Significance
In mitosis, microtubules extend and shrink before the bilateral attachment is established. However, which molecules regulate this activity for spindle formation is not fully elucidated. Using two in-house developed small molecules that target the Aurora kinases, we show that hepatoma up-regulated protein (HURP) is highly dynamic, trafficking between centrosome and kinetochore driven by Aurora A-dependent phosphorylation and protein phosphatase 1/2A-associated dephosphorylation. These compounds demonstrate a spatial hierarchical preference of HURP in the attachment of microtubules extending from the mother to the daughter centrosome. These findings help explain the biology of mitosis and may lead to the development of anticancer compounds.
Abstract
The overexpression of Aurora kinases in multiple tumors makes these kinases appealing targets for the development of anticancer therapies. This study identified two small molecules with a furanopyrimidine core, IBPR001 and IBPR002, that target Aurora kinases and induce a DFG conformation change at the ATP site of Aurora A. Our results demonstrate the high potency of the IBPR compounds in reducing tumorigenesis in a colorectal cancer xenograft model in athymic nude mice. Human hepatoma up-regulated protein (HURP) is a substrate of Aurora kinase A, which plays a crucial role in the stabilization of kinetochore fibers. This study used the IBPR compounds as well as MLN8237, a proven Aurora A inhibitor, as chemical probes to investigate the molecular role of HURP in mitotic spindle formation. These compounds effectively eliminated HURP phosphorylation, thereby revealing the coexistence and continuous cycling of HURP between unphosphorylated and phosphorylated forms that are associated, respectively, with microtubules emanating from centrosomes and kinetochores. Furthermore, these compounds demonstrate a spatial hierarchical preference for HURP in the attachment of microtubules extending from the mother to the daughter centrosome. The finding of inequality in the centrosomal microtubules revealed by these small molecules provides a versatile tool for the discovery of new cell-division molecules for the development of antitumor drugs.
The overexpression of Aurora kinases is closely associated with tumorigenesis (1, 2). Small molecules that inhibit the kinase activity of Aurora have attracted considerable attention for their applicability in cancer treatment, and a number of Aurora kinase inhibitors have been assessed in clinical trials (1, 3–6). Aurora kinases are serine/threonine kinases, which regulate mitotic progression, centrosome maturation, and spindle assembly. Therefore, small molecules capable of inhibiting Aurora kinases also can be used as chemical probes to determine the interplay of Aurora kinases and their substrates in spindle formation.
To ensure fidelity of segregation, duplicated chromatids need to be properly attached by mitotic spindles at the kinetochores (7). At onset of mitosis, microtubules that emanate from the duplicated centrosomes gradually extend to reach the kinetochores. The formation of robust spindles relies on the cooperation between two assembly pathways: the kinetochore capture by microtubule spindles originating from centrosomes, and the ras-related nuclear GTP (RanGTP)-mediated microtubule nucleation and organization in the vicinity of chromosomes (8–13). Human hepatoma up-regulated protein (HURP) is an Aurora A substrate up-regulated in hepatomas (14, 15). HURP stabilizes kinetochore fibers (K-fibers) and promotes nucleation and crosslinking of microtubules (16–19). In Xenopus egg extract, anti-HURP antibodies disrupt the formation of chromosome- and centrosome-induced spindles (16), suggesting the involvement of HURP in both mechanisms. HURP also has been characterized as a direct cargo of importin β, involved in RanGTP-regulated spindle (Ran spindle) assembly in the vicinity of chromosomes (17–19). Because the kinase activity of Aurora A is essential to the formation of Ran spindles (16), HURP has been proposed to be phosphorylated at the spindle poles by Aurora A, thereby allowing its translocation to RanGTP-dependent K-fibers (17).
Because HURP expression is cell-cycle dependent and limited to prophase through anaphase, investigating how HURP is temporally regulated by phosphorylation would require rapid inhibition of the kinase activity of Aurora A, which is not achievable using RNAi or other genetic methods (15, 19). Here we use the Aurora kinase inhibitors we developed in house to dissect the Aurora–HURP pathway in the formation of spindles. This study reports the identification and characterization of two Aurora inhibitors, IBPR001 and IBPR002, that efficiently eliminate HURP phosphorylation in mitosis. The efficacy of the two IBPR compounds in HURP dephosphorylation is better than that of MLN8237 and VX-680. The rapid elimination of HURP phosphorylation supports the notion of a dynamic equilibrium between the two forms of HURP regulated by Aurora A-mediated phosphorylation, each playing a role in the differential assembly of centrosomal and kinetochore microtubules. These results also suggest that the symmetric distribution of HURP to centrosomal microtubules requires kinase activity of Aurora A.
Results
Synthesis and Characterization of IBPR Compounds Targeting Aurora Kinases.
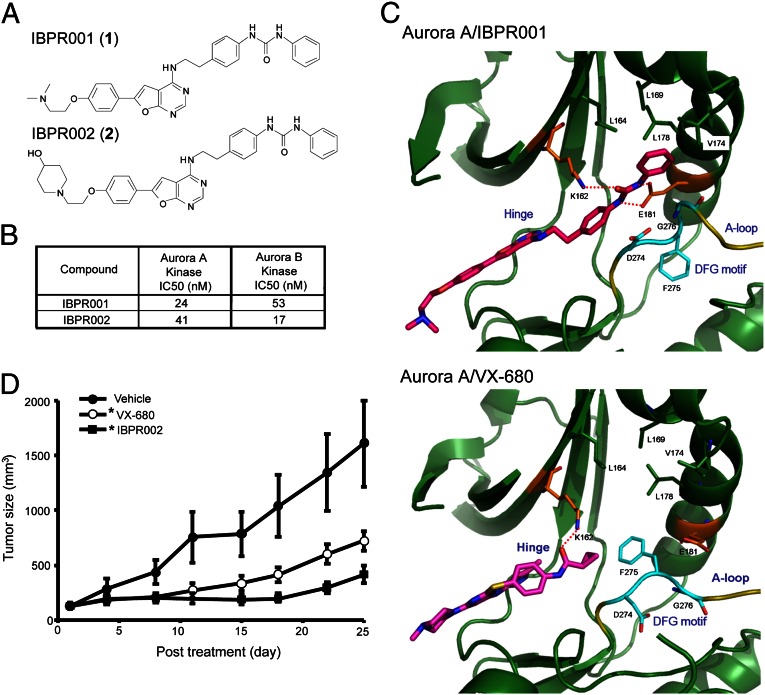
We have reported a lead compound with a furanopyrimidine core capable of inhibiting Aurora kinase activity (20, 21). Using this structure as a scaffold, we synthesized (Fig. S1) more than 200 analogs and identified two compounds, IBPR001 and IBPR002 (Fig. 1A), that demonstrate potent Aurora inhibitory activity (Fig. 1B).
Fig. 1.
Structures of IBPR compounds that induce a change in DFG conformation in Aurora A. (A) Chemical structures of IBPR001 (1) and IBPR002 (2). (B) IC50 of the compounds from the in vitro Aurora A and Aurora B activity assay. (C) Active sites of Aurora A in complex with IBPR001 and VX-680. The interacting residues with the inhibitors are shown in stick representation and are labeled. The DFG motif (cyan) at the activation loop (A-loop, yellow) adopts a different conformation to accommodate IBPR001 from VX-680. Hydrogen bonds between the inhibitors and Aurora A are shown as red dashed lines. For figure presentation clarity, hydrogen bonds between the inhibitors and the hinge region are omitted. (D) Athymic nude mice xenograft with HCT116 cancer cells were injected i.v. with control vehicle or 50 mg/kg of VX-680 or IBPR002. Mean tumor volumes (in cubic millimeters) ± SEM (n = 10 per group) are shown from the initiation of treatment (∼100 mm3). *P < 0.05 compared with vehicle.
To determine the specificity of these IBPR compounds, we performed in vitro activity profiling for 57 kinases associated with cancer. Of the kinases tested, IBPR002 demonstrated the strongest inhibitory activity against Aurora A. The inhibition of IBPR002 at 1.0 μM is listed in Table S1. All but 12 (including Aurora A) of the profiled kinases showed <50% inhibition at 1.0 μM. The effectiveness of IBPR002 as an inhibitor of mitosis-associated kinases polo-like kinase 1 (PLK1) (−14% inhibition at 1.0 μM) and NIMA-related kinase 2 (NEK2) (61% inhibition at 1.0 μM, IC50 = 0.532 μM) was less pronounced than the inhibition of Aurora A (101% inhibition at 1.0 μM, IC50 = 41 nM) (Fig. 1B and Table S1).
Crystal Structures of Aurora A in Complex with IBPR Compounds and VX-680.
VX-680 is a first-generation small molecule that inhibits the catalytic activity of Aurora kinases through competitive interactions with the ATP-binding site (3). Structures of the Aurora A kinase domain (amino acids 123–401) in complex with IBPR001 [Protein Data Bank (PDB) ID: 4JBO] and VX-680 (PDB ID: 4JBQ) were solved to provide insight into the interaction of the compounds with the protein (X-ray data and structure refinement are summarized in Table S2). Although both compounds were well suited to the ATP-binding site and formed conserved hydrogen bonds with Glu211 and Ala213 in the hinge region (22), the diphenylurea moiety of IBPR001 extends into the Aurora A back pocket, which was unoccupied in the Aurora A/VX-680 complex (Fig. 1C). Moreover, the Aurora A kinase domain adopts the DFG (Asp-Phe-Gly)-in conformation to accommodate the phenylurea group of IBPR001, which forms two hydrogen bonds with the conserved Glu181 of Aurora A. On the other hand, Phe275 of the DFG motif (23) at the activation loop points toward the cyclopropyl group of VX-680.
The structure of the Aurora A kinase domain with IBPR002 also was solved (PDB ID: 4JBP) to a resolution of 2.45Å (Table S2). The structure of IBPR002 was well superimposed with IBPR001, except that the additional piperidinol group extended toward the solvent-exposed area.
IBPR Compounds Reduce Tumorigenesis in Mice.
Aurora kinases have emerged as promising chemotherapeutic targets for cancer because of their pivotal role in mitosis and their overexpression in malignant cells (5). To evaluate whether IBPR compounds are able to reduce tumorigenesis in vivo, we used a colorectal cancer xenograft model in athymic nude mice. Ten male mice in each group were inoculated s.c. with HCT116 colorectal cancer cells overexpressing Aurora A. When the tumor size reached ≥100 mm3, mice were administrated IBPR002 or VX-680 i.v. via the tail vein, at a dosage of 50 mg⋅kg−1⋅d−1, five daily doses per week, for two consecutive weeks. Tumor size was observed for an additional 13 d following the final injection. IBPR002 significantly (P < 0.05) inhibited the growth of xenograft colorectal cancer cells, in a manner similar toVX-680 (Fig. 1D).
IBPR Compounds Eliminate HURP Phosphorylation.
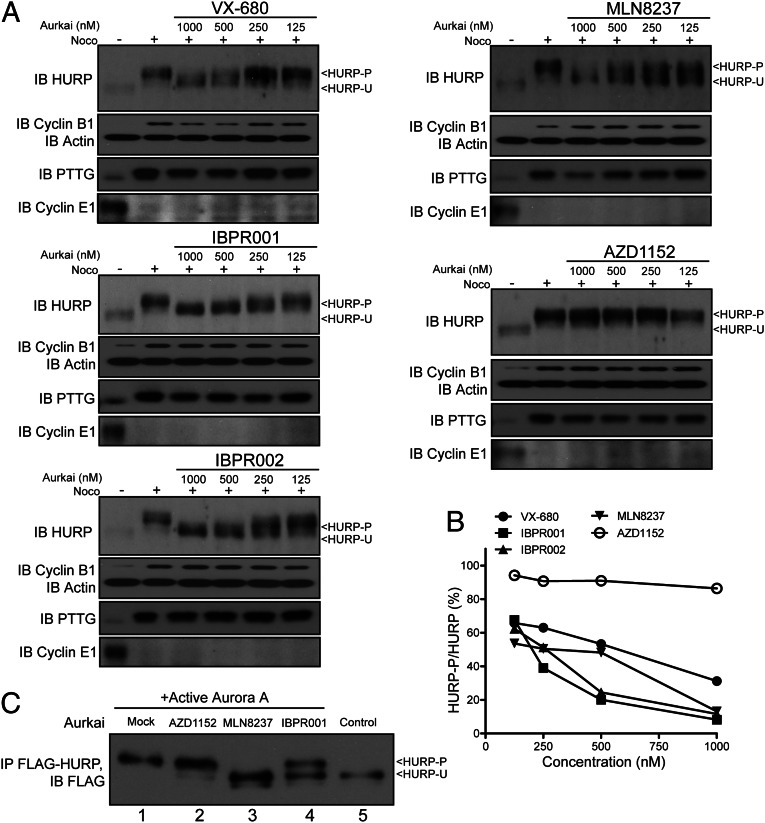
The expression of HURP is cell-cycle regulated (Fig. S2A and refs. 15 and 19). HURP promotes the nucleation and crosslinking of microtubules (16, 17, 24), but whether this activity requires phosphorylation remains unclear. We examined whether VX-680 inhibits HURP phosphorylation and found that the efficacy is less than optimal (Fig. 2A). The results of Western blotting indicate that the anti-HURP phosphorylation activity of IBPR001 and IBPR002 exceed that of VX-680 (Fig. 2 A and B). We also tested HURP phosphorylation in cells treated with a reported Aurora A-selective inhibitor, MLN8237. Like the IBPR compounds, MLN8237 eliminated HURP phosphorylation, albeit with reduced effectiveness. In contrast, AZD1152 (a reported Aurora B inhibitor) failed to reduce HURP phosphorylation (Fig. 2 A and B). The inhibition of HURP phosphorylation was verified further by an in vitro kinase assay (Fig. 2C), suggesting that IBPR001 and MLN8237 play a direct role in the inhibition of Aurora A-mediated HURP phosphorylation. In cells the depletion of Aurora A (using RNA silencing), but not of other mitotic kinases such as Aurora B, cyclin-dependent kinase 1 (CDK1), or NEK2, eliminated the expression of phosphorylated HURP (HURP-P) (Fig. S2 B and C). These results indicate that IBPR compounds inhibit Aurora A-mediated HURP phosphorylation in cells.
Fig. 2.
IBPR001 and IBPR002 efficiently eliminate HURP phosphorylation. (A) Immunoblot of HURP from HeLa cells treated with increasing concentrations of the Aurora inhibitors VX-680, IBPR001, IBPR002, MLN8237, or AZD1152. Cells were arrested in the M phase with nocodazole (400 nM) for 24 h followed by 1-h cotreatment with the compounds and MG132 (5.0 μg/mL). Actin immunoblot was included as a loading control. Cyclin B1 (expressed in late G2-metaphase), pituitary tumor transforming gene (PTTG) (expressed in prophase-metaphase), and cyclin E1 (expressed in G1-S phase) (Fig. S2A) immunoblots are shown for verification of the M-phase synchronization. The Western signals of HURP-P and HURP-U are denoted. (B) Ratios of HURP-P/total HURP were quantified from the Western blot results in A. (C) The kinase activity of Aurora A for HURP phosphorylation was assessed in an in vitro kinase assay. Full-length FLAG-tagged HURP was expressed and immunoprecipitated from 293T cells as a substrate for Aurora A. The addition of active Aurora A increased the molecular size of HURP (compare lanes 1 and 5). The molecular size of HURP failed to increase in the reactions that contain MLN8237 (lane 3) and IBPR001 (lane 4) but increased in the reaction that contained AZD1152 (lane 2).
IBPR001 and MLN8237 Disrupt Nucleation and Bundling of K-Fibers.
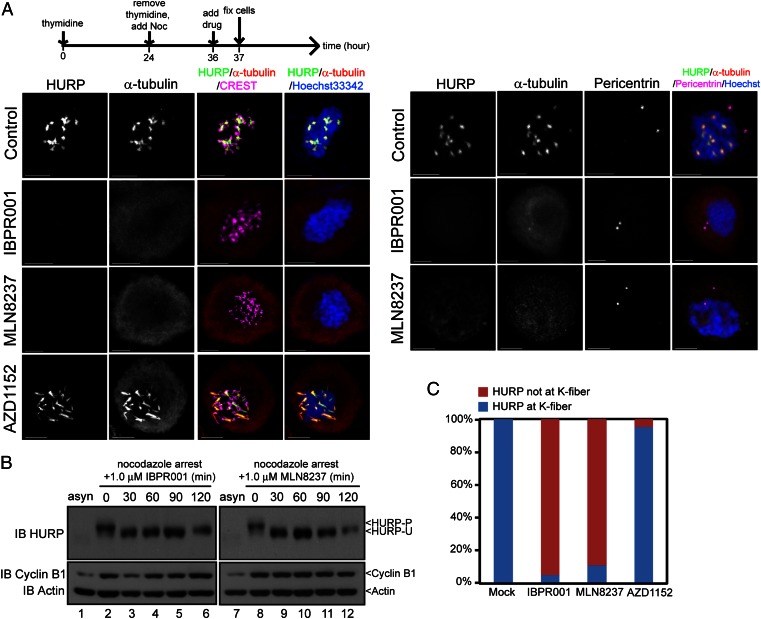
We used IBPR compounds and MLN8237 as chemical probes to gain insight into the association of the Aurora A–HURP pathway in spindle formation. Nocodazole inhibits mitotic progression by disrupting the assembly of microtubules. Under treatment with 300–400 nM nocodazole (25), microtubules form kinetochore-associated bundles but fail to nucleate at centrosomes, suggesting different nucleation pathways exist for centrosomal and kinetochore microtubules (Fig. 3A). We found that HURP is >95% phosphorylated under this condition (Fig. 3B, lanes 2 and 8) and associated exclusively with kinetochore [labeled with CREST (calcinosis, Raynaud's phenomenon, esophageal dismotility, sclerodactyly, telangiectasia; a centromere marker); Fig. 3A, Left] but not centrosomal (labeled with pericentrin; Fig. 3A, Right) microtubules. The addition of IBPR001 or MLN8237 to nocodazole-treated cells efficiently converted HURP-P to unphosphorylated HURP (HURP-U) within 1 h of treatment (Fig. 3B, lanes 3–6 and 9–12), accompanied by depolymerization of the kinetochore microtubule bundles (Fig. 3 A and C). It appears that the reduction of HURP intensity was not a result of degradation (26) because the total level of HURP expression had not altered with the treatment of MG132, a proteosome inhibitor (Fig. 3B). In contrast to IBPR001 and MLN8237, assembly of K-fibers was not interrupted by an Aurora B-selective inhibitor AZD1152 (Fig. 3 A and C). The colocalization of HURP-P with kinetochore microtubule bundles (Fig. 3A) and the disassembly of kinetochore microtubules in conjunction with HURP-P dephosphorylation (Fig. 3 A and C) suggest that HURP-P, rather than HURP-U, is required for the nucleation of the kinetochore microtubules.
Fig. 3.
IBPR compounds and MLN8237 disrupt nucleation of kinetochore microtubules. (A) HeLa cells were treated as shown in the upper scheme, followed by coimmunofluorescence staining with two combinations of antibodies: HURP/α-tubulin/CREST (a centromere marker) or HURP/α-tubulin/Pericentrin (a centrosome marker). Tubulins were nucleated as thick bundles that link to kinetochores (i.e., K-fibers) in control, but not in IBPR001 or MLN8237 treated cells. DNA was stained with Hoechst33342. Noc, nocodazole. All images are summations of z-stacks. (Scale bars: 5 μm.) (B) Immunoblot of HURP from nocodazole-arrested HeLa cells treated with 1.0 μM of IBPR001 or MLN8237 and 5 μg/mL MG132. Total cell lysates were collected at 0, 30, 60, 90, and 120 min after the compound treatment. (C) Statistics of HURP that form K-fibers in A. (Scale bars: 5 μm.)
HURP Cycles Between Centrosomes and Kinetochores Through Phosphorylation.
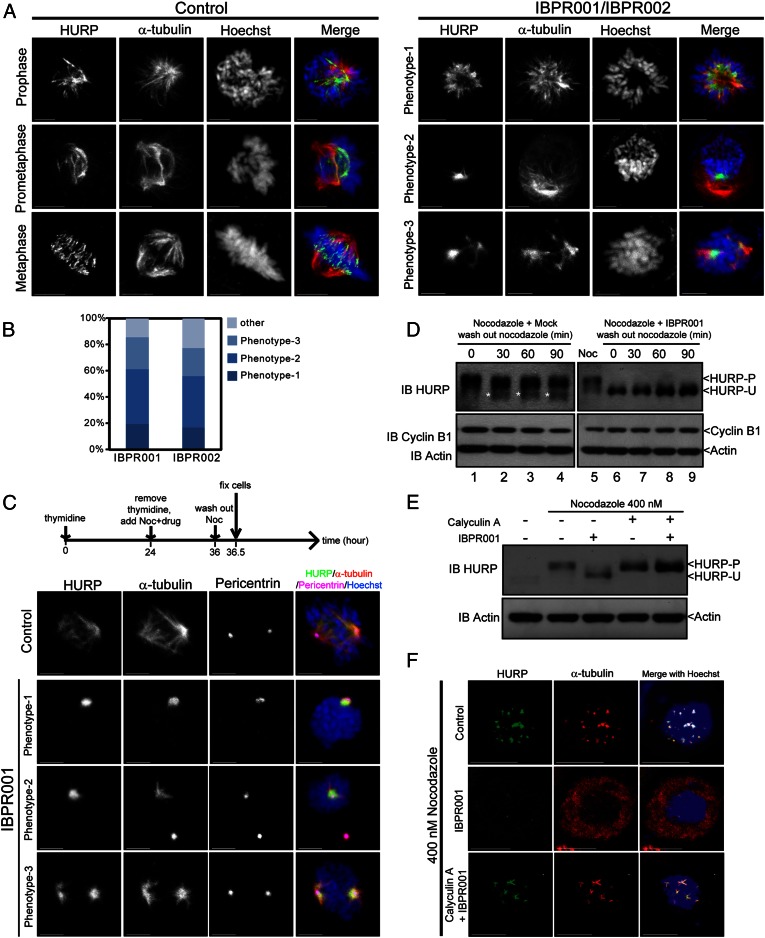
Establishing robust mitotic spindles requires the cooperation of both K-fibers and centrosomal microtubules (9, 27). In control cells, HURP colocalized with mitotic spindles in early prophase and gradually moved to the plus end of the spindles in the vicinity of mitotic chromosomes along with mitotic progression (Fig. 4A, Left) (28). A lack of adequate equipment for detecting HURP-P in cells prevented us from determining the phosphorylation status of HURP at the plus and minus ends of mitotic spindles. Nevertheless, blocking of Aurora kinase activity by IBPR compounds and MLN8237 clearly demonstrates that HURP-P can be converted to HURP-U (Fig. 3B). In the compound-treated cells, HURP resided primarily at the minus end of the microtubules, close to the centrosomes (Fig. 4A, Right and Fig. S2 D and E). It should be noted that nocodazole was not added to these cells to maintain tubulin polymerization. Nearly 20% of the cells treated with IBPR001 were monoastral (Fig. 4A, phenotype 1), similar to the phenotype observed in Aurora A-depleted cells (29, 30). The remainder of the cells treated with IBPR compounds exhibited microtubules emanating from separated poles, with HURP localized to one (Fig. 4A, phenotype 2) or both (Fig. 4A, phenotype 3) poles. Similar phenotypes were observed in cells treated with MLN8237 (Fig. S2D). The spatial relationship between HURP and centrosomes also was demonstrated by immunofluorescence staining of γ-tubulin (Fig. S2E). Unlike normally progressing cells which form biorientated spindles in metaphase (Fig. S2E, Left), IBPR001 and IBPR002 disrupted bipolarity (Fig. S2E, Right). HURP may surround the unseparated centrosomes (Fig. S2E, phenotype 1), associate with centrosomal microtubules projecting toward chromosomes (Fig. S2E, phenotype 2), or wrap around one (Fig. S2E, phenotype 3) or two (Fig. S2E, phenotype 4) of the separated centrosomes.
Fig. 4.
IBPR compounds restrict the association of HURP with centrosomal microtubules. (A) Representative HURP morphological phenotypes in HeLa cells treated with DMSO control (Left) or 1.0 μM of IBPR001/IBPR002 (Right) for 13 h following thymidine release. Cells were coimmunostained with rabbit anti-HURP and mouse anti–α-tubulin antibodies. DNA was stained with Hoechst33342. (B) Statistics of the representative HURP morphological phenotypes presented in A. (C) Cells were treated as shown in the upper scheme. HURP is associated with centrosomal microtubules (stained with α-tubulin) emanating from centrosomes (stained with Pericentrin) in IBPR001-treated cells upon nocodazole removal. Images are summations of z-stacks. (Scale bars: 5 μm.) (D) Cells were treated as in the upper scheme in C. Cell lysates were harvested every 30 min after the removal of nocodazole. Immunoblots of cyclin B1 and actin were included to indicate the cell-cycle status and serve as a loading control, respectively. The immunoblot of HURP shows that HURP remained unphosphorylated upon nocodazole removal (lanes 7–9). On the other hand, part of HURP was converted to the unphosphorylated form upon nocodazole removal in control cells (lanes 2–4, denoted by asterisks). (E) Immunoblot of HURP in nocodazole-arrested cells treated with 1.0 μM of IBPR001, 100 nM of Calyculin A, or both for 1 h. Immunoblot of actin was included as a loading control. (F) Cellular localization of HURP (green) and α-tubulin (red) in nocodazole-arrested cells treated with 1.0 μM of IBPR001 or cotreated with 1.0 μM of IBPR001 and 100 nM of Calyculin A. Cells were fixed after 1 h of drug treatment. A control cell (DMSO) is shown for comparison. Images are summation of z-stacks. (Scale bars: 5 μm.)
HURP is associated with the minus end of centrosomal microtubules when treated with IBPR compounds and MLN8237 (Fig. 4A and Fig. S2 D and E). However, in the presence of nocodazole, treatment with IBPR compounds caused HURP to disperse into the cytoplasm instead of accumulating around the centrosomes (Fig. 3). This result raises the question of whether tubulin polymerization at centrosomes is essential to this process. By removing nocodazole from cells pretreated with IBPR001, centrosomal microtubules were reestablished in conjunction with the accumulation of HURP at the minus ends (Fig. 4C), similar to the phenotypes observed in Fig. 4A. This localization differed from the control cells, in which HURP was located at both the plus and minus ends of microtubules (Fig. 4C). We also observed the expression of HURP-U in control cells upon nocodazole removal (Fig. 4D, lanes 1–4, marked by an asterisk). In contrast, treatment with IBPR001 led to the preservation of HURP-U (Fig. 4D, lanes 6–9). These results verified the notion that HURP-U is indeed associated with centrosomal microtubules. Moreover, they suggest that polymerization of centrosomal microtubules precedes HURP-U association.
These data imply the existence of an underlying mechanism associated with HURP dephosphorylation. The protein phosphatase family targets multiple mitotic structures such as chromosomes, centrosomes, and spindles in assisting mitotic progression (31). For example, Aurora B and protein phosphatase 1 (PP1) act antagonistically for the phosphorylation of histone H3 serine 10 in chromosome condensation (31, 32). To determine whether protein phosphatase is responsible for HURP dephosphorylation, we compared HURP Western profiles in nocodazole-arrested cells treated with IBPR001 alone or cotreated with Calyculin A, an inhibitor of PP1 and protein phosphatase 2A (PP2A) (33). The IBPR001-induced HURP dephosphorylation was eliminated by Calyculin A (Fig. 4E), suggesting that the dephosphorylation of HURP is associated with protein phosphatase 1/2A (PP1/PP2A) activity. Consistently, cotreatment with Calyculin A and IBPR001 enables HURP to associate with the nucleated microtubule bundles, similar to control cells (Fig. 4F).
Collectively, these results suggest that the two forms of HURP (HURP-P and HURP-U) cycle between centrosomes and kinetochores through Aurora A-dependent phosphorylation and protein phosphatase-regulated dephosphorylation in the establishment of mitotic spindles.
IBPR001 and MLN8237 Result in an Asymmetric Association of HURP to Centrosomal Microtubules.
During quantification of the HURP morphology resulting from treatment with the IBPR compounds, we observed the association of HURP with one of the spindle poles that resides closer to the chromosomes in ∼10% of cells (Fig. S2E, phenotype 3). This observation raises the question of whether HURP preferentially associates with the mother or daughter centrosome. [the mother centrosome contains the centriole of the eldest mother as well as that of the newborn daughter; conversely, the daughter centrosome contains the centriole of the second oldest mother and that of the newborn daughter (34, 35).] Mitotic spindles are perceived as a symmetric structure connecting the kinetochore of the duplicated chromatids with equal tension before separation (11, 27). Conversely, asymmetric cell divisions were observed in neural and male germ stem cells in which the mother centrosome oriented toward the stem cell niche and the daughter centrosome migrated through chromosomes to the opposite side of the mother centrosome (36, 37).
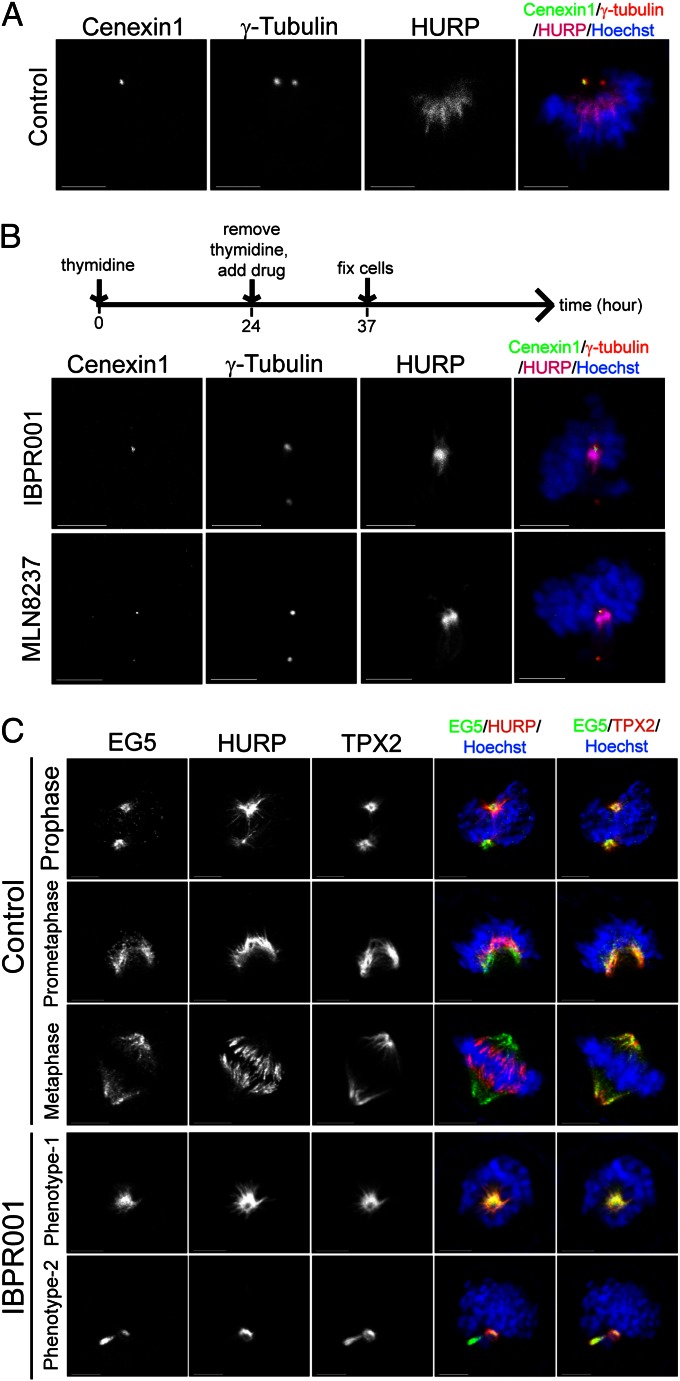
Outer dense fiber 2 (Odf2) was identified as a major component of the sperm tail cytoskeleton, which is a component of the centrosomal scaffold preferentially associated with the appendages of the mother centriole in somatic cells (38). Human Cenexin1 is an Odf2-related protein preferentially associated with the centrosome that contains the mother centriole (Fig. 5A) (39). To verify whether a spatial hierarchy exists in the HURP–centrosome association, we monitored the localization of HURP and Cenexin1 in cells treated with IBPR001 or MLN8237. As a result, the centrosome that was stained positive for Cenexin1 always (in 30 of 30 IBPR001- or MLN8237-treated asymmetric cells) was associated with HURP (Fig. 5B).
Fig. 5.
HURP-U is preferentially associated with the mother centrosome. (A) The eldest mother centriole was stained positive for Cenexin1. Centrosomes were stained positive for γ-tubulin. DNA was stained with Hoechst33342. (B) HURP preferentially resides with the mother centrosome that was stained positive (or stronger) for Cenexin1 in cells treated with 1.0 μM IBPR001 or MLN8237. Note that the γ-tubulin antibody stained both centrosomes. (C) Cells were treated with DMSO control or IBPR001 as shown in the scheme in B and were coimmunofluorescence stained for EG5, HURP, and TPX2. Images are maximum projections of z-stacks. (Scale bars: 5 μm.)
The formation and function of the complex containing HURP, kinesin-related motor protein 5 (EG5), and targeting protein for Xklp2 (TPX2) depends on Aurora A for the conversion of aster-like to spindle-like structures (16). We coimmunostained IBPR001-treated cells with HURP, EG5, and TPX2 to determine whether the asymmetric distribution of HURP is unique or is a phenomenon common to other Aurora A-regulated proteins. Unlike EG5 or TPX2, HURP appears to locate unequally to centrosomal esters in prophase cells; however, distribution became symmetric when the cell cycle entered prometaphase (Fig. 5C). Similar phenotypes were observed in cells treated with IBPR001. Thus, the phenotype of asymmetry was not generated by the Aurora kinase inhibitors. Rather, these compounds assisted in revealing the asymmetric nature of HURP (as expressed by its association with microtubules emanating from the mother centrosome; Fig. 5B) through the inhibition of Aurora A kinase activity and subsequent HURP dephosphorylation.
Discussion
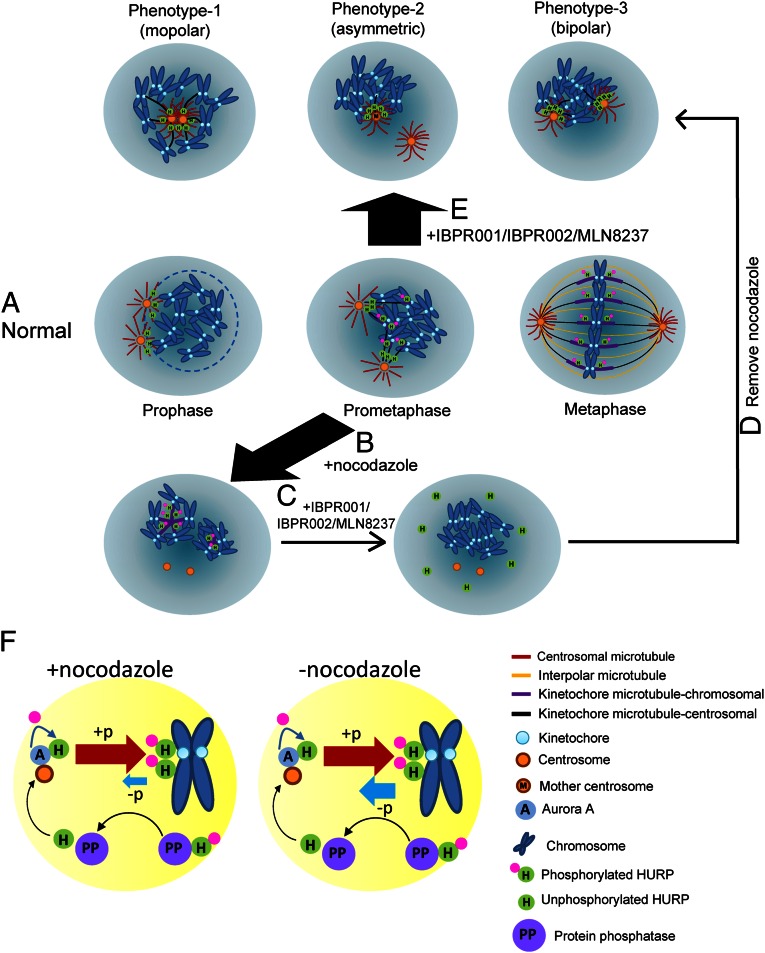
Small molecules that inhibit Aurora kinase activity have been extensively developed for their potential use in inhibiting tumor growth (3, 5, 6). Most attention has focused on the correlation between the kinase activity of Aurora A and mitotic progression (3, 4, 6). However, how these compounds influence the molecular properties of the substrate remains unclear. Our use of small molecules demonstrates that Aurora A-mediated HURP phosphorylation is required to initiate and stabilize microtubules emanating from the kinetochore but is not required for those originating at the centrosome. We also identified HURP’s preferential association with the mother centrosome. These findings provide direct experimental evidence correlating HURP phosphorylation and microtubule nucleation between centrosomes and kinetochores in mammalian cells. These findings are summarized in Fig. 6 A–E.
Fig. 6.
Models for nucleation of centrosomal and kinetochore microtubule fibers by Aurora A-regulated HURP phosphorylation during spindle formation. (A) As a cell enters mitosis, the nuclear envelope (light blue circle) breaks down; HURP is expressed and associated with the minus end of centrosomal microtubules that project toward chromosomes. As the cell cycle proceeds to prometaphase and metaphase, HURP gradually is phosphorylated and translocated to the vicinity of chromosomes to assist nucleation and stabilization of kinetochore fibers. Finally, bipolarity is established. Phosphorylated HURP forms a rod-like structure (purple bar) that links to kinetochore. (B) Treatment with 300–400 nM nocodazole enriched the phosphorylated HURP that nucleates kinetochore microtubules. (C) Adding IBPR001/IBPR002/MLN8237 disrupts nucleation of the kinetochore microtubule. (D) Removing nocodazole under the treatment with IBPR001/IBPR002/MLN8237 reinitiates tubulin polymerization from centrosomes but not from kinetochores. HURP goes to the minus end of centrosomal microtubules that face toward the chromosomes. (E) Inhibiting HURP phosphorylation by IBPR001/IBPR002/MLN8237 abolishes nucleation of HURP in the vicinity of chromosomes. The unphosphorylated HURP distributes restrictively to the minus end of centrosomal microtubules. Because IBPR001/IBPR002/MLN8237 do not completely block the separation of the duplicated centrosomes, HURP is preferentially associated with the microtubules that emanate from the mother centrosome, which resides in proximity to the chromosomes. (F) Models for HURP phosphorylation and dephosphorylation with and without nocodazole. In the presence of nocodazole, the force that drives HURP phosphorylation (by Aurora A) overrides dephosphorylation (through a PP1/PP2A-dependent pathway). Conversely, the ratio of phosphorylation/dephosphorylation is decreased in the absence of nocodazole.
Rapid inhibition of kinase activities is necessary to identify the functional role of protein phosphorylation in spindle formation, which generally reaches completion within 1 h. VX-680 and MLN8237 inhibit Aurora A kinase activity more effectively than IBPR compounds in vitro (Fig. 2C and references 3 and 40); however, our results suggest that IBPR compounds are more effective inhibitors of HURP phosphorylation in cells (Fig. 2 A and B). The discrepancies between cellular and in vitro inhibition can be attributed to compound pharmacokinetics and/or cofactors binding to Aurora A in cells. The crystal structures reveal that the Aurora A kinase alters the conformation of the activation loop in accommodating IBPR001 from VX-680. The activation loop in protein kinases is important for activity regulation and substrate binding (41). The difference in the conformation of the activation loop between Aurora A/IBPR001 and Aurora A/VX-680 may confer Aurora A’s sensitivity to HURP phosphorylation. Thus, the two IBPR compounds identified in this study could help reveal the molecular mechanism underlying spindle formation and perhaps lead to more effective clinical treatments of cancer in the future.
During spindle formation, microtubules emanating from the duplicated centrosomes continuously extend and shrink before the bilateral attachment is established. Our results suggest the phosphorylation status of HURP is different at the two extreme ends (i.e., centrosome and kinetochore) of the mitotic spindles and exhibits distinct functions in tubulin nucleation (Figs. 3 and 4). These results raised additional questions regarding how HURP migrates between these two loci during the dynamic process of spindle formation. If the process of HURP phosphorylation is unidirectional, then with nocodazole treatment HURP should remain phosphorylated and always associate with kinetochore microtubules (Fig. 3A). In this setting, the inhibition of Aurora A activity by IBPR compounds or MLN8237 should not influence HURP phosphorylation, but this was not the case (Fig. 3B), suggesting that the Aurora A-mediated HURP phosphorylation can be reversed. We showed that when cells are released from nocodazole arrest, HURP dephosphorylation resumes (Fig. 4D, lanes 1–4), and HURP reassociates with the centrosomal microtubules (Fig. 4C, control). These results imply, first, the existence of an underlying mechanism that is associated with HURP dephosphorylation, which was shown to involve the PP1/PP2A activity (Fig. 4 E and F). Second, HURP is highly dynamic, trafficking between centrosomes and kinetochores driven by the mechanisms of Aurora A-dependent phosphorylation and PP1/PP2A-associated dephosphorylation (Fig. 6F). Third, although tubulin flux is not required for HURP phosphorylation or dephosphorylation, tubulin polymerization may facilitate HURP dephosphorylation to generate a balanced HURP phosphorylation status and consequently the establishment of robust bipolar spindles between centrosomes and kinetochores (Figs. 4 C and D and 6F). In addition, to provide a mechanistic explanation for HURP in spindle establishment, the assays developed in this study enable the enrichment of HURP in its phosphorylated form at the kinetochore and in its unphosphorylated form at the centrosomes. This methodology may help reveal new biomolecules that participate in the nucleation and bundling of kinetochore or centrosomal microtubules.
Although centrosome-derived microtubules are radial initially, they begin growing with directional bias so that the density of microtubules between centrosomes and the mitotic chromosomes is greater than between centrosomes and the cell cortex (42). These observations suggest a lack of equality in the extension of microtubules to the cortex or to chromosomes. It remains unclear what elements, including microtubule-associated proteins or motor proteins, cause this directional activity (42). HURP is one element that participates in this asymmetry through its association with microtubules that grow toward the mitotic chromosomes rather than the cortex (Fig. 4A and ref. 17). Using IBPR001 or MLN8237 to block Aurora A activity, we identified a phenotype in which HURP preferentiality resides in the microtubules initiated from the mother centrosome (Fig. 5 B and C). Like HURP, TPX2 is a Ran-regulated spindle assembly factor, which is enriched near the spindle poles and required for K-fiber formation (16, 17, 28). However, TPX2 does not present asymmetric distribution (Fig. 5C). This phenotype suggests that HURP plays a unique role in generating an additional dimension of asymmetry associated with mitotic centrosomal microtubules.
Asymmetry has been observed in many forms of cell division in which spindles organize asters with various dynamics, associate with various molecules or subcellular domains, and perform various functions (43). For example, in budding yeast, one spindle pole nucleates more stable microtubules than the other (43). In the zygote of Caenorhabditis elegans, the anterior aster of the asymmetrically positioned spindle is large and has many microtubules, whereas the posterior aster appears flattened and smaller with fewer astral microtubules (44, 45). In Drosophila neuroblasts, the astral microtubules on the basal spindle pole are induced to depolymerize, whereas those of the apical aster are stabilized, resulting in a larger apical and smaller basal aster that together constitute an asymmetric spindle (44, 46). Intriguingly, Aurora A is required for the asymmetric localization of atypical protein kinase C to prevent it from localizing to the basal cortex in Drosophila neuroblasts (47, 48). In aurora-A loss-of-function mutants, supernumerary self-renewal neuroblasts are produced, whereas neuronal differentiation is reduced (47). As with Aurora A, HURP allows efficient sorting of the microtubule-organizing center into distinct poles, efficient congression of chromosomes, and the establishment of bipolarity in mouse oocytes, by promoting microtubule stability in the central domain of the spindle (49). In summary, our results establish the involvement of both Aurora A kinase activity and HURP in microtubule nucleation and the symmetry of mitotic spindles in cultured mammalian cells. Whether cell-division molecules other than Aurora A and HURP contribute to this process warrants future investigation (50).
Materials and Methods
Cell Culture, Plasmids and Transfection.
HeLa and 293T cells were maintained in high-glucose DMEM (Invitrogen) supplemented with 10% (wt/vol) FBS (Biological Industries), 2 mM L-glutamine, and antibiotics. The human colorectal cancer HCT116 cell line was obtained from American Type Culture Collection. Cells were maintained in McCoy’s 5A Medium (Gibco) containing 10% FBS (Gibco).
Antibodies and Reagents.
Antibodies were obtained from the following sources. Abcam: mouse anti–γ-tubulin (ab11316) and mouse anti-TPX2 (ab32795); Sigma-Aldrich: mouse anti–α-tubulin (T5168) and mouse anti-actin (A1978); Santa Cruz Biotechnology: goat anti-HURP (sc-68540); Covance: rabbit anti-Pericentrin (PRB-432C); Cell Signaling: mouse anti-cyclin E1 (4129); BD Transduction Laboratories: mouse anti-Aurora A (610939); Invitrogen: rabbit anti-Aurora B (36-5200) and rabbit anti-PTTG (34-1500); Bethyl Laboratories: rabbit anti-HURP (A300-853A); ProteinTech Group: rabbit anti-ODF2/Cenexin1 and rabbit anti-NEK2 (629402); Cortex Biochem: CREST antiserum; Epitomics: rabbit anti-EG5 (S1765) and rabbit anti-CDK1 (3787-1). Thymidine (T1895) and nocodazole (M1404) were from Sigma-Aldrich; MG132 was from Calbiochem (474790). MLN8237 and AZD1152 were purchased from Selleckchem.
RNAi.
Experimentally verified FlexiTube double-stranded siRNAs of AURKA (SI02223305), AURKB (SI02622032), HURP (SI02654169), CDK1 (SI00299719), and NEK2 (SI00605640 and SI00605647) were from QIAGEN. siRNAs were introduced into HeLa cells using the Lipofectamine RNAiMax transfection reagent (Invitrogen) following the manufacturer’s protocol.
Immunofluorescence and Confocal Microscopy.
Cells were fixed in 4% paraformaldehyde in PBS for 30 min and permeabilized with 0.1% Triton X-100 for 5 min at room temperature. For γ-tubulin, Pericentrin, and Cenexin1 staining, cells were fixed in methanol for 10 min at −20 °C. Cells were incubated with 1% BSA in PBS for 30 min to block nonspecific binding. Primary antibodies were added at dilutions of 1:100–1:1,000 and were incubated for 1.5 h at room temperature. After three washes with PBS, cells were probed with corresponding fluorescent (Alexa 488, Alexa 594, or Alexa 647)-conjugated secondary antibodies (Invitrogen). Cell nuclei were counterstained with Hoechst33342 (Invitrogen). Cells were mounted onto glass slides with ProLong Gold antifade reagent (Invitrogen) and were visualized using a Leica TCS SP5 confocal microscope. Images were processed by the Imaris 7.2.1 software (Bitplane).
Western Blotting.
Total cell lysates were extracted with ice-cold RIPA buffer [50 mM Hepes (pH 7.3), 150 mM NaCl, 2 mM EDTA, 20 mM β-glycerophosphate, 0.1 mM Na3VO4, 1 mM NaF, 0.5 mM DTT, and protease inhibitor mixture (Roche)] containing 1% Nonidet P-40 plus mild sonication. Phosphatase inhibitors were not supplied for shrimp alkaline phosphatase (SAP; Fermentas) treatment. Lysates were analyzed by SDS/PAGE, transferred to PVDF membrane (GE Healthcare), and blotted with antibodies. Alkaline phosphatase-conjugated secondary antibodies (Sigma-Aldrich) were added, and the blots were developed by chemiluminescence following the manufacturer’s protocol (Perkin-Elmer).
Supplementary Material
Acknowledgments
We thank the staff at beamline BL13B1 and BL13C1 (National Synchrotron Radiation Research Centre, Taiwan) and the staff at beamline SP12B2 (SPring-8, Japan) for technical assistance, Dr. Chang-Tze Rickey Yu for technical suggestions, and Dr. Lindsay Sawyer, Dr. Kuan-Teh Jeang, Dr. Jeng-Jiann Chiu, Dr. Tang K. Tang, and Dr. Yung-Chi Cheng for critical comments on this manuscript. The specificity of compound IBPR002 to kinases was determined in a contract with Ricerca Biosciences. This work was supported by National Health Research Institutes Grants 00A1-CSPP11-014 and 01A1-CSPP13-014 (to Y.-H.C.), BP-100-PP-01 and NSC-96-2113-M-400-001-MY3 (to S.-Y.W.), and NSC-95-2113-M-400-001-MY3 and NSC-100-2325-B-400-003 (to H.-P.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4JBO, 4JBP, and 4JBQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220523110/-/DCSupplemental.
References
- 1.Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10(12):825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- 2.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18(6):658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Harrington EA, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10(3):262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 4.Manfredi MG, et al. Antitumor activity of MLN8054, an orally active small-molecule inhibitor of Aurora A kinase. Proc Natl Acad Sci USA. 2007;104(10):4106–4111. doi: 10.1073/pnas.0608798104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katayama H, Sen S. Aurora kinase inhibitors as anticancer molecules. Biochim Biophys Acta. 2010;1799(10-12):829–839. doi: 10.1016/j.bbagrm.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Görgün G, et al. A novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell-cycle arrest in multiple myeloma. Blood. 2010;115(25):5202–5213. doi: 10.1182/blood-2009-12-259523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinsky BA, Biggins S. The spindle checkpoint: Tension versus attachment. Trends Cell Biol. 2005;15(9):486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Tillement V, et al. Spindle assembly defects leading to the formation of a monopolar mitotic apparatus. Biol Cell. 2009;101(1):1–11. doi: 10.1042/BC20070162. [DOI] [PubMed] [Google Scholar]
- 9.Maiato H, Rieder CL, Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol. 2004;167(5):831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Connell CB, Khodjakov AL. Cooperative mechanisms of mitotic spindle formation. J Cell Sci. 2007;120(Pt 10):1717–1722. doi: 10.1242/jcs.03442. [DOI] [PubMed] [Google Scholar]
- 11.Rieder CL. Kinetochore fiber formation in animal somatic cells: Dueling mechanisms come to a draw. Chromosoma. 2005;114(5):310–318. doi: 10.1007/s00412-005-0028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gadde S, Heald R. Mechanisms and molecules of the mitotic spindle. Curr Biol. 2004;14(18):R797–R805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 13.Waters JC, Salmon E. Pathways of spindle assembly. Curr Opin Cell Biol. 1997;9(1):37–43. doi: 10.1016/s0955-0674(97)80149-4. [DOI] [PubMed] [Google Scholar]
- 14.Tsou AP, et al. Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene. 2003;22(2):298–307. doi: 10.1038/sj.onc.1206129. [DOI] [PubMed] [Google Scholar]
- 15.Yu CT, et al. Phosphorylation and stabilization of HURP by Aurora-A: Implication of HURP as a transforming target of Aurora-A. Mol Cell Biol. 2005;25(14):5789–5800. doi: 10.1128/MCB.25.14.5789-5800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koffa MD, et al. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006;16(8):743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 17.Silljé HH, Nagel S, Körner R, Nigg EA. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol. 2006;16(8):731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 18.Wilde A. “HURP on” we’re off to the kinetochore! J Cell Biol. 2006;173(6):829–831. doi: 10.1083/jcb.200605150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong J, Lerrigo R, Jang CY, Fang G. Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol Biol Cell. 2008;19(5):2083–2091. doi: 10.1091/mbc.E07-10-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coumar MS, et al. Identification, SAR studies, and X-ray co-crystallographic analysis of a novel furanopyrimidine aurora kinase A inhibitor. ChemMedChem. 2010;5(2):255–267. doi: 10.1002/cmdc.200900339. [DOI] [PubMed] [Google Scholar]
- 21.Coumar MS, et al. Fast-forwarding hit to lead: Aurora and epidermal growth factor receptor kinase inhibitor lead identification. J Med Chem. 2010;53(13):4980–4988. doi: 10.1021/jm1000198. [DOI] [PubMed] [Google Scholar]
- 22.Cheetham GM, et al. Crystal structure of aurora-2, an oncogenic serine/threonine kinase. J Biol Chem. 2002;277(45):42419–42422. doi: 10.1074/jbc.C200426200. [DOI] [PubMed] [Google Scholar]
- 23.Martin MP, et al. A novel mechanism by which small molecule inhibitors induce the DFG flip in Aurora A. ACS Chem Biol. 2012;7(4):698–706. doi: 10.1021/cb200508b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santarella RA, Koffa MD, Tittmann P, Gross H, Hoenger A. HURP wraps microtubule ends with an additional tubulin sheet that has a novel conformation of tubulin. J Mol Biol. 2007;365(5):1587–1595. doi: 10.1016/j.jmb.2006.10.064. [DOI] [PubMed] [Google Scholar]
- 25.Jordan MA, Thrower D, Wilson L. Effects of vinblastine, podophyllotoxin and nocodazole on mitotic spindles. Implications for the role of microtubule dynamics in mitosis. J Cell Sci. 1992;102(Pt 3):401–416. doi: 10.1242/jcs.102.3.401. [DOI] [PubMed] [Google Scholar]
- 26.Song L, Rape M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol Cell. 2010;38(3):369–382. doi: 10.1016/j.molcel.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka TU. Kinetochore-microtubule interactions: Steps towards bi-orientation. EMBO J. 2010;29(24):4070–4082. doi: 10.1038/emboj.2010.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong J, Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol. 2006;173(6):879–891. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ducat D, Zheng Y. Aurora kinases in spindle assembly and chromosome segregation. Exp Cell Res. 2004;301(1):60–67. doi: 10.1016/j.yexcr.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 30.Hannak E, Kirkham M, Hyman AA, Oegema K. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J Cell Biol. 2001;155(7):1109–1116. doi: 10.1083/jcb.200108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ceulemans H, Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev. 2004;84(1):1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- 32.Murnion ME, et al. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J Biol Chem. 2001;276(28):26656–26665. doi: 10.1074/jbc.M102288200. [DOI] [PubMed] [Google Scholar]
- 33.Wakimoto T, Matsunaga S, Takai A, Fusetani N. Insight into binding of calyculin A to protein phosphatase 1: Isolation of hemicalyculin a and chemical transformation of calyculin A. Chem Biol. 2002;9(3):309–319. doi: 10.1016/s1074-5521(02)00118-7. [DOI] [PubMed] [Google Scholar]
- 34.Januschke J, Llamazares S, Reina J, Gonzalez C. Drosophila neuroblasts retain the daughter centrosome. Nat Commun. 2011;2:243. doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315(5811):518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rebollo E, et al. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12(3):467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 37.Cabernard C, Doe CQ. Stem cell self-renewal: Centrosomes on the move. Curr Biol. 2007;17(12):R465–R467. doi: 10.1016/j.cub.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 38.Ishikawa H, Kubo A, Tsukita S, Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol. 2005;7(5):517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- 39.Soung NK, et al. Requirement of hCenexin for proper mitotic functions of polo-like kinase 1 at the centrosomes. Mol Cell Biol. 2006;26(22):8316–8335. doi: 10.1128/MCB.00671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sloane DA, et al. Drug-resistant aurora A mutants for cellular target validation of the small molecule kinase inhibitors MLN8054 and MLN8237. ACS Chem Biol. 2010;5(6):563–576. doi: 10.1021/cb100053q. [DOI] [PubMed] [Google Scholar]
- 41.Endicott JA, Noble ME, Johnson LN. The structural basis for control of eukaryotic protein kinases. Annu Rev Biochem. 2012;81:587–613. doi: 10.1146/annurev-biochem-052410-090317. [DOI] [PubMed] [Google Scholar]
- 42.Duncan T, Wakefield JG. 50 ways to build a spindle: The complexity of microtubule generation during mitosis. Chromosome Res. 2011;19(3):321–333. doi: 10.1007/s10577-011-9205-8. [DOI] [PubMed] [Google Scholar]
- 43.Barral Y, Liakopoulos D. Role of spindle asymmetry in cellular dynamics. Int Rev Cell Mol Biol. 2009;278:149–213. doi: 10.1016/S1937-6448(09)78004-9. [DOI] [PubMed] [Google Scholar]
- 44.Kaltschmidt JA, Brand AH. Asymmetric cell division: Microtubule dynamics and spindle asymmetry. J Cell Sci. 2002;115(Pt 11):2257–2264. doi: 10.1242/jcs.115.11.2257. [DOI] [PubMed] [Google Scholar]
- 45.Keating HH, White JG. Centrosome dynamics in early embryos of Caenorhabditis elegans. J Cell Sci. 1998;111(Pt 20):3027–3033. doi: 10.1242/jcs.111.20.3027. [DOI] [PubMed] [Google Scholar]
- 46.Giansanti MG, Gatti M, Bonaccorsi S. The role of centrosomes and astral microtubules during asymmetric division of Drosophila neuroblasts. Development. 2001;128(7):1137–1145. doi: 10.1242/dev.128.7.1137. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, et al. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 2006;20(24):3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135(1):161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Breuer M, et al. HURP permits MTOC sorting for robust meiotic spindle bipolarity, similar to extra centrosome clustering in cancer cells. J Cell Biol. 2010;191(7):1251–1260. doi: 10.1083/jcb.201005065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inaba M, Yamashita YM. Asymmetric stem cell division: Precision for robustness. Cell Stem Cell. 2012;11(4):461–469. doi: 10.1016/j.stem.2012.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.