: Isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci USA. 1993;90(17):8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaDu MJ, et al. Isoform-specific binding of apolipoprotein E to beta-amyloid. J Biol Chem. 1994;269(38):23403–23406. [PubMed] [Google Scholar]
- 26.LaDu MJ, et al. Preferential interactions between ApoE-containing lipoproteins and Aβ revealed by a detection method that combines size exclusion chromatography with non-reducing gel-shift. Biochim Biophys Acta. 2012;1821(2):295–302. doi: 10.1016/j.bbalip.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koudinov AR, Koudinova NV, Kumar A, Beavis RC, Ghiso J. Biochemical characterization of Alzheimer’s soluble amyloid beta protein in human cerebrospinal fluid: Association with high density lipoproteins. Biochem Biophys Res Commun. 1996;223(3):592–597. doi: 10.1006/bbrc.1996.0940. [DOI] [PubMed] [Google Scholar]
- 28.Petrlova J, et al. A differential association of Apolipoprotein E isoforms with the amyloid-β oligomer in solution. Proteins. 2011;79(2):402–416. doi: 10.1002/prot.22891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Z, Smith JD, Greengard P, Gandy S. Alzheimer amyloid-beta peptide forms denaturant-resistant complex with type epsilon 3 but not type epsilon 4 isoform of native apolipoprotein E. Mol Med. 1996;2(2):175–180. [PMC free article] [PubMed] [Google Scholar]
- 30.Sanan DA, et al. Apolipoprotein E associates with beta amyloid peptide of Alzheimer’s disease to form novel monofibrils. Isoform apoE4 associates more efficiently than apoE3. J Clin Invest. 1994;94(2):860–869. doi: 10.1172/JCI117407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bentley NM, Ladu MJ, Rajan C, Getz GS, Reardon CA. Apolipoprotein E structural requirements for the formation of SDS-stable complexes with beta-amyloid-(1-40): The role of salt bridges. Biochem J. 2002;366(Pt 1):273–279. doi: 10.1042/BJ20020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan W, Fornwald J, Brawner M, Wetzel R. Native complex formation between apolipoprotein E isoforms and the Alzheimer’s disease peptide A beta. Biochemistry. 1996;35(22):7123–7130. doi: 10.1021/bi952852v. [DOI] [PubMed] [Google Scholar]
- 33.Yang DS, Smith JD, Zhou Z, Gandy SE, Martins RN. Characterization of the binding of amyloid-beta peptide to cell culture-derived native apolipoprotein E2, E3, and E4 isoforms and to isoforms from human plasma. J Neurochem. 1997;68(2):721–725. doi: 10.1046/j.1471-4159.1997.68020721.x. [DOI] [PubMed] [Google Scholar]
- 34.Tokuda T, et al. Lipidation of apolipoprotein E influences its isoform-specific interaction with Alzheimer’s amyloid beta peptides. Biochem J. 2000;348(Pt 2):359–365. [PMC free article] [PubMed] [Google Scholar]
- 35.Hayek T, Oiknine J, Brook JG, Aviram M. Role of HDL apolipoprotein E in cellular cholesterol efflux: Studies in apo E knockout transgenic mice. Biochem Biophys Res Commun. 1994;205(2):1072–1078. doi: 10.1006/bbrc.1994.2775. [DOI] [PubMed] [Google Scholar]
- 36.Dietschy JM, Turley SD. Cholesterol metabolism in the brain. Curr Opin Lipidol. 2001;12(2):105–112. doi: 10.1097/00041433-200104000-00003. [DOI] [PubMed] [Google Scholar]
- 37.De Strooper B. Loss-of-function presenilin mutations in Alzheimer disease. Talking Point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 2007;8(2):141–146. doi: 10.1038/sj.embor.7400897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fagan AM, et al. Decreased cerebrospinal fluid Abeta(42) correlates with brain atrophy in cognitively normal elderly. Ann Neurol. 2009;65(2):176–183. doi: 10.1002/ana.21559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morikawa M, et al. Production and characterization of astrocyte-derived human apolipoprotein E isoforms from immortalized astrocytes and their interactions with amyloid-beta. Neurobiol Dis. 2005;19(1-2):66–76. doi: 10.1016/j.nbd.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 40.DeMattos RB, et al. Purification and characterization of astrocyte-secreted apolipoprotein E and J-containing lipoproteins from wild-type and human apoE transgenic mice. Neurochem Int. 2001;39(5-6):415–425. doi: 10.1016/s0197-0186(01)00049-3. [DOI] [PubMed] [Google Scholar]
- 41.Fagan AM, et al. Differences in the Abeta40/Abeta42 ratio associated with cerebrospinal fluid lipoproteins as a function of apolipoprotein E genotype. Ann Neurol. 2000;48(2):201–210. [PubMed] [Google Scholar]
- 42.Rellin L, Heeren J, Beisiegel U. Recycling of apolipoprotein E is not associated with cholesterol efflux in neuronal cells. Biochim Biophys Acta. 2008;1781(5):232–238. doi: 10.1016/j.bbalip.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Cruchaga C, et al. Alzheimer’s Disease Neuroimaging Initiative Cerebrospinal fluid APOE levels: An endophenotype for genetic studies for Alzheimer’s disease. Hum Mol Genet. 2012;21(20):4558–4571. doi: 10.1093/hmg/dds296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shankar GM, Welzel AT, McDonald JM, Selkoe DJ, Walsh DM. Isolation of low-n amyloid β-protein oligomers from cultured cells, CSF, and brain. Methods Mol Biol. 2011;670:33–44. doi: 10.1007/978-1-60761-744-0_3. [DOI] [PubMed] [Google Scholar]
- 45.Tamamizu-Kato S, et al. Interaction with amyloid beta peptide compromises the lipid binding function of apolipoprotein E. Biochemistry. 2008;47(18):5225–5234. doi: 10.1021/bi702097s. [DOI] [PubMed] [Google Scholar]
- 46.Magde D, Elson EL, Webb WW. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13(1):29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 47.Basak JM, Verghese PB, Yoon H, Kim J, Holtzman DM. Low-density lipoprotein receptor represents an apolipoprotein E-independent pathway of Aβ uptake and degradation by astrocytes. J Biol Chem. 2012;287(17):13959–13971. doi: 10.1074/jbc.M111.288746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wyss-Coray T, et al. Adult mouse astrocytes degrade amyloid-beta in vitro and in situ. Nat Med. 2003;9(4):453–457. doi: 10.1038/nm838. [DOI] [PubMed] [Google Scholar]
- 49.Koistinaho M, et al. Apolipoprotein E promotes astrocyte colocalization and degradation of deposited amyloid-beta peptides. Nat Med. 2004;10(7):719–726. doi: 10.1038/nm1058. [DOI] [PubMed] [Google Scholar]
- 50.Kanekiyo T, et al. Heparan sulphate proteoglycan and the low-density lipoprotein receptor-related protein 1 constitute major pathways for neuronal amyloid-beta uptake. J Neurosci. 2011;31(5):1644–1651. doi: 10.1523/JNEUROSCI.5491-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sagare A, et al. Clearance of amyloid-beta by circulating lipoprotein receptors. Nat Med. 2007;13(9):1029–1031. doi: 10.1038/nm1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castellano JM, et al. Low-density lipoprotein receptor overexpression enhances the rate of brain-to-blood Aβ clearance in a mouse model of β-amyloidosis. Proc Natl Acad Sci USA. 2012;109(38):15502–15507. doi: 10.1073/pnas.1206446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deane R, Sagare A, Zlokovic BV. The role of the cell surface LRP and soluble LRP in blood-brain barrier Abeta clearance in Alzheimer’s disease. Curr Pharm Des. 2008;14(16):1601–1605. doi: 10.2174/138161208784705487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katsouri L, Georgopoulos S. Lack of LDL receptor enhances amyloid deposition and decreases glial response in an Alzheimer’s disease mouse model. PLoS ONE. 2011;6(7):e21880. doi: 10.1371/journal.pone.0021880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong N, Ramaswamy G, Weisgraber KH. Apolipoprotein E4 domain interaction induces endoplasmic reticulum stress and impairs astrocyte function. J Biol Chem. 2009;284(40):27273–27280. doi: 10.1074/jbc.M109.014464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cash JG, et al. Apolipoprotein E4 impairs macrophage efferocytosis and potentiates apoptosis by accelerating endoplasmic reticulum stress. J Biol Chem. 2012;287(33):27876–27884. doi: 10.1074/jbc.M112.377549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cirrito JR, et al. In vivo assessment of brain interstitial fluid with microdialysis reveals plaque-associated changes in amyloid-beta metabolism and half-life. J Neurosci. 2003;23(26):8844–8853. doi: 10.1523/JNEUROSCI.23-26-08844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mawuenyega KG, et al. Decreased clearance of CNS beta-amyloid in Alzheimer’s disease. Science. 2010;330(6012):1774. doi: 10.1126/science.1197623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bateman RJ, et al. Human amyloid-beta synthesis and clearance rates as measured in cerebrospinal fluid in vivo. Nat Med. 2006;12(7):856–861. doi: 10.1038/nm1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biere AL, et al. Amyloid beta-peptide is transported on lipoproteins and albumin in human plasma. J Biol Chem. 1996;271(51):32916–32922. doi: 10.1074/jbc.271.51.32916. [DOI] [PubMed] [Google Scholar]
- 61.Takamura A, et al. Dissociation of β-amyloid from lipoprotein in cerebrospinal fluid from Alzheimer’s disease accelerates β-amyloid-42 assembly. J Neurosci Res. 2011;89(6):815–821. doi: 10.1002/jnr.22615. [DOI] [PubMed] [Google Scholar]
- 62.Shuvaev VV, Siest G. Interaction between human amphipathic apolipoproteins and amyloid beta-peptide: Surface plasmon resonance studies. FEBS Lett. 1996;383(1-2):9–12. doi: 10.1016/0014-5793(96)00206-2. [DOI] [PubMed] [Google Scholar]
- 63.Bell RD, et al. Transport pathways for clearance of human Alzheimer’s amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system. J Cereb Blood Flow Metab. 2007;27(5):909–918. doi: 10.1038/sj.jcbfm.9600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deane R, et al. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J Clin Invest. 2008;118(12):4002–4013. doi: 10.1172/JCI36663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iliff JJ, et al. A Paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid beta. Sci Transl Med. 2012;4(147):147ra111. doi: 10.1126/scitranslmed.3003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim J, et al. Haploinsufficiency of human APOE reduces amyloid deposition in a mouse model of amyloid-β amyloidosis. J Neurosci. 2011;31(49):18007–18012. doi: 10.1523/JNEUROSCI.3773-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bien-Ly N, Gillespie AK, Walker D, Yoon SY, Huang Y. Reducing human apolipoprotein E levels attenuates age-dependent Aβ accumulation in mutant human amyloid precursor protein transgenic mice. J Neurosci. 2012;32(14):4803–4811. doi: 10.1523/JNEUROSCI.0033-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winkler K, et al. Competition of Abeta amyloid peptide and apolipoprotein E for receptor-mediated endocytosis. J Lipid Res. 1999;40(3):447–455. [PubMed] [Google Scholar]
- 69.Ho YY, et al. Endogenously expressed apolipoprotein E has different effects on cell lipid metabolism as compared to exogenous apolipoprotein E carried on triglyceride-rich particles. Biochemistry. 2000;39(16):4746–4754. doi: 10.1021/bi992294a. [DOI] [PubMed] [Google Scholar]
- 70.Chen Y, Durakoglugil MS, Xian X, Herz J. ApoE4 reduces glutamate receptor function and synaptic plasticity by selectively impairing ApoE receptor recycling. Proc Natl Acad Sci USA. 2010;107(26):12011–12016. doi: 10.1073/pnas.0914984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heeren J, et al. Impaired recycling of apolipoprotein E4 is associated with intracellular cholesterol accumulation. J Biol Chem. 2004;279(53):55483–55492. doi: 10.1074/jbc.M409324200. [DOI] [PubMed] [Google Scholar]
- 72.Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: Pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10(5):333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deane R, et al. LRP/amyloid beta-peptide interaction mediates differential brain efflux of Abeta isoforms. Neuron. 2004;43(3):333–344. doi: 10.1016/j.neuron.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 74.Bell RD, et al. SRF and myocardin regulate LRP-mediated amyloid-beta clearance in brain vascular cells. Nat Cell Biol. 2009;11(2):143–153. doi: 10.1038/ncb1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanekiyo T, Liu CC, Shinohara M, Li J, Bu G. LRP1 in brain vascular smooth muscle cells mediates local clearance of Alzheimer’s amyloid-β. J Neurosci. 2012;32(46):16458–16465. doi: 10.1523/JNEUROSCI.3987-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.DeMattos RB, et al. Peripheral anti-A beta antibody alters CNS and plasma A beta clearance and decreases brain A beta burden in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 2001;98(15):8850–8855. doi: 10.1073/pnas.151261398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.