Abstract
Memory is initially labile and gradually consolidated over time through new protein synthesis into a long-lasting stable form. Studies of odor-shock associative learning in Drosophila have established the mushroom body (MB) as a key brain structure involved in olfactory long-term memory (LTM) formation. Exactly how early neural activity encoded in thousands of MB neurons is consolidated into protein-synthesis–dependent LTM remains unclear. Here, several independent lines of evidence indicate that changes in two MB vertical lobe V3 (MB-V3) extrinsic neurons are required and contribute to an extended neural network involved in olfactory LTM: (i) inhibiting protein synthesis in MB-V3 neurons impairs LTM; (ii) MB-V3 neurons show enhanced neural activity after spaced but not massed training; (iii) MB-V3 dendrites, synapsing with hundreds of MB α/β neurons, exhibit dramatic structural plasticity after removal of olfactory inputs; (iv) neurotransmission from MB-V3 neurons is necessary for LTM retrieval; and (v) RNAi-mediated down-regulation of oo18 RNA-binding protein (involved in local regulation of protein translation) in MB-V3 neurons impairs LTM. Our results suggest a model of long-term memory formation that includes a systems-level consolidation process, wherein an early, labile olfactory memory represented by neural activity in a sparse subset of MB neurons is converted into a stable LTM through protein synthesis in dendrites of MB-V3 neurons synapsed onto MB α lobes.
Keywords: CPEB, CREB, fragile X mental retardation, STAU, PUM
Long-term memory (LTM) and long-term synaptic plasticity require de novo protein synthesis, which is regulated at transcriptional and/or translational levels in a synapse-specific manner (1–3). Synapse-specific plasticity during LTM formation in some contexts may involve local regulation of protein translation by a family of RNA-binding proteins, the cytoplasmic polyadenylation element-binding proteins (CPEBs) (2). Neuronal CPEBs have two conformational states. The inactive state predominates at low levels of CPEB expression and represses translation from nascent mRNAs. The active state is achieved either via a self-perpetuating prion-like state when expression levels surpass a threshold or via Ca2+/calmoduline-dependent protein kinase II (CaMKII)-mediated phosphorylation, and translation is initiated by elongation of an mRNA’s poly-A tail (4–6). In other species, CPEB1 has been shown to contribute to long-term facilitation or potentiation (5, 7). In Drosophila, oo18 RNA-binding protein 2 (ORB2) appears required for long-term memory formation after courtship conditioning (8, 9). Any role for ORB in fruit fly memory formation, however, remains unclear.
Drosophila can learn to associate an odor (conditioned stimulus, CS) with foot-shock punishment (unconditioned stimulus, US). This odor–shock association initially is labile, lasting for only about a day after one training session. With repetitive, spaced training (ST) sessions (rest intervals between each session), a protein synthesis-dependent, LTM is formed. With repetitive, massed training (MT) sessions (no rest intervals between sessions), LTM is not formed. The consolidation of odor–shock memories presumably involves multiple nodes in the underlying neuronal network, including antenna lobes (10), mushroom body (MB) (11–13), ellipsoid body (14), and two dorsal anterior lateral (DAL) neurons (15). The MB in each hemisphere consists of ∼2,500 intrinsic neurons that can be classified into at least five major types: γ, α′/β′, pioneer α/β, early α/β, and late α/β (16). Surprisingly, inhibition of protein synthesis or disruption of cAMP response element binding protein 2 (CREB2) activity in MB neurons does not impair LTM; rather, these hallmarks of memory consolidation are required in DAL neurons (15). Exactly how early labile memory encoded in the MB neurons is converted into a stable LTM in DAL neurons and whether such experience-dependent transcription and translation during LTM formation involves additional neurons remain unclear. Here, we show that LTM formation requires CREB2-independent protein synthesis and ORB-dependent translational regulation in two MB output neurons.
Results
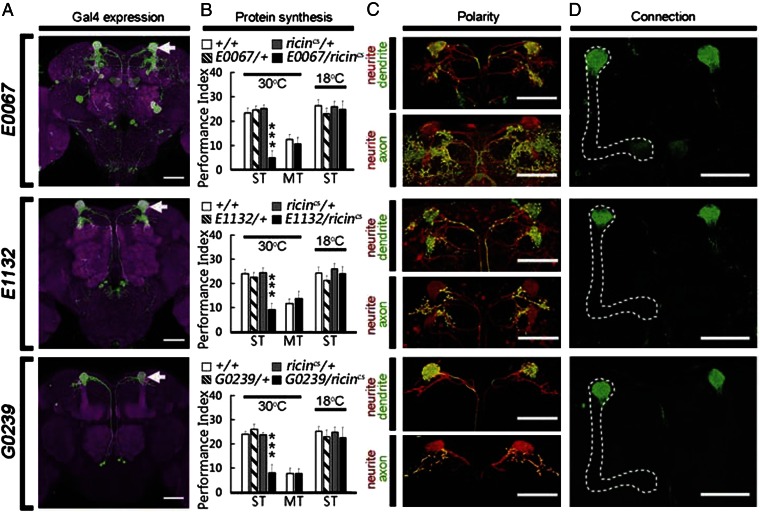
To address how early, neural activity in MB neurons is consolidated into a stable protein-synthesis–dependent LTM outside of MB, we searched individual MB output neurons for evidence of de novo protein synthesis after odor–shock training. Using targeted expression of a cold-sensitive RICINcs, a potent toxic protein that inactivates ribosomes at 30 °C but not 18 °C (15), we found that de novo protein synthesis in E0067-, E1132- and G0239-Gal4 neurons after spaced training was necessary for LTM formation (Fig. 1 A and B). In control experiments, expression of inactivated RICINcs after spaced training, or activated RICINcs after massed training, did not affect 1-d memory (Fig. 1B). Two pairs of MB extrinsic neurons, called MB-V3 neurons (17), were common to the expression patterns of these three Gal4 lines (Fig. S1). In particular, G0239-Gal4 expressed only in MB-V3 neurons (Fig. 1A) and, thus, was selected for further behavioral study.
Fig. 1.
Inhibiting protein synthesis in MB-V3 neurons impairs LTM formation. (A) Expression patterns of E0067-, E1132- and G0239-Gal4s. GFP labeled two pairs of MB-V3 neurons innervating α-lobe tips (arrow). Brain structures were immunostained with antidiscs large antibody (magenta). E0067- and E1132-Gal4s also expressed in brain surface glia (digitally removed). (Scale bars, 50 μm.) (B) Effects of RICINcs inhibition in E0067-, E1132-, and G0239-Gal4 neurons. One-day memory is impaired by inhibiting protein synthesis after spaced training (ST), but not massed training (MT), with active RICINcs (30 °C). One-day memory after spaced training was normal with inactive RICINcs (18 °C). Values are means ± SEM (***P < 0.001; n = 8 for each group). (C) The MB-V3 neuron projects Dscam-positive dendrites (green) exclusively within the entire α-lobe tip and syt-positive axons (green) at superior dorsofrontal protcerebrum outside of the MB. Fibers are labeled by a monomeric Kusabira Orange (mKO) protein (red). (D) Structural connections between MB-V3 dendrites and axons of MB neurons (L0124-LexA, dotted line) were visualized by GRASP labeling (green, Lower). (Scale bars, 50 μm.) For more details, please see SI Methods.
Labeling MB-V3 dendrites with Down syndrome cell adhesion molecule fused with GFP (Dscam::GFP) established that they projected to the tip of the MB α lobe. Conversely, labeling MB-V3 axons with synaptotagmin fused with GFP (syt::GFP) showed that they projected to the superior dorsofrontal protocerebrum (Fig. 1C). To assess potential connections between MB-V3 dendrites and axons of MB neurons, we used the GFP reconstitution across synaptic partners (GRASP) technique (18). One half of the split-GFP GRASP reporter was expressed in the MB-V3 neurons, whereas the other half of the split-GFP GRASP reporter was expressed in distinct types of MB neurons using an appropriate LexA driver (Fig. S2). We found that MB-V3 dendrites are in close contact with axons of three types of MB α/β neurons, including pioneer, early, and late α/β neurons (Fig. 1D and Fig. S2). They do not make contact with MB α′/β′ neurons, however (Fig. 1A).
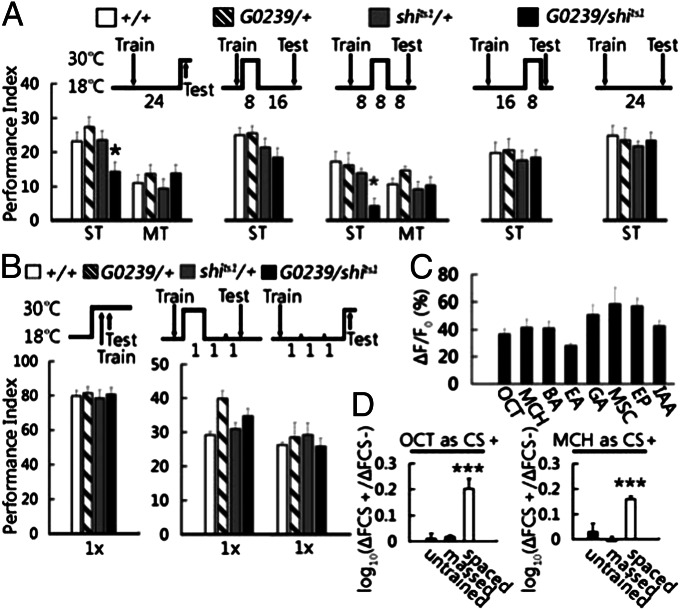
Next, we evaluated the role of neural activity from MB-V3 by temporally blocking neurotransmssion with UAS-shits1, a temperature-sensitive mutant dynamin protein SHIBIRE that blocks neurotransmission at 30 °C but not at 18 °C (19). Blocking neurotransmssion from MB-V3 neurons during LTM retrieval impaired 1-d memory after spaced training but not after massed training (Fig. 2A). Intriguingly, normal 24-h memory retention required neurotransmission from MB-V3 neurons only during the second, but not the first or the third, 8-h period after spaced training. This requirement for synaptic transmission 8–16 h after spaced training was specific to LTM formation, because the same disruption did not affect 1-d memory after massed training (Fig. 2A) and was corroborated with an independent MB-V3–expressing Gal4 driver, E0067-Gal4 (Fig. S3). Synpatic transmission from MB-V3 neurons also was not required for learning or for 3-h memory after a single training session (Fig. 2B). Together, these data establish that neural activity from MB-V3 is required specifically for LTM consolidation and retrieval.
Fig. 2.
Neurotransmission and functional response of MB-V3 neurons during LTM formation. (A) Roles of MB-V3 neurotransmission on LTM. Blocking neurotransmission from MB-V3 neurons with temperature sensitive shits1 protein (30 °C) during retrieval (P = 0.015; n = 12) or 8–16 h after training (P = 0.012; n = 8) impaired 1-d memory after spaced training, but not after massed training. Blocking neurotransmission during 0–8 or 16–24 h after spaced training or keeping the shits1 flies in permissive temperature (18 °C) had no effect on 1-d memory. (B) Blocking neurotransmission from MB-V3 neurons during acquisition or 3 h after a single training session had no effects on memory retention. Values are means ± SEM (n = 8 for each group). (C) Neural activity in MB-V3 neurons in response to eight different odors (OCT, MCH, BA, EA, GA, MSC, EP, and IAA). Values are means ± SEM (n ≧ 8 for each odor). (D) Enhanced neural activity in MB-V3 neurons in response to conditioned odors after spaced, but not massed, training. Values are means ± SEM (P < 0.001; n ≧ 8 for each group). For more details, please see SI Methods.
The GRASP technology can signal only close proximity between neurons (18), so we confirmed direct synaptic connectivity using EM labeling. A membrane-fused horseradish peroxidase (HRP::CD2) was expressed in the pioneer α/β MB neurons and/or in MB-V3 neurons. We found that small HRP-positive MB-V3 dendrites synpased with HRP-positive MB neurons peripherally at the tip of the α lobe and with large HRP-negative boutons at the core α lobe (Fig. S4 A–E). These EM observations suggest a direct synaptic connection between MB-V3 dendrites and axonal terminals of several different types of α/β MB neurons. Blocking neurotransmission from the pioneer α/β neurons in c708a-Gal4 impaired 24-h memory during retrieval, but not during consolidation (Fig. S4F). Also, neurotransmission outputs from core α/β neurons (20) and MB-V3 neurons are both necessary during a delayed time period after spaced training for LTM formation. Altogether, these data show direct structural connectivity and are suggestive of potential functional connectivity between α/β MB neurons and MB-V3 neurons.
To visualize functional responses to conditioned odors in MB-V3 dendrites synapsing at the tip of α lobe, we used the calcium-sensitive fluorescent protein, UAS-GCaMP1.6 (21). In naïve flies, MB-V3 neurons (Fig. 2C), but not ellipsoid body neurons (Fig. S5), showed strong responses to eight different odors [3-octanol (OCT), 4-methylcyclohexanol (MCH), benzaldehyde (BA), ethyl acetate (EA), geranyl acetate (GA), methyl salicylate (MSC), ethyl propionate (EP), and isoamyl acetate (IAA)]. Thus, the MB-V3 neurons appear to be odor generalists. In response to conditioned odors (using either OCT or MCH), MB-V3 neurons exhibited an elevated GCaMP intensity 24 h after spaced but not after massed training compared with naïve (untrained) flies (Fig. 2D).
The observations that (i) MB-V3 neurons respond generally to odors and (ii) show an experience-dependent increase in neural activity prompted us to ascertain whether these neurons were capable of experience-dependent structural changes, a cellular mechanism often attributed to LTM formation (22–25). In naïve flies, MB-V3 neurons showed intact morphology (Fig. S6 A, E, and I). About 3 wk after removal of both antennae from newly eclosed adults, in contrast, some MB-V3 neurons showed dramatic changes in morphology, including unrestricted dendrite arborization (Fig. S6 B and G), cell body displacement (Fig. S6 C and H), extended dendrite innervation further into the α lobe (Fig. S6C) and extra branching to innervate the tip of α lobe (Fig. S6D), whereas structural changes in axons appeared relatively minor (Fig. S6 F–H). In the alpha-lobe-absence (ala) mutant, MB-V3 neurons failed to arborize in the absence of MB vertical lobes but grew normally in the absence of MB horizontal lobes (Fig. S6 J–L). Thus, a previous conclusion that LTM must reside within the MB because LTM was impaired in the ala mutant (11) now can be reinterpreted: Disconnection of MB-V3 neurons from the α lobes prevents the proper induction of protein synthesis within MB-V3 neurons (Fig. 1) during LTM formation. We also tried to investigate if any morphological change in MB-V3 neurons could be detected after conditioning, but as expected any putative dendritic plasticity induced by spaced training was too subtle to visualize (Fig. S6 M and N). Together, these results indicate that MB-V3 neurons are specialized for olfactory information processing, exhibit experience-dependent structural plasticity, and are required for LTM consolidation and retrieval.
LTM formation requires the transcription factor, CREB, and the N-methyl-D-aspartate (NMDA) receptor in many animals, including Drosophila (12, 14, 26). Recently, we showed that down-regulation of either CREB2 or NMDA receptor in two DAL neurons is sufficient to impair LTM, which is consistent with our finding that LTM is impaired when protein synthesis is inhibited in DAL neurons (15). Unexpectedly, we found that (i) overexpressing a CREB repressor protein (UAS-dcreb2-b; Fig. 3A) or (ii) knockdown of CREB2 with UAS-creb2RNAi (Fig. 3B) or (iii) of NMDA receptor with UAS-dsNR2;UAS-dsNR1 in the MB-V3 neurons did not impair 1-d memory after spaced training (Fig. 3C). Also, 1-d memory after spaced training remains intact after RNAi-mediated knockdown of Drosophila ORB2 in MB-V3 neurons (Fig. 3D), even though inhibition of protein synthesis in these neurons impairs LTM (Fig. 1). In contrast, RNAi-mediated knockdown in the MB-V3 neurons of either Drosophila ORB (Fig. 3 E and F and Figs. S7 and S8), or CaMKII (Fig. 3G and Fig. S8), which regulates CPEB phosphorylation bidirectionally (6), impaired 1-d memory after spaced, but not massed, training.
Fig. 3.
Molecular machinery for LTM formation in the MB-V3 neurons. The G0239-Gal4-driven UAS-RNAi constructs (or CREB repressor) were inhibited by activated Gal80ts (18 °C) throughout development and then were expressed by inactivating Gal80ts (30 °C) 3 d before training. Control flies were kept constantly at 18 °C (activated Gal80ts). (A–D) Induced knockdown of CREB with either UAS-dcreb2-b transgenic overexpression (A) or UAS-creb2RNAi (B), of NMDAR1 and NMDAR2 with UAS-dsNR2;UAS-dsNR1 (C), or of ORB2 (D) with UAS-orb2RNAi did not affect 1-d memory after spaced training. (E–K) In contrast, 1-d memory was impaired after spaced, but not massed, training with induced knockdown of ORB with UAS-orbRNAi(R1) (P = 0.013) (E) or UAS-orbRNAi(R5) (P = 0.01) (F), of CaMKII with UAS-CaMKIIhpn (P = 0.003) (G), of FMR with UAS-fmrRNAi (1–7) (P < 0.001) (H) or UAS-fmrRNAi (2–1) (P = 0.011) (I), of STUAFEN with UAS-stauRNAi (P = 0.015) (J) or of PUMILIO with UAS-pumRNAi (P = 0.009) (K). Values are means ± SEM (n ≧ 8 for each group). For more details, please see SI Methods.
We have previously shown that in vivo disruptions of fragile X mental retardation (FMR), STAUFEN or PUMULIO proteins, additional molecular machinery involved in local control of mRNA translation, also yield defective LTM (27, 28). Using targeted RNAi-mediated knockdown in the MB-V3 neurons, we found that disruptions of FMR (Fig. 3 H and I), STAUFEN (Fig. 3J), or PUMULIO (Fig. 3K) protein again impaired 1-d memory after spaced training, but not after massed training. These transgenic flies nonetheless exhibited normal learning (Fig. S9), implying that acquisition and the sensorimotor responses that subserve it are normal. The efficiency of RNAi knockdown was determined via immunostaining; protein expressions were significantly decreased in the targeted MB-V3 neurons but not in other brain regions (Fig. S8). The specificity of RNAi knockdowns was addressed by misexpressing the RNAi transgenes in mushroom body M3 (MB-M3) neurons, a different set of MB extrinsic neurons required for 3-h memory (29); 1-d memory after spaced training was normal in these cases (Fig. S10). Together, these results suggest that ORB-dependent protein synthesis might be regulated by mRNA translation in MB-V3 neurons, a process quite different from the CREB-dependent regulation of transcription in DAL neurons.
Discussion
Drosophila MB-V3 neurons are similar to honey bee pedunculus extrinsic (PE1) neurons, which are considered to act as the conditioned response neurons when training increases activity in the output synapses of simultaneously activated Kenyon cells (30). Structurally, MB-V3 and PE1 neurons both are MB efferent neurons connecting many Kenyon cells in the α lobe, which is crucial for LTM formation, to the superior dorsofrontal protocerebrum. Functionally, both types of neurons are responsive to multiple odors and show enhanced neural activity to conditioned odor after repetitive training. During olfactory conditioning, an odor CS signal is encoded in sparse subsets of MB neurons (31), whereas the aversive US signal is delivered to most, if not all, Kenyon cells via dopaminergic neurons (29, 32, 33). The striking dendritic plasticity observed after antennal removal (Fig. S6) suggests that MB-V3 neurons are integrally involved in processing information from antennal sensory inputs. A previous model suggests that MB-V3 neurons act as odor generalists that receive synapses from all three types of α/β Kenyon cells (Fig. S2) and respond to all tested odors (Fig. 2C). Conditioning to specific odors then induces increased neural activity and de novo protein synthesis (Fig. 1), rendering MB-V3s as “conditioned response neurons” that anticipate the US (30).
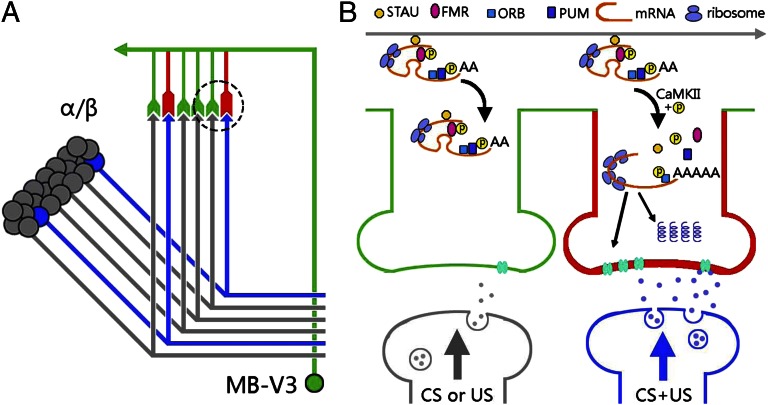
A wealth of evidence suggests that aversive olfactory memory exists as a persistent neural activity (“trace”) in a sparse subset of MB neurons—initially in the γ lobes lasting for several minutes and then spreading to α/β lobes for several hours (34–36). This physiological trace drives a protein-synthesis–dependent process in postsynaptic MB-V3 neurons, yielding structural change at MB::MB-V3 synapses (Fig. 4A). Recent advances in several animal models have suggested two distinct components of synaptic capture that stabilize long-term functional and structural changes at specific synapse: one requires CREB-regulated transcription and NMDA receptor/PKA activities, whereas the other requires CPEB-regulated translation and local protein synthesis (3). Our results suggest that the latter molecular mechanism might occur in MB-V3 neurons (Fig. 4B). LTM was normal after disruption of CREB2 or NMDA but was impaired after down-regulation of ORB (a CPEB variant that lacks the “prion domain”) and several RNA-binding proteins (RBPs), including FMR, STAUFEN, and PUMULIO that suppress and stabilize mRNA distributed among synapses (Fig. 3). LTM also was impaired by down-regulation of CaMKII (Fig. 3G), which contains in its 3′ UTR cis-acting localization elements for RBPs and prevents their degradation and transportation (2, 10, 27, 37, 38). Consequently, local protein synthesis at the MB-V3 dendrites is capable of providing a mechanism permitting rapid synaptic changes in response to presynaptic CS/US signals. Interestingly, despite the role for ORB2 (a CPEB variant that contains the prion domain) in long-term courtship memory (8, 9), our results showed that down-regulation of ORB2 in MB-V3 neurons did not affect olfactory LTM (Fig. 3D). These observations imply that different CPEB proteins (i.e., ORB and ORB2) may function in different neurons underlying different forms of long-term memory, although our negative results cannot completely exclude ORB2 function in MB-V3 neurons.
Fig. 4.
A model of local mRNA translation in Drosophila olfactory memory. (A) Schematic representation of memory transformation from physiological coding in sparse α/β Kenyon cells (blue) to structural coding (red) in preexisting postsynaptic components of a MB-V3 neuron. (B) At activated MB-V3 synapses (red), presynaptic CS+US signals (blue), but not CS alone or US alone (gray), an CREB-independent mechanism relieves the translational block of RNA granules, which involves (i) STAUFEN detachment (38, 41), (ii) FMR and PUMILIO dephosphorylations (38, 42, 43), and (iii) ORB phosphorylation by activated CaMKII, leading to the poly-A tail elongation (4–6). Consequently, these events recruit additional ribosomes to up-regulate mRNA translation of LTM-related proteins (42), which eventually lead to an enhanced response to the CS+. Note that the subcellular locations of proteins and their cofunctions in the model have not been experimentally determined.
Our results suggest that spaced training induces coincident neural activity between subsets of MB axons and MB-V3 dendrites. This coincident synaptic activity disinhibits translation of nascent mRNAs, presumably contained in neural granules along with ORB, FMR, PUMILIO, and STAUFEN, in a CaMKII-dependent fashion (38). A new memory trace then emerges from the functional and structural changes induced at these coincident MB::MB-V3 synapses. Thus, a single MB-V3 neuron is capable of performing selective synaptic plasticity similar to the glomerulus-specific plasticity of a single olfactory local neuron in the antenna lobe (39). Our model also predicts that a different subset of MB-V3 dendrites will be used to encode conditioned responses for different odors. Several obvious gaps remain: (i) Are these translation-related molecules located in postsynaptic compartment in MB-V3 neurons, and do they function together during LTM formation? (ii) What effector proteins are synthesized during LTM consolidation? (iii) Are they causally related to the observed increase in neural activity in MB-V3 neurons during LTM retrieval? Elucidation in MB-V3 neurons of this differential synaptic coding—and the neurotransmitters involved—promises to reveal further how synaptic plasticity encodes in memory consolidation and retrieval at a systems level (Fig. 2).
By identifying specific neuronal types involved in olfactory long-term memory formation in Drosophila, we have begun to deconvolute two molecular mechanisms, regulation of transcription and regulation of translation, both of which have been implicated in several model systems. Previously, we have shown that CREB-dependent transcription is involved with LTM formation in DAL neurons, whereas here we show instead that ORB-dependent translation is involved with LTM formation in MB-V3 neurons. Thus, two molecular mechanisms of protein synthesis may be partitioned into different neurons outside of MB but nonetheless within a common memory circuit.
MB-V3 and DAL neurons also appear to play different roles at the systems level. The former encodes odor specificity among the synapses connecting to MB (Fig. 4), whereas the latter is recruited to the memory circuit presumably to strengthen a conditioned response. Interestingly, this notion predicts that LTM is likely to be enhanced by activation of DAL but not MB-V3 neurons. It remains to be addressed whether MB-V3 neurons connect directly or indirectly to DAL neurons (15), which then might serve to form a neural circuit loop with MB during LTM retrieval. Alternatively, MB-V3 neurons may modulate presynaptic MB activity directly through retrograde feedback, as in the Aplysia model system (40).
Methods
Fly Strains and Behavior.
Fly stocks were maintained on standard corn meal/yeast/agar medium at 25 ± 1 °C or 18 ± 1 °C and 70% relative humidity on a 12h:12h light:dark cycle. Memory behaviors were performed as previously described (15). For detailed fly strains, please see SI Methods.
In Vivo GCaMP Imaging.
In vivo GCaMP imaging is as previously described in Wang et al. (1). Please see SI Methods for more details.
Statistics.
Raw data were analyzed with SigmaPlot 10.0 and SigmaStat 3.5. The data were evaluated via one-way ANOVAs followed by planned comparisons among the relevant groups with a Tukey’s honestly significant difference test. The P values were evaluated via one-way ANOVAs.
Supplementary Material
Acknowledgments
We thank T. Lee, D. Reiff, S. Benzer, C. J. O’Kane, K. Scott, L. Davis, S. Kunes, K. Si, the Bloomington Drosophila Stock Center, the Drosophila Genomics Resource Center, the Vienna Drosophila RNAi Center, and the National Institute of Genetics for fly stocks. This work was funded by grants from Dart Neurosciences, LLC, the National Science Council, and the Ministry of Education (to A.-S.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216336110/-/DCSupplemental.
References
- 1.Wang Y, Mamiya A, Chiang AS, Zhong Y. Imaging of an early memory trace in the Drosophila mushroom body. J Neurosci. 2008;28(17):4368–4376. doi: 10.1523/JNEUROSCI.2958-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loya CM, Van Vactor D, Fulga TA. Understanding neuronal connectivity through the post-transcriptional toolkit. Genes Dev. 2010;24(7):625–635. doi: 10.1101/gad.1907710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kandel ER. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol Brain. 2012;5(1):14. doi: 10.1186/1756-6606-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16(1):102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140(3):421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Atkins CM, Nozaki N, Shigeri Y, Soderling TR. Cytoplasmic polyadenylation element binding protein-dependent protein synthesis is regulated by calcium/calmodulin-dependent protein kinase II. J Neurosci. 2004;24(22):5193–5201. doi: 10.1523/JNEUROSCI.0854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alarcon JM, et al. Selective modulation of some forms of schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn Mem. 2004;11(3):318–327. doi: 10.1101/lm.72704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keleman K, Krüttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 2007;10(12):1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- 9.Majumdar A, et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148(3):515–529. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124(1):191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 11.Pascual A, Préat T. Localization of long-term memory within the Drosophila mushroom body. Science. 2001;294(5544):1115–1117. doi: 10.1126/science.1064200. [DOI] [PubMed] [Google Scholar]
- 12.Yu D, Akalal DB, Davis RL. Drosophila alpha/beta mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron. 2006;52(5):845–855. doi: 10.1016/j.neuron.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis RL. Traces of Drosophila memory. Neuron. 2011;70(1):8–19. doi: 10.1016/j.neuron.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu CL, et al. Specific requirement of NMDA receptors for long-term memory consolidation in Drosophila ellipsoid body. Nat Neurosci. 2007;10(12):1578–1586. doi: 10.1038/nn2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CC, et al. Visualizing long-term memory formation in two neurons of the Drosophila brain. Science. 2012;335(6069):678–685. doi: 10.1126/science.1212735. [DOI] [PubMed] [Google Scholar]
- 16.Lin HH, Lai JS, Chin AL, Chen YC, Chiang AS. A map of olfactory representation in the Drosophila mushroom body. Cell. 2007;128(6):1205–1217. doi: 10.1016/j.cell.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka NK, Tanimoto H, Ito K. Neuronal assemblies of the Drosophila mushroom body. J Comp Neurol. 2008;508(5):711–755. doi: 10.1002/cne.21692. [DOI] [PubMed] [Google Scholar]
- 18.Feinberg EH, et al. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57(3):353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 19.Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47(2):81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 20.Huang C, et al. A permissive role of mushroom body α/β core neurons in long-term memory consolidation in Drosophila. Curr Biol. 2012;22(21):1981–1989. doi: 10.1016/j.cub.2012.08.048. [DOI] [PubMed] [Google Scholar]
- 21.Reiff DF, et al. In vivo performance of genetically encoded indicators of neural activity in flies. J Neurosci. 2005;25(19):4766–4778. doi: 10.1523/JNEUROSCI.4900-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey CH, Chen M. Long-term memory in Aplysia modulates the total number of varicosities of single identified sensory neurons. Proc Natl Acad Sci USA. 1988;85(7):2373–2377. doi: 10.1073/pnas.85.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glanzman DL, Kandel ER, Schacher S. Target-dependent structural changes accompanying long-term synaptic facilitation in Aplysia neurons. Science. 1990;249(4970):799–802. doi: 10.1126/science.2389145. [DOI] [PubMed] [Google Scholar]
- 24.Yang G, Pan F, Gan WB. Stably maintained dendritic spines are associated with lifelong memories. Nature. 2009;462(7275):920–924. doi: 10.1038/nature08577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts TF, Tschida KA, Klein ME, Mooney R. Rapid spine stabilization and synaptic enhancement at the onset of behavioural learning. Nature. 2010;463(7283):948–952. doi: 10.1038/nature08759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akalal DB, Yu D, Davis RL. A late-phase, long-term memory trace forms in the γ neurons of Drosophila mushroom bodies after olfactory classical conditioning. J Neurosci. 2010;30(49):16699–16708. doi: 10.1523/JNEUROSCI.1882-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubnau J, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13(4):286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 28.Bolduc FV, Bell K, Cox H, Broadie KS, Tully T. Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat Neurosci. 2008;11(10):1143–1145. doi: 10.1038/nn.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aso Y, et al. Specific dopaminergic neurons for the formation of labile aversive memory. Curr Biol. 2010;20(16):1445–1451. doi: 10.1016/j.cub.2010.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heisenberg M. Mushroom body memoir: From maps to models. Nat Rev Neurosci. 2003;4(4):266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 31.Honegger KS, Campbell RA, Turner GC. Cellular-resolution population imaging reveals robust sparse coding in the Drosophila mushroom body. J Neurosci. 2011;31(33):11772–11785. doi: 10.1523/JNEUROSCI.1099-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claridge-Chang A, et al. Writing memories with light-addressable reinforcement circuitry. Cell. 2009;139(2):405–415. doi: 10.1016/j.cell.2009.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krashes MJ, et al. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell. 2009;139(2):416–427. doi: 10.1016/j.cell.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blum AL, Li W, Cressy M, Dubnau J. Short- and long-term memory in Drosophila require cAMP signaling in distinct neuron types. Curr Biol. 2009;19(16):1341–1350. doi: 10.1016/j.cub.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee PT, et al. Serotonin-mushroom body circuit modulating the formation of anesthesia-resistant memory in Drosophila. Proc Natl Acad Sci USA. 2011;108(33):13794–13799. doi: 10.1073/pnas.1019483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin H, et al. Gamma neurons mediate dopaminergic input during aversive olfactory memory formation in Drosophila. Curr Biol. 2012;22(7):608–614. doi: 10.1016/j.cub.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richter JD, Lorenz LJ. Selective translation of mRNAs at synapses. Curr Opin Neurobiol. 2002;12(3):300–304. doi: 10.1016/s0959-4388(02)00318-5. [DOI] [PubMed] [Google Scholar]
- 38.Doyle M, Kiebler MA. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 2011;30(17):3540–3552. doi: 10.1038/emboj.2011.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCann C, et al. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc Natl Acad Sci USA. 2011;108(36):E655–E662. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai D, Chen S, Glanzman DL. Postsynaptic regulation of long-term facilitation in Aplysia. Curr Biol. 2008;18(12):920–925. doi: 10.1016/j.cub.2008.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miki T, Takano K, Yoneda Y. The role of mammalian Staufen on mRNA traffic: A view from its nucleocytoplasmic shuttling function. Cell Struct Funct. 2005;30(2):51–56. doi: 10.1247/csf.30.51. [DOI] [PubMed] [Google Scholar]
- 42.Bassell GJ, Warren ST. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60(2):201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller MA, Olivas WM. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip Rev RNA. 2011;2(4):471–492. doi: 10.1002/wrna.69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.