Abstract
The current limitations in genome sequencing technology require the construction of physical maps for high-quality draft sequences of large plant genomes, such as that of Aegilops tauschii, the wheat D-genome progenitor. To construct a physical map of the Ae. tauschii genome, we fingerprinted 461,706 bacterial artificial chromosome clones, assembled contigs, designed a 10K Ae. tauschii Infinium SNP array, constructed a 7,185-marker genetic map, and anchored on the map contigs totaling 4.03 Gb. Using whole genome shotgun reads, we extended the SNP marker sequences and found 17,093 genes and gene fragments. We showed that collinearity of the Ae. tauschii genes with Brachypodium distachyon, rice, and sorghum decreased with phylogenetic distance and that structural genome evolution rates have been high across all investigated lineages in subfamily Pooideae, including that of Brachypodieae. We obtained additional information about the evolution of the seven Triticeae chromosomes from 12 ancestral chromosomes and uncovered a pattern of centromere inactivation accompanying nested chromosome insertions in grasses. We showed that the density of noncollinear genes along the Ae. tauschii chromosomes positively correlates with recombination rates, suggested a cause, and showed that new genes, exemplified by disease resistance genes, are preferentially located in high-recombination chromosome regions.
Keywords: single nucleotide polymorphism, synteny, gene density, Oryza, BAC contig coassembly
Many plants have large genomes with vast amounts of repeated DNA. An example is Aegilops tauschii, the diploid progenitor of the D genome of hexaploid wheat (Triticum aestivum). The estimates of its genome size range from 4.02 (1) to 4.98 Gb (2), and 90% of its genome was estimated to be repetitive DNA (3). The Ae. tauschii genome and the D genome of hexaploid wheat are closely related due to the recent origin of hexaploid wheat (4). Ae. tauschii is therefore an important resource for wheat breeding, and its genome is an invaluable reference for wheat genomics, as illustrated by the utility of its sequences in the analysis of the wheat gene space (5). The utility of Ae. tauschii for wheat genetics and genomics would be further enhanced by a high-quality draft sequence of its genome. With current technology, the only approach to produce a high-quality de novo draft sequence for a genome of this size and complexity is the ordered-clone sequencing approach, which requires a physical map.
Physical map construction necessitates fingerprinting multiple genome equivalents of bacterial artificial chromosome (BAC) clones, assembling them into contigs, and anchoring the contigs on a genetic map (6–8). Great strides have been made in BAC fingerprinting techniques (7, 9–12) and software for fingerprint editing and contig assembly (13–16). It is now possible with these technological advances to fingerprint and assemble contigs from hundreds of thousands of BAC clones (7, 8, 17–19). In contrast, contig anchoring remains a weakness in physical mapping of large plant genomes because of their low gene density, extensive gene duplication, and abundance of repetitive DNA. BAC end sequences (BESs) are an effective means of contig anchoring in small genomes (11). In large genomes, however, hundreds of thousands of BESs are needed. DNA hybridization and PCR-based anchoring (6, 7, 20, 21) is laborious and often produces equivocal results. Contig anchoring with highly multiplexed Illumina GoldenGate SNP assays overcomes some of these limitations (22), but its relatively high cost limits the number of markers that can be used.
Here we report the construction of a physical map of the Ae. tauschii genome using the SNaPshot BAC fingerprinting technology (9, 11) and the Illumina Infinium SNP array technology in contig anchoring. To make the physical map comparative, we use SNPs in sequences greatly enriched for genes (23) in designing the array and then extend the mapped SNP markers with whole genome shotgun (WGS) reads. We use the Ae. tauschii comparative map to reassess collinearity between the Ae. tauschii genome and the Brachypodium distachyon, rice (Oryza sativa), and sorghum (Sorghum bicolor) genomes and report insights into several aspects of grass genome evolution.
Results
Physical Map Construction.
We fingerprinted 461,706 BAC clones (Table S1) of Ae. tauschii accession AL8/78, edited them, and assembled 399,448 clones into 3,153 BAC contigs with Fingerprinted Contig (FPC) software (14) (SI Text; Table 1); 15,683 clones remained as singletons. To construct a genetic map for contig anchoring, we built a 10K Infinium iSelect SNP oligonucleotide array (SI Text) using 10,000 SNPs selected from 195,631 SNPs between Ae. tauschii accessions AL8/78 and AS75, the parents of our F2 mapping population, mostly located in genes (23). We genotyped 1,102 F2 plants of a single biparental mapping population (Fig. S1) and constructed a 7,185-marker genetic map (Table 2; SI Text; http://probes.pw.usda.gov/WheatDMarker/downloads/ComparativeMapData.xls).
Table 1.
Statistics of FPC contig assembly and editing (disjoining of chimeric contigs)
| Contigs | No. of contigs | Total length (Mb) | Average length (kb) | N50 (kb) |
| After FPC assembly | 3,153 | 4,756 | 1,509 | 2,665 |
| After editing | 3,578 | 4,792 | 1,339 | 2,092 |
| Anchored contigs | 2,263 | 4,030 | 1,778 | 2,385 |
| Unanchored contigs | 1,315 | 757 | 578 | 945 |
Table 2.
Maps of the Ae. tauschii chromosomes
| Chromosome | Genetic map |
Physical map |
|||
| No. of markers | Length (cM) | No. of markers | No. of contigs anchored | Length (Mb) | |
| 1D | 973 | 175.6 | 943 | 295 | 520 |
| 2D | 1,326 | 235.1 | 1,282 | 385 | 672 |
| 3D | 1,101 | 204.1 | 1,062 | 356 | 633 |
| 4D | 821 | 143.9 | 799 | 267 | 520 |
| 5D | 1,034 | 215.4 | 1,001 | 319 | 577 |
| 6D | 771 | 172.2 | 746 | 267 | 466 |
| 7D | 1,159 | 228.1 | 1,159 | 374 | 642 |
| Total | 7,185 | 1,374.4 | 6,992 | 2,263 | 4,030 |
To anchor BAC contigs, we constructed five-dimensional (5-D) BAC pools (22) (SI Text). To avoid nonspecific DNA amplification in negative pools, we added AS75 genomic DNA to all pools. The Infinium assays failed to produce clear-cut clustering of positive and negative genotypes in the GenomeStudio graphs (Fig. S1), which we overcame as described in SI Text. Genotyping data for 356 (4.96%) low-deconvolution confidence markers were deconvoluted manually and were either confirmed or eliminated from the physical map. Ultimately, we assigned 6,992 (97.3%) of the 7,185 SNP markers to contigs.
We then examined contigs for false joins by using (i) contig anchoring on the genetic map (http://probes.pw.usda.gov/WheatDMarker/downloads/ComparativeMapData.xls), (ii) analyses of FPC consensus band (CB) maps (Fig. S2), and (iii) coassembly of contigs using Ae. tauschii BAC clones and clones of wheat D-genome subgenomic BAC libraries (24) (Fig. S3; SI Text). The contig coassembly was made possible by the close phylogenetic proximity of the Ae. tauschii genome to the wheat D genome.
We disjoined 425 (13.5%) chimeric contigs and obtained a 4,030-Mb physical map (Fig. 1) consisting of 2,263 anchored contigs (Tables 1 and 2). Contig disjoining increased the number of contigs and decreased their average and N50 lengths (Table 1).
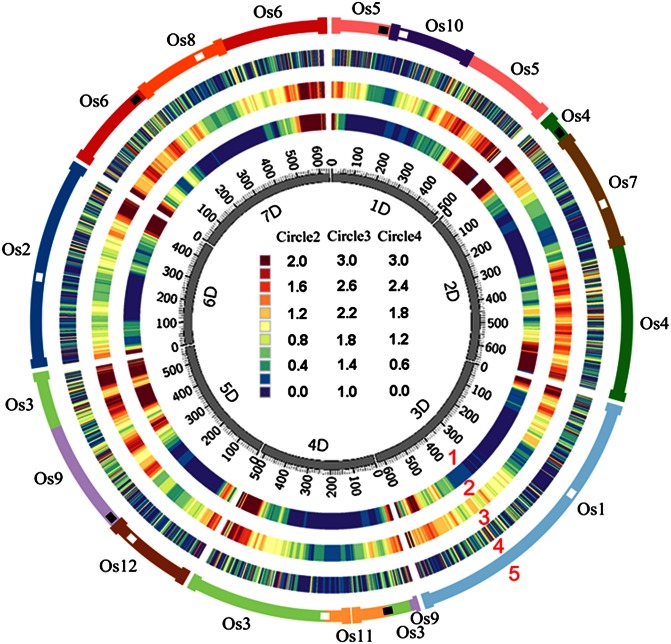
Fig. 1.
Ae. tauschii genome circle maps. The inner circle (#1) contains the physical maps of the Ae. tauschii chromosomes each with its short arm tip at 0 Mb. Circle #2 contains heat maps of recombination rates, circle #3 contains gene density heat maps, circle #4 contains heat maps of only genes collinear with the B. distachyon, rice, or sorghum pseudomolecules, and circle #5 shows global synteny with the rice chromosomes symbolizing 12 ancestral chromosomes. In circle #5, the active Ae. tauschii centromeres are white, inferred extinct centromeres are black, and the locations of current and inferred ancient telomeres are diagrammed by thick bars. Thirty-megabase windows sliding by 1 Mb were used in the heat map construction. The heat map units for circle #2 are cM/Mb and for circles #3 and #4 are numbers of genes and gene fragments discovered in the extended sequences per megabase of the physical maps.
The 4,030-Mb physical map (http://probes.pw.usda.gov/WheatDMarker/) represented 84.2% of 4,792 Mb, the total length of the 3,578 BAC contigs (Table 1). The total contig length was within the range of published Ae. tauschii genome size estimates. However, 4,792 Mb must be an overestimate because undetected overlaps between contigs were necessarily counted twice. The remaining 15.8% of the total length were unanchored, mostly short contigs of average length = 578 kb (Table 1). The minimal tiling path (MTP) across all anchored and unanchored contigs consisted of 42,822 clones.
Marker Sequence Extension.
We extended the sequences of the 7,185 mapped SNP markers with 3.1× and 50× genome equivalent of Roche GS FLX Titanium and Illumina HiSeq WGS reads, respectively (SI Text). The sequence associated with each marker was on average extended to 7,869 bp with an N50 of 10,830 bp (Table 3).
Table 3.
Gene prediction in extended sequences anchored on the physical map
| Category | Measure |
| Extended sequence contigs (number) | 7,185 |
| Mean extended length (bp) | 7,869 |
| N50 (bp) | 10,830 |
| Genes/gene fragments (number) | 17,093 |
| Complete genes (number) | 9,716 |
| Genes aligned to ESTs (percentage) | 84.0 |
| Genes aligned to proteins (percentage) | 95.4 |
| Averaged gene length (bp) | 2,772 |
| Average exon number per gene | 5.5 |
| Median exon length (bp) | 245 |
| Average exon length (bp) | 266 |
| Median intron length (bp) | 151 |
| Average intron length (bp) | 156 |
| Gene fragments (number) | 7,377 |
To assess the accuracy of sequence extension, we aligned extended sequences to 37 homologous AL8/78 BAC clone sequences in the National Center for Biotechnology Information (NCBI) database. Of the 37 extended sequences, the alignment of 22 and 27 exceeded 99% and 95% of the sequence length, respectively, and the alignment of all sequences exceeded 81% sequence length. A reduced alignment length of two sequences was caused by gaps in the BAC sequence scaffolds. Nucleotide identity, except for one BAC clone, exceeded 98% of aligned nucleotides.
The 7,185 extended sequences (http://probes.pw.usda.gov/WheatDMarker/) contained 17,093 genes and gene fragments (http://probes.pw.usda.gov/WheatDMarker/downloads/GeneList.xls). Of these, 9,716 (56.8%) were complete genes (defined in SI Text), and 7,377 (43.2%) were gene fragments based on alignment with wheat and barley expressed sequence tags (ESTs) (4,866) or, in the absence of EST evidence, with proteins (2,511).
Chromosome Structure.
We previously hypothesized that the seven Ae. tauschii chromosomes originated from 12 ancestral chromosomes by five nested chromosome insertions (NCIs) (25) (Fig. 1). Our data showed that during the NCIs that produced Ae. tauschii chromosomes 1D, 2D, 5D, and 7D, a telomere of the inserted chromosome was inserted near the centromere, in a gene-containing region (Fig. 1). That centromere was lost, and the centromere of the inserted chromosome became the active centromere in each compound chromosome (Fig. 1).
Chromosome 5D originated by NCI of a chromosome corresponding to Os12 near the centromere of Os9, but we did not find evidence of the expected homology between the 5D short arm and the 1-Mb tip of the short arm of Os9. However, the small size of the region may have precluded its detection. In addition to this putative NCI, chromosome 5D acquired a segment corresponding to a distal portion of the short arm of Os3, which was attached to a segment corresponding to the long arm of Os9, making up the arm 5DL (Fig. 1). We detected the reciprocal product of this translocation. The tip of the long arm of Os9 forms the tip of the 4D short arm; hence, the translocation between chromosomes corresponding to Os3 and Os9 was reciprocal.
Rate of Genome Evolution.
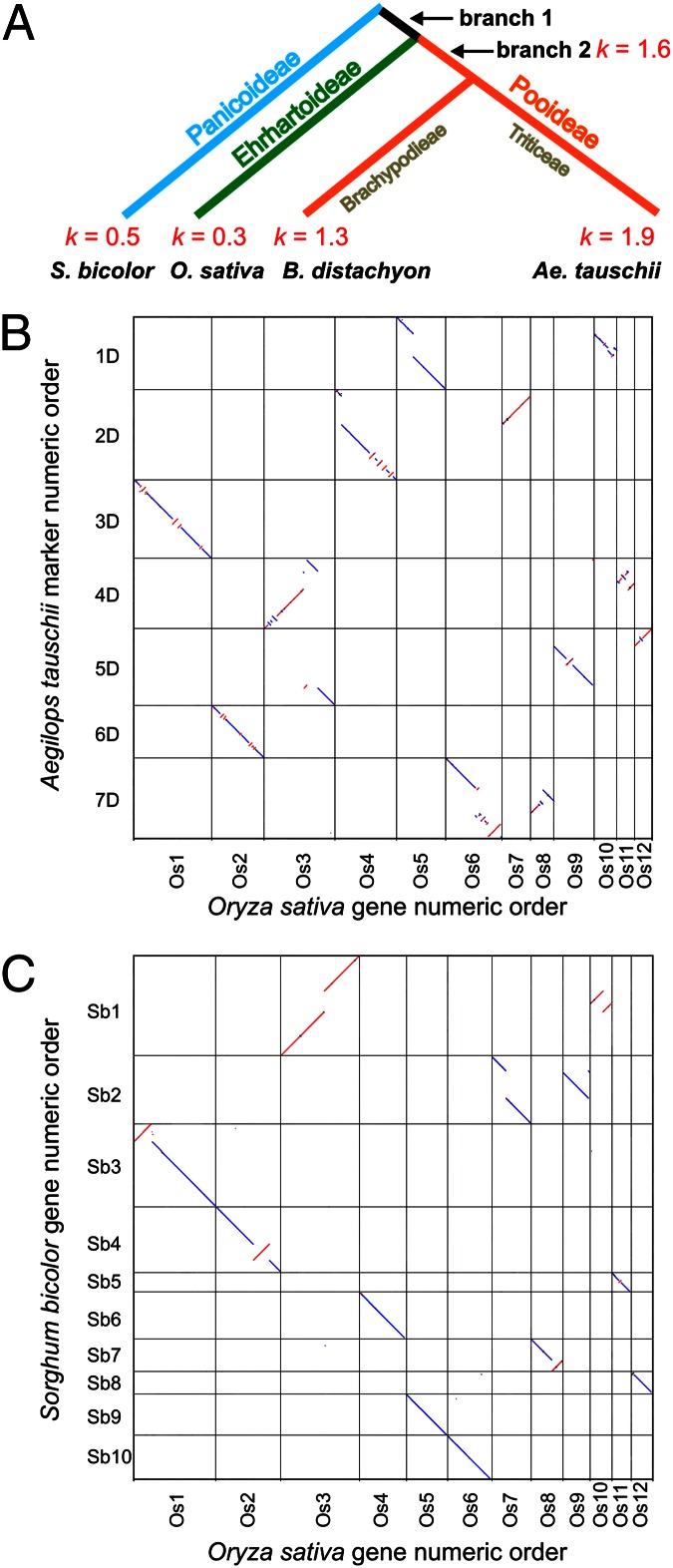
We assigned inversions, translocations, and NCIs (http://probes.pw.usda.gov/WheatDMarker/downloads/ComparativeMapData.xls) to each branch of a grass phylogenetic tree (Fig. 2A) as described earlier (25). Using divergence time estimates (26), we estimated the rates (k) of structural genome evolution (Fig. 2A). We confirmed the highest rate of genome evolution in the Ae. tauschii branch (67 inversions, translocations, and NCIs) and the slowest rates in the rice and sorghum branches (respectively 16 and 25 inversions, translocations, and NCIs). The rate was also high in the B. distachyon branch (48 inversions, translocations, and NCIs) and internal branch 2 (18 inversions and translocations), showing that all investigated branches of subfamily Pooideae had elevated rates of genome evolution compared with those of Panicoideae and Ehrhartoideae (P = 0.01). The fast rate of genome evolution in Ae. tauschii is illustrated by dot plots (see SI Text for methodology). The Ae. tauschii–rice dot-plot shows more chromosome rearrangements than the sorghum–rice dot plot (Fig. 2 B and C). The Ae. tauschii–B. distachyon dot plot (Fig. S4A) shows the largest number of rearrangements illustrating the fast rates of genome evolution in both lineages.
Fig. 2.
Rates of structural genome evolution in the grass family. (A) Rates (k; numbers of inversions, translocations and NCIs per million years) during the evolution of grass subfamilies Panicoideae, Ehrhartoideae, and Pooideae. (B and C) Pairwise dot-plot comparisons of gene order along Ae. tauschii physical maps (B) and along sorghum pseudomolecules (C) relative to that along the rice pseudomolecules. Chromosomes and pseudomolecules on the y axis have the tips of the short arms at 0 Mb, respectively, at the top. Blue dots indicate parallel order and red dots indicate antiparallel order of chromosomes and pseudomolecules on the x and y axes. Only orthologous loci are shown for the sake of clarity.
The orthologous and paralogous relationships of the Ae. tauschii chromosomes relative to rice pseudomolecules are shown by a dot plot in Fig. S4B. The paralogous relationships reflect the pan-grass whole-genome duplication (27).
Recombination Rates and Gene Density.
The average recombination rate was 0.32 cM/Mb and ranged from nearly zero in the proximal chromosome regions to about 1.5–2.0 cM/Mb in the distal regions (Fig. S5). Recombination rates dropped more precipitously in the short arms than in the long arms (Fig. 1; Fig. S5).
Genes were distributed across the entire spans of the physical maps of the Ae. tauschii chromosomes including the low-recombination pericentromeric regions (Fig. 1; Fig. S5). Gene density computed for nonoverlapping 30-Mb intervals along Ae. tauschii chromosomes based on the distribution of 17,093 genes and gene fragments fluctuated two- to threefold (Fig. S5). Gene density was correlated with recombination rate (r = 0.53, P < 0.0001). An insertion of a telomeric region into the centromeric region during NCI juxtaposes a high-gene-density terminal region and a low-gene-density centromeric region in the nascent compound chromosome. These gene-density juxtapositions have been observed in the B. distachyon genome (26). They appear to be absent from the Ae. tauschii genome (Fig. 1; Fig. S5).
Noncollinear Genes.
We selected among the extended sequences of 7,185 SNP markers a nonredundant set of 5,901 Ae. tauschii genes or gene fragments with one or more homologs in at least one of the B. distachyon, rice, and sorghum genomes. If there were more than one homolog in a compared genome, we always considered only the homolog with the lowest E value. We then counted the numbers of homologs in collinear locations in the B. distachyon, rice, and sorghum genomes and used their percentage as a measure of collinearity of genes between the Ae. tauschii genome and the three grass genomes (Table 4). Collinearity of Ae. tauschii genes with B. distachyon, rice, and sorghum genes was proportional to the phylogenetic distance of compared genomes (Table 4).
Table 4.
Nonredundant Ae. tauschii complete genes and gene fragments in collinear positions relative to homologous genes in B. distachyon, rice, and sorghum
| Genome | Total (no.) | Collinear (no.) | Percentage |
| B. distachyon | 5,901* | 3,624‡ | 61.4 |
| 5,272† | 3,523§ | 66.6 | |
| Rice | 5,901* | 3,209‡ | 54.4 |
| 5,272† | 3,136§ | 59.5 | |
| Sorghum | 5,901* | 3,008‡ | 51.0 |
| 5,272† | 2,942§ | 55.4 |
Complete genes plus gene fragments.
Complete genes only.
Estimates followed by the same footnote symbol differ significantly from each other (χ2 test with Yates correction, P < 0.001).
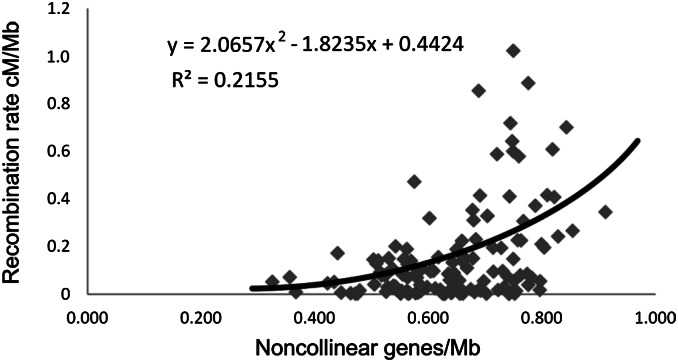
Of the 5,901 Ae. tauschii genes, 1,540 (26.1%) were complete genes having homologs in collinear locations in none of the three grass genomes (Table S2) and were therefore most likely genes transposed or translocated to new locations in the Ae. tauschii lineage after its divergence from the B. distachyon lineage. The following indicated that these genes were preferentially located in the distal, high-recombination regions of the Ae. tauschii chromosomes. The elevated gene density in the distal regions of the Ae. tauschii chromosomes (circle #3 in Fig. 1) largely disappeared when we excluded the noncollinear genes from the physical map (circle #4 in Fig. 1). Dot plots of individual Ae. tauschii chromosomes compared with the 12 rice chromosomes showed dense clouds of rice homologs in noncollinear locations in the distal, high-recombination regions of each of the Ae. tauschii chromosomes (Fig. S6). Finally, the density of Ae. tauschii noncollinear genes per megabase correlated with recombination rate (r = 0.464, P < 0.0001; Fig. 3).
Fig. 3.
Relationship between recombination rate and density of noncollinear genes in the Aegilops tauschii genome.
Recombination and Location of Actively Evolving Genes.
We selected the average recombination rate as an arbitrary boundary to divide each Ae. tauschii chromosome arm into high- and low-recombination regions. We then selected 4,134 Ae. tauschii complete genes in 23 gene ontology (GO) categories and allocated them into four groups: collinear high recombination, collinear low recombination, noncollinear high recombination, and noncollinear low recombination (Table S3). The distribution of collinear genes in each of the 23 GO categories with respect to high- and low-recombination regions did not significantly differ (2 × 2 contingency tables). However, the distribution of noncollinear genes in GO category “receptor activity” (GO: 0004872) was significantly (P = 0.002) overrepresented in the high-recombination group (Table S3). This overrepresentation pattern was also observed for Ae. tauschii disease resistance genes, which were identified among the 4,134 genes by homology search against the plant disease resistance (R) gene database (28) (Table 5) (noncollinear R genes and their GOs are listed in http://probes.pw.usda.gov/WheatDMarker/downloads/RGeneGOClass.xls). This overrepresentation is typified by the distribution of NB-LRR disease resistance protein genes (NB-ARC domain-containing proteins), which accounted for 4.7% of the noncollinear genes in the high-recombination regions but only for 0.7% of noncollinear genes in the low-recombination region.
Table 5.
Numbers of Ae. tauschii genes homologous to plant disease resistance genes in four indicated groups
| Class | Recom-bination rate | Genes | Genes homologous to R genes | Percent of genes in the class |
| Collinear | High | 1,202 | 56 | 3.7 |
| Collinear | Low | 1,930 | 94 | 3.8 |
| Noncollinear | High | 620 | 43 | 4.7 |
| Noncollinear | Low | 382 | 8 | 1.4 |
| Total | 4,134 | 201 |
P < 0.0001 between noncollinear classes (Fisher exact test). P = 0.049 between collinear and noncollinear classes in high recombination regions (Fisher exact test).
Discussion
Physical Mapping.
The most significant technical advance we report here is contig anchoring with an Infinium SNP array. Prior physical mapping of other medium size or large grass genomes used multiple marker systems, some of them requiring intensive laboratory work, to anchor contigs (7, 8, 21). Contig anchoring with an Infinium SNP array opened the door to the use of essentially unlimited numbers of SNP assays in contig anchoring with minimal labor beyond that needed for the construction of 5-D BAC pools. Another important technical advance, which greatly reduced the ambiguity of contig anchoring, was a computer script that used the locations of positive BAC clones in contigs to discriminate between true-positive and false-positive BAC clones during the deconvolution of the 5-D BAC pools (29). These two advances made it possible to include unequivocally 6,992 of the 7,185 markers (97.4%) on the genetic map into BAC contigs for contig anchoring. The construction of the physical map also benefitted from the use of an Ae. tauschii-wheat D-genome contig coassembly as one of the criteria for detecting Ae. tauschii chimeric contigs.
The results of our contig assembly and contig anchoring compare favorably with other Triticeae physical mapping endeavors. We assembled 399,448 edited clones into 3,578 edited contigs with an N50 contig length of 2,092 kb. During the physical mapping of wheat chromosome 3B, 56,952 fingerprinted clones were assembled into 1,036 edited contigs of an average size of 783 kb (21). The likely reasons for the relatively greater number of contigs and shorter average contig length in physical mapping of chromosome 3B were the use of fewer BAC libraries, a shorter average insert length, and a smaller number of restriction fragments in fingerprints caused by the use of shorter size standard during capillary electrophoresis (LIZ500 rather than LIZ1200). During the physical mapping of the barley genome, 517,202 edited BAC clones were assembled into 9,265 contigs with a N50 contig length of 904 kb (8). Because of the greater number of clones, the barley assembly should have generated a smaller number of contigs with greater N50 contig length than our assembly. However, the reverse was obtained. Because the two assemblies used similar stepwise strategies and similar stringencies, the reasons for the different outcomes likely lie in some undetermined technical factor.
Gene Sequences.
We encountered 17,093 gene and gene fragment sequences in the extended sequences of the 7,185 SNP markers, which is seemingly incongruous with genes representing as little as 2.5% of the Ae. tauschii genome (3). We attribute this apparent contradiction to two factors: most of the 7,185 SNP markers were already located in genes (23) and clustering of Ae. tauschii genes (30).
The structural characteristics of the 9,716 complete genes we found in the extended sequences, such as the average number of exons per gene (5.5), the average exon length (245 bp), and the average gene length (2.8 kb), were similar to those reported for B. distachyon genes (5.5, 268 bp, and 2.6 kb, respectively) and rice genes (4.8, 364 bp, and 2.5 kb, respectively) (26). For unknown reasons, our data agree less closely with those reported for barley (7.6, 454 bp, and 3.0 kb, respectively) (8).
Centromere Fate During NCI.
In each Ae. tauschii chromosome that originated by NCI whereby a telomere of the inserted chromosome was inserted into the vicinity of the centromere of the recipient chromosome, the centromere of the recipient chromosome became extinct, whereas the centromere of the inserted chromosome remained active. We observed the same pattern in the seven NCIs in the B. distachyon genome (26) and two NCIs in the sorghum genome (25). Because the centromere of the recipient chromosome has a telomeric region in its neighborhood, we speculate that such NCIs impair centromere functioning and generate functionally monocentric nascent chromosomes. A corollary is that NCIs taking place far away from the centromere may generate functionally dicentric chromosomes that are lost, which could explain why all NCIs observed to date in grasses took place in the vicinity of the centromere.
Genome Evolution Rate and Genome Size.
A previous study (25) that did not include B. distachyon suggested a possible relationship between a large genome size and a fast rate of genome evolution among grasses. Here we show that genomes in all investigated branches of the subfamily Pooideae have been evolving fast, including the Brachypodieae branch. Because B. distachyon has a small genome (26), the fast rate of evolution of its genome contradicts the previous study. Genera Melica, Glyceria, Nardus, and Lygeum that diverged from the Brachypodieae lineage before the divergence of the Brachypodieae and Triticeae lineages (31) have 1C genome sizes >1.5 Gb (The C-value database, Kew Royal Botanical Garden, http://data.kew.org/cvalues/). We therefore speculate that the B. distachyon genome evolved from a larger ancestral genome by size contraction to account for the relatively fast rate of Brachypodieae genome evolution.
Noncollinear Genes and Evolution of New Genes.
Transposition or translocation of a gene into a new location in a genome creates polymorphism for a paralogous gene pair. If both paralogues are retained and are not in tandem they appear as dispersed duplicated genes (32). The ancestral gene in the paralogous pair is in a collinear location relative to related genomes, whereas the derived gene is in a noncollinear location. Ae. tauschii collinear genes were distributed more-or-less uniformly across Ae. tauschii chromosomes, but noncollinear genes were concentrated in the distal regions, and their density correlated with recombination rates along chromosomes.
We propose the following hypothesis that may at least partially account for the concentration of noncollinear genes in distal, high-recombination regions of Ae. tauschii chromosomes. Neutral SNP and restriction fragment length polymorphism (RFLP) were shown to have a greater rate of loss in low-recombination regions of Ae. tauschii chromosomes than in high-recombination regions (33, 34), presumably due to the effects of selection sweeps and background selection. Polymorphic noncollinear genes must be affected by the same processes as other neutral polymorphism and hence have a greater rate of loss in low-recombination regions compared with high-recombination regions. Noncollinear genes therefore have a greater chance of survival in distal high-recombination regions of Ae. tauschii chromosomes than in proximal low-recombination regions.
The population of noncollinear genes is an arena for evolution of new gene functions, and the high-recombination regions of Ae. tauschii chromosomes should therefore be enriched for new actively evolving genes, including R genes, which are rapidly evolving genes in plants (35). Ae. tauschii noncollinear genes in distal, high-recombination chromosome regions were indeed enriched for R genes, as predicted by the hypothesis. In agreement with enrichment for R genes, high-recombination regions were also enriched for signal transduction genes (GO class “receptor activity” GO: 0004872), which play important roles in the innate immunity in plants (36). This observation supports the hypothesis that the evolution of new genes preferentially takes place in the high-recombination regions of Triticeae chromosomes, as previously hypothesized on the basis of other evidence (37, 38).
Materials and Methods
BAC Fingerprinting and Contig Assembly.
We used nine large-insert libraries (Table S1, SI Text) and fingerprinted 461,706 clones of average length 120.5 kb (Table S1), as described earlier (9, 11). We edited fingerprints with FPMiner software (11) (SI Text) and assembled 399,448 edited clones into contigs with FPC (version 9.3; www.agcol.arizona.edu/software/fpc/) at an initial Sulston cutoff of 1 × 10−70, stepwise reduction of stringency accompanied by contig joining allowing a maximum of 15% Q clones, and terminated at the Sulston cutoff of 1 × 10−22 (SI Text). We converted FPC length metrics CB units into kilobases by estimating the insert lengths in 100 BAC clones per library by pulse field electrophoresis and dividing the total length by the number of restriction fragments in the fingerprints. The conversion factor was 1.5 kb/CB unit.
Ae. tauschii 10K Infinium Array.
We used an algorithm similar to that described previously (39) in selection of SNPs to maximize the likelihood of having a single SNP marker per gene. Based on the evaluation of 10,000 sequences containing SNPs with Illumina’s Assay Design Tool, we obtained 9,485 functional assays in the 10K Infinium array. They included 515 SNPs located in wheat ESTs (labeled by GenBank accession numbers) that have been previously mapped on an AL8/78 × AS75 map (25). The 10K Infinium database can be found at http://probes.pw.usda.gov/WheatDMarker/al878_gene_10000_snps_order_070410.csv.
Genetic and Physical Map Construction.
F2 mapping population, DNA isolation, 10K Infinium genotyping of the F2 plants, and genetic map construction are described in SI Text. The genetic map database is available at http://probes.pw.usda.gov/WheatDMarker/downloads/ComparativeMapData.xls. The construction of the Ae. tauschii physical map consisted of the following steps: (i) 5D pooling of BAC clones, (ii) genotyping of the pools with the Ae. tauschii 10K SNP Infinium assay, (iii) deconvolution of the 5D BAC pool genotyping data to identify BAC clones bearing marker genes, (iv) assigning each positive BAC clone to a marker locus on the genetic map, and (v) manual editing BAC contigs and disjoining chimeric contigs. Steps i–v are described in SI Text.
Marker Sequence Extension.
We constructed contigs from the 3.1× Roche 454 reads, extended them with 50× Illumina contigs, and generated a set of gene predictions in the 7,185 mapped extended sequences as described in SI Text. We assumed that 9,716 of the genes found in the extended sequences that were without any gap in the coding sequence and aligned fully with ESTs or annotated proteins were complete genes. We also obtained sequences that were incomplete but showed partial alignment to wheat and barley ESTs or proteins if no EST evidence was obtained. The output of MAKER (http://gmod.org/wiki/MAKER) was used to create a .gff file for our Gbrowse web interface (available at http://probes.pw.usda.gov/WheatDMarker/). A matrix of 17,093 genes and gene fragments including name, location on the genetic map, locations of homologous genes in B. distachyon, rice, and sorghum, and GO is available at http://probes.pw.usda.gov/WheatDMarker/downloads/GeneList.xls.
Recombination Rate and Gene Density.
We computed the cumulative lengths of contigs along the physical maps of the Ae.tauschii chromosomes. For Fig. 1, we used a 30-Mb window sliding by 1 Mb at a time to compute a rate. For Fig. S5 and statistics, we used a 30-Mb nonoverlapping window to compute a rate.
Collinearity Between Ae. tauschii and B. distachyon, Rice, and Sorghum.
We searched for homology between the nucleotide sequences of Ae. tauschii genes and gene fragments in the B. distachyon, rice, and sorghum pseudomolecules (builds Brachypodium 1.2 from www. brachypodium.org, Osativa_120 from www.phytozome.net/, and Sorghum 1.0 from http://genome.jgi-psf.org/Sorbi1/Sorbi1.info.html). If we detected more than one homologous gene in the B. distachyon, rice, and sorghum genome, we selected the gene with the lowest E value. We used the progressive increase or decrease of gene starts along the pseudomolecule as evidence of collinearity and changes in this progression as evidence of inversions or translocations. We color-coded each change in the progression along a pseudomolecule relative to the order of genes along the Ae. tauschii genetic map (http://probes.pw.usda.gov/WheatDMarker/downloads/ComparativeMapData.xls). Using maximum parsimony, we assigned each inversion or translocation to a lineage. Because we did not have an outgroup, we could not decide if a change in gene order took place in the sorghum lineage or in branch 1 of the phylogenetic tree (Fig. 2A). We arbitrarily assigned all changes in these two branches to the sorghum branch.
GO.
We used gene sequences in a BLASTX search against the UniProt knowledgebase (UniProtKB) database (www.uniprot.org) to assign each gene a functional GO term (BLASTX cutoff E value < 1E−7). GOs were assigned on the basis of biological, functional, and molecular annotation available from GO (www.geneontology.org). We also used gene sequences in a BLAST search against the plant resistance gene database (PRGdb; http://prgdb.crg.eu/wiki/Main_Page).
Supplementary Material
Acknowledgments
We thank C. Soderlund, W. Nelson, J. Messing, and P. Langridge for their service as advisors to the DBI-0701916 project and E. Ghiban at Cold Spring Harbor Laboratory for assistance with Illumina sequencing for the IOS-1032105 project. This work was supported by National Science Foundation Plant Genome Research Program Grants DBI-0701916 (Principal Investigator, J. Dvorak), DBI-0822100 (Principal Investigator, S.F.K.), and IOS-1032105 (Principal Investigator, W.R.M.), US Agricultural Research Service Projects 5325-21000-014 and 1907-21000-030, and Czech Science Foundation Grant P501/12/2554 (Principal Investigator, H.Š.). This paper is a contribution to the International Wheat Genome Sequencing Consortium.
Footnotes
Conflict of interest statement: W.R.M. has participated in Illumina-sponsored meetings (past 4 years) and received travel reimbursement and honorarium for presenting at these events (Illumina had no role in decisions relating to this study and the decision to publish), has participated in Pacific Biosciences-sponsored meetings (past 3 years) and received travel reimbursement for presenting at these events, and is a founder and shared holder of Orion Genomics, which focuses on plant genomics and cancer genetics.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov/) (accession nos. SRP012566, SRA052214.1, SRX129979, SRX125241, SRX125233, SRX124436, SRX116546, SRX037891, and SRX037088.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219082110/-/DCSupplemental.
References
- 1.Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9(3):208–218. [Google Scholar]
- 2.Rees H, Walters MR. Nuclear DNA and evolution of wheat. Heredity. 1965;20(1):73–82. [Google Scholar]
- 3.Li W, Zhang P, Fellers JP, Friebe B, Gill BS. Sequence composition, organization, and evolution of the core Triticeae genome. Plant J. 2004;40(4):500–511. doi: 10.1111/j.1365-313X.2004.02228.x. [DOI] [PubMed] [Google Scholar]
- 4. Nesbitt M, Samuel D (1996) From staple crop to extinction? The archaeology and history of hulled wheats. Hulled Wheats. Promoting the Conservation and Use of Underutilized and Neglected Crops. Proceedings of the First International Workshop on Hulled Wheats, eds Padulosi S, Hammer K, Keller J (International Plant Genetic Resources Institute, Rome, Italy), pp 41–100.
- 5.Brenchley R, et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature. 2012;491(7426):705–710. doi: 10.1038/nature11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein PE, et al. A high-throughput AFLP-based method for constructing integrated genetic and physical maps: Progress toward a sorghum genome map. Genome Res. 2000;10(6):789–807. doi: 10.1101/gr.10.6.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei F, et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007;3(7):e123. doi: 10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.International Barley Genome Sequencing Consortium A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491(7426):711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- 9.Luo MC, et al. High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics. 2003;82(3):378–389. doi: 10.1016/s0888-7543(03)00128-9. [DOI] [PubMed] [Google Scholar]
- 10.Nelson WM, et al. Whole-genome validation of high-information-content fingerprinting. Plant Physiol. 2005;139(1):27–38. doi: 10.1104/pp.105.061978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu YQ, et al. A BAC-based physical map of Brachypodium distachyon and its comparative analysis with rice and wheat. BMC Genomics. 2009;10:496. doi: 10.1186/1471-2164-10-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Oeveren J, et al. Sequence-based physical mapping of complex genomes by whole genome profiling. Genome Res. 2011;21(4):618–625. doi: 10.1101/gr.112094.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soderlund C, Longden I, Mott R. FPC: A system for building contigs from restriction fingerprinted clones. Comput Appl Biosci. 1997;13(5):523–535. doi: 10.1093/bioinformatics/13.5.523. [DOI] [PubMed] [Google Scholar]
- 14.Soderlund C, Humphray S, Dunham A, French L. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 2000;10(11):1772–1787. doi: 10.1101/gr.gr-1375r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.You FM, et al. GenoProfiler: batch processing of high-throughput capillary fingerprinting data. Bioinformatics. 2007;23(2):240–242. doi: 10.1093/bioinformatics/btl494. [DOI] [PubMed] [Google Scholar]
- 16.Frenkel Z, Paux E, Mester D, Feuillet C, Korol A. LTC: A novel algorithm to improve the efficiency of contig assembly for physical mapping in complex genomes. BMC Bioinformatics. 2010;11:584. doi: 10.1186/1471-2105-11-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo MC, et al. 2003. Construction of contigs of Ae. tauschii genomic DNA fragments cloned in BAC and BiBAC vectors. Proceedings of the Tenth International Wheat Genetics Symposium, eds Pogna NE, Romano M, Pogna EA, Galterio G (S.IM.I., Rome Italy), pp 293–296.
- 18.Messing J, et al. Sequence composition and genome organization of maize. Proc Natl Acad Sci USA. 2004;101(40):14349–14354. doi: 10.1073/pnas.0406163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng SHS, et al. A physical map of the genome of Atlantic salmon, Salmo salar. Genomics. 2005;86(4):396–404. doi: 10.1016/j.ygeno.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Cai WW, Reneker J, Chow CW, Vaishnav M, Bradley A. An anchored framework BAC map of mouse chromosome 11 assembled using multiplex oligonucleotide hybridization. Genomics. 1998;54(3):387–397. doi: 10.1006/geno.1998.5620. [DOI] [PubMed] [Google Scholar]
- 21.Paux E, et al. A physical map of the 1-gigabase bread wheat chromosome 3B. Science. 2008;322(5898):101–104. doi: 10.1126/science.1161847. [DOI] [PubMed] [Google Scholar]
- 22.Luo MC, et al. A high-throughput strategy for screening of bacterial artificial chromosome libraries and anchoring of clones on a genetic map constructed with single nucleotide polymorphisms. BMC Genomics. 2009;10:28. doi: 10.1186/1471-2164-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.You FM, et al. Annotation-based genome-wide SNP discovery in the large and complex Aegilops tauschii genome using next-generation sequencing without a reference genome sequence. BMC Genomics. 2011;12:59. doi: 10.1186/1471-2164-12-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo MC, et al. Feasibility of physical map construction from fingerprinted bacterial artificial chromosome libraries of polyploid plant species. BMC Genomics. 2010;11:122. doi: 10.1186/1471-2164-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo MC, et al. Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc Natl Acad Sci USA. 2009;106(37):15780–15785. doi: 10.1073/pnas.0908195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International Brachypodium Initiative Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463(7282):763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- 27.Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA. 2004;101(26):9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanseverino W, et al. PRGdb 2.0: Towards a community-based database model for the analysis of R-genes in plants. Nucleic Acids Res. 2013;41(Database issue) D1:D1167–D1171. doi: 10.1093/nar/gks1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You FM, et al. A new implementation of high-throughput five-dimensional clone pooling strategy for BAC library screening. BMC Genomics. 2010;11:692. doi: 10.1186/1471-2164-11-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gottlieb A, et al. Insular organization of gene space in grass genomes. PLoS ONE. 2013;8(1):e54101. doi: 10.1371/journal.pone.0054101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aliscioni S, et al. Grass Phylogeny Working Group II New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 2012;193(2):304–312. doi: 10.1111/j.1469-8137.2011.03972.x. [DOI] [PubMed] [Google Scholar]
- 32.Akhunov ED, Akhunova AR, Dvorak J. Mechanisms and rates of birth and death of dispersed duplicated genes during the evolution of a multigene family in diploid and tetraploid wheats. Mol Biol Evol. 2007;24(2):539–550. doi: 10.1093/molbev/msl183. [DOI] [PubMed] [Google Scholar]
- 33.Dvorák J, Luo M-C, Yang Z-L. Restriction fragment length polymorphism and divergence in the genomic regions of high and low recombination in self-fertilizing and cross-fertilizing Aegilops species. Genetics. 1998;148(1):423–434. doi: 10.1093/genetics/148.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang JR, et al. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013;198(3):925–937. doi: 10.1111/nph.12164. [DOI] [PubMed] [Google Scholar]
- 35.Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8(11):1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 36.Vakhrusheva OA, Nedospasov SA. System of innate immunity in plants. Mol Biol. 2011;45(1):16–23. [PubMed] [Google Scholar]
- 37.Dvorak J, Akhunov ED. Tempos of deletions and duplications of gene loci in relation to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics. 2005;171:323–332. doi: 10.1534/genetics.105.041632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.See DR, et al. Gene evolution at the ends of wheat chromosomes. Proc Natl Acad Sci USA. 2006;103(11):4162–4167. doi: 10.1073/pnas.0508942102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mammadov JA, et al. Development of highly polymorphic SNP markers from the complexity reduced portion of maize [Zea mays L.] genome for use in marker-assisted breeding. Theor Appl Genet. 2010;121(3):577–588. doi: 10.1007/s00122-010-1331-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.