Abstract
Lateral gene transfer events between bacteria and animals highlight an avenue for evolutionary genomic loss/gain of function. Herein, we report functional lateral gene transfer in animal parasitic nematodes. Members of the Nematoda are heme auxotrophs, lacking the ability to synthesize heme; however, the human filarial parasite Brugia malayi has acquired a bacterial gene encoding ferrochelatase (BmFeCH), the terminal step in heme biosynthesis. BmFeCH, encoded by a 9-exon gene, is a mitochondrial-targeted, functional ferrochelatase based on enzyme assays, complementation, and inhibitor studies. Homologs have been identified in several filariae and a nonfilarial nematode. RNAi and ex vivo inhibitor experiments indicate that BmFeCH is essential for viability, validating it as a potential target for filariasis control.
The phylum Nematoda is estimated to comprise up to one hundred million species (1), including free-living worms (such as the widely used experimental organism, Caenorhabditis elegans) as well as parasitic species that threaten the health of agriculturally important plants, wildlife, domesticated animals, and humans. Comparative genomics suggests that lateral gene transfer (LGT) may have played an important role in the evolution of plant parasitism by facilitating adoption of a parasitic lifestyle (2, 3). For animal parasitic nematodes, such as the human filarial nematode Brugia malayi, several key metabolic processes have been lost (4). For example, the absence of heme biosynthesis appears to be a peculiar feature of the Phylum Nematoda (5). In filarial nematodes such as B. malayi, essential compounds may be provided by the obligate endosymbiotic bacteria Wolbachia (6). LGT events have been documented between the Wolbachia endosymbiont and its nematode hosts, but to date no functionality has been determined for the transferred DNA (7–9). Here, we describe both the functionality and essentiality of a bacterial gene acquired by filarial nematodes via LGT: that of the last step of heme biosynthesis [ferrochelatase (FeCH)] (Fig. S1). Because of its essentially, FeCH represents a potential drug target against the causative agents of lymphatic filariasis and onchocerciasis (river blindness). These diseases affect more than 150 million individuals with over 1 billion people at risk for infection (10, 11). No new classes of drugs directly targeting filariae with good efficacy and safety profiles have been developed in over 20 y, and suboptimal response/emerging resistance to currently used antifilarial drugs is apparent (12, 13).
Results
BmFeCH Origin and Features.
The draft genomic sequence of B. malayi (4) revealed two adjacent ORFs (Bm1_14315 and Bm1_14320) that appeared to be components of the same ferrochelatase (FeCH) gene. We established that these ORFs are, in fact, part of a single transcript with strong similarity to FeCH genes in the GenBank database. Genome annotation did not identify any additional heme biosynthesis genes, and, even when using low stringency search/match parameters, we were unable to identify any other genes in this pathway. PCR using generic primers based on conserved regions of other heme biosynthesis pathway genes also failed to give any specific products.
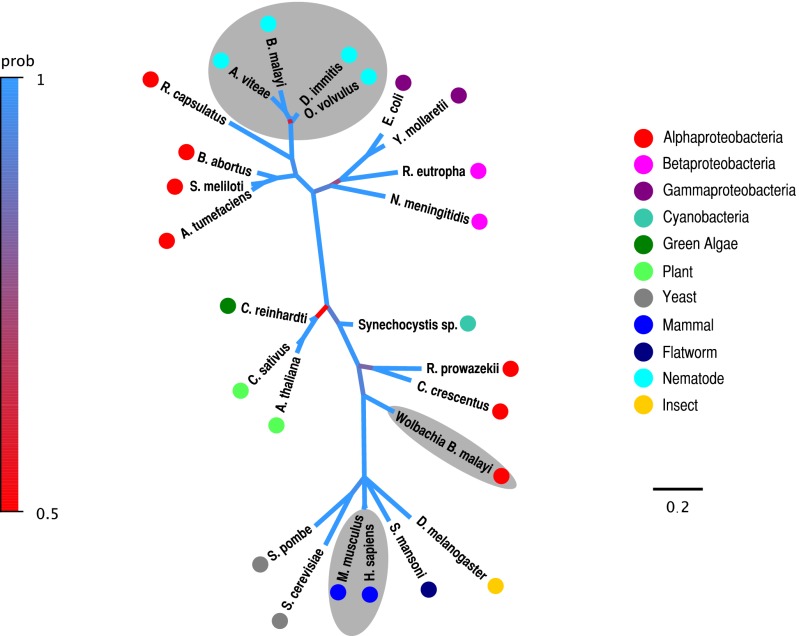
The B. malayi ferrochelatase protein (BmFeCH) clusters with a subset of α-proteobacterial sequences within the Orders Rhizobiales and Rhodobacterales with high probability but is more distantly related to orthologs in Wolbachia or mammals, as shown by a consensus tree produced from Bayesian phylogenetic analysis of aligned FeCH protein sequences (Fig. 1, Fig. S2). BmFeCH also clusters closely with other nematode FeCH sequences. Evolutionarily, FeCH proteins are poorly conserved across diverse taxa (14); however, all, including the predicted BmFeCH, have invariant residues in the active site pocket. BmFeCH does not possess the C-terminal extension found in eukaryotic counterparts or the signature for the [2Fe-2S] cluster found in Actinobacteria and metazoan FeCHs (Fig. S2). It does possess the proposed membrane-binding loop absent in Actinobacteria and Firmicutes (14).
Fig. 1.
Consensus tree produced from Bayesian phylogenetic analysis of aligned FeCH protein sequences. Nematode, mammalian and Wolbachia leaf nodes are highlighted with light gray shading. Taxonomic classifications are indicated with small colored circles next to the organism names. The scale bar units are residue changes per site. Branches are colored based on their posterior probability using the color scale at left. Organisms and FeCH accessions: B. malayi ADI33748, D. immitis ADI33752, A. viteae ADI33750, O. volvulus ADI33751, Agrobacterium tumefaciens NP 356851, Sinorhizobium meliloti AAL67571, Brucella abortus YP 222888, Rhodobacter capsulatus JC4752, Neisseria meningitides Q9K097, Ralstonia eutropha YP 295260, E. coli P23871, Yersinia mollaretii ZP 00826183, Wolbachia of B. malayi AAW71307, Caulobacter crescentus NP 422556, Rickettsia prowazekii Q9ZC84, Arabidopsis thaliana P42043, Cucumis sativus T10246, Chlamydomonas reinhardtii AAK16728, Synechocystis sp. PCC6803 NP 442453, Saccharomyces cerevisiae NP 014819, Schizosaccharomyces pombe O59786, Schistosoma mansoni CAZ31135, Drosophila melanogaster AAC26225, Mus musculus NP 032024, and Homo sapiens XP 008784.
Despite its apparent bacterial origin, sequence of the ∼4.5-kb BmFeCH gene revealed a eukaryotic gene organization, containing nine putative exons and eight introns. The deduced protein includes an N-terminal extension (encoded by part of the first exon), predicted to include a mitochondrial targeting domain (Fig. S2). PCR was used to amplify full-length FeCH cDNAs from various filarial species: B. malayi, Dirofilaria immitis, Onchocerca volvulus, and Acanthocheilonema viteae. The latter lacks the Wolbachia endosymbiont, but harbored it in its evolutionary past (9). Genomic databases support the occurrence of FeCH in other filariae: Wuchereria bancrofti, Loa loa and Litomosoides sigmodontis, as well as the nonfilarial nematodes, Strongyloides ratti and Strongyloides venezuelensis (this study and refs. 15 and 16). All nematode homologs have an N-terminal extension that is not present in bacterial FeCH enzymes. The presence of FeCH homologs in filarial nematodes (Spirurina, clade III) as well as in nonfilarial worms of the genus Strongyloides (Tylenchina, clade IV) suggests that acquisition of FeCH predated the divergence of clades III and IV or that independent acquisitions occurred in the lineages leading to the Onchocercidae and Strongyloididae (15). We note that the nine-exon gene structure is conserved in all six filarial genera for which draft/ongoing genomic sequence data are available (4) (www.nematodes.org/nematodegenomes/index.php/Main_Page). Although S. ratti (www.sanger.ac.uk/resources/software/blast/) has only one intron, the splice junction is conserved with one of those in the filariae, strongly indicating a monophyletic origin of nematode FeCH but with different rates of intron evolution in the distinct lineages. In addition, whereas most of these draft genome sequences still consist of many small contigs, we observe that, in the two most completely assembled genomes (B. malayi and L. loa), there is conserved synteny of FeCH and its flanking genes.
BmFeCH Mitochondrial Targeting.
To confirm the function of the predicted BmFeCH mitochondrial-targeting domain, we compared localization of full-length BmFeCH::GFP to a version lacking 28 amino acids at the N terminus (IP633; BmFeCHΔ28::GFP) in transgenic C. elegans lines. Using MitoTracker Red CMXRos (which specifically stains mitochondria), we determined that, in lines carrying a full-length BmFeCH::GFP translational fusion (IP637 and IP634), the GFP signal localizes to mitochondria (Fig. 2). In contrast, in the transgenic C. elegans line carrying BmFeCHΔ28::GFP (IP633), the mitochondrial localization pattern is lost and the GFP signal is detected in the cytoplasm and nuclei. These results confirm our in silico localization prediction as well as previous studies in mammalian cells (17).
Fig. 2.
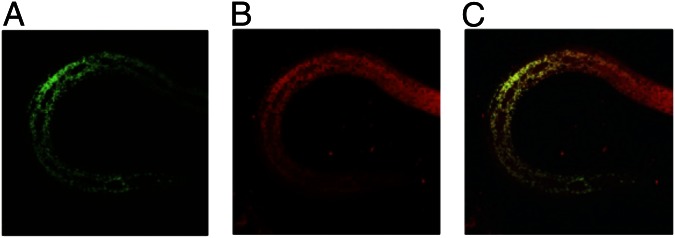
Confocal micrographs of transgenic third stage larva of C. elegans (IP634) carrying the full length BmFeCH::GFP construct. (A) GFP signal resulting from the BmFeCH::GFP transgene, (B) mitochondrial signal from MitoTracker CMXRos and (C) merged fluorescence signals.
In Vivo Localization.
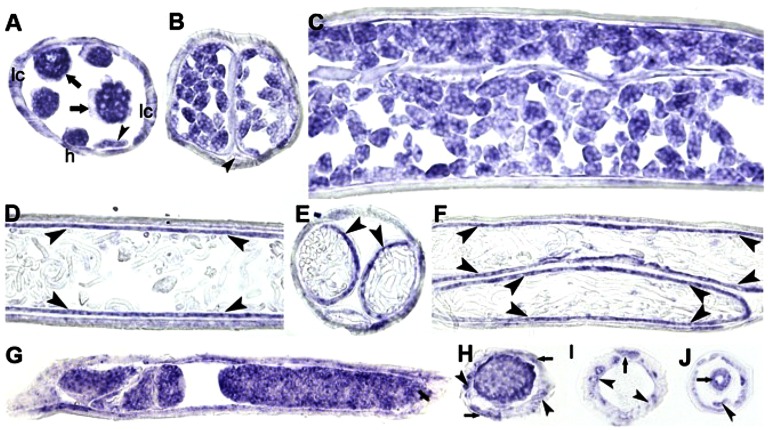
To localize BmFeCH in B. malayi, a digoxigenin-labeled 467-bp BmFeCH RNA probe was used for in situ hybridization (ISH). BmFeCH appears to be ubiquitously expressed in both male and female tissues, except for female late-stage embryos and male late-stage sperm cells (Fig. 3). In adult female worms, hybridization signals were detected in oocytes within the ovary (Fig. 3A) and early stage embryos, such as morula stage (Fig. 3 B and C), but not in late-stage embryos, like curved microfilariae (Fig. 3D) and stretched microfilariae (Fig. 3 E and F). However, the uterine epithelial cells that contain late-stage embryos showed strong hybridization signals (Fig. 3 D–F). Hybridization signals were also observed in somatic tissues such as lateral cords, intestine, and hypodermis (Fig. 3A). In adult male worms, BmFeCH transcripts were mainly detected in early stages of developing sperm such as spermatocytes (Fig. 3 G and H), but not in late stages such as spermatids (Fig. 3I). BmFeCH transcripts were also detected in lateral cords, muscles (Fig. 3 H and I), intestine, and vas deferens (Fig. 3J).
Fig. 3.
In situ hybridization localization of FeCH expression in adult B. malayi females (A–F) and males (G–J). (A) Cross section of a female worm at the region of the ovary (arrows). Arrowhead indicates intestine. h, hypodermis, lc, lateral cords. (B) Cross section of female worm at the region of the uteri containing early embryos (morula stage). Arrowhead indicates intestine. (C) Longitudinal section at region of female uteri containing early embryos (morula stage). (D) Longitudinal section of uterine region containing curved microfilariae. (E) Cross section of uteri containing stretched microfilariae. (F) Longitudinal section of uterine region containing stretched microfilariae, Arrowheads in D–F indicate uterine epithelial cells. (G) Longitudinal section at anterior region of male testis with developing sperm, mostly spermatogonia. (H) Cross section through anterior region of male testis with early stage sperm, mostly spermatocytes. (I) Cross-section through male at end of testis showing spermatids. Arrows and arrowheads in H and I indicate lateral cords and muscle, respectively. (J) Cross section through male to show vas deferens (arrow). Arrowhead indicates intestine.
BmFeCH Functionality.
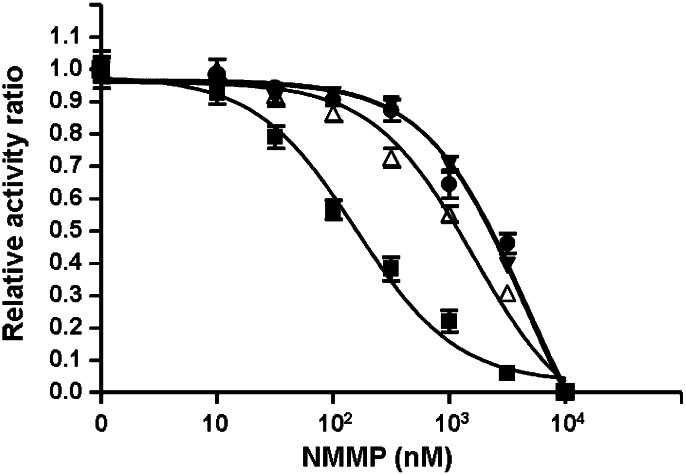
We used a variety of approaches to verify the functionality of BmFeCH. The gene was cloned, and recombinant protein was expressed and purified. The clone produces active ferrochelatase with a specific activity of 30.5 nmol/mg per h Zinc (II) protoporphyrin IX formation (Zn-PPIX). For comparative purposes, recombinant FeCHs from A. viteae (AvFeCH; 25 nmol/mg per h), the Wolbachia endosymbiont of B. malayi (wBmFeCH; 43.5 nmol/mg per h), and human (HsFeCH; 90.6 nmol/mg per h) were produced. Functionality was also assayed by addition of the competitive FeCH inhibitor N-methyl mesoporphyrin (NMMP) (18), which reduced or eliminated FeCH activity (Fig. 4). The IC50 values for NMMP inhibition of BmFeCH and AvFeCH were ∼4.5 μM whereas wBmFeCH and HsFeCH were ∼1.6 μM and 0.16 μM, respectively.
Fig. 4.
Inhibition of FeCH activity by NMMP. Activity of recombinant BmFeCH (filled inverted triangles), AvFeCH (filled circles), HsFeCH (filled squares) and wBmFeCH (open triangles) is set at 1.0 for Zn-PPIX formation in the absence of NMMP.
The in vivo functionality of BmFeCH was assessed by using genetic complementation of an Escherichia coli FeCH deletion mutant, ΔhemH (FeCH), mutant that is unable to grow unless hemin is supplied to the media (19). Supplementation with PPIX, the substrate for FeCH, failed to support growth of the mutant strain. However, the E. coli ΔhemH strain containing the BmFeCH cDNA grew in the absence of supplemented hemin. This result occurred with full-length BmFeCH and an N-terminally truncated version, as well as with wBmFeCH and HsFeCH.
To evaluate functionality in intact nematodes, we generated transgenic C. elegans lines that carried the full-length BmFeCH cDNA fused to the green fluorescent protein (GFP) DNA sequence (BmFeCH::GFP) integrated into the genome. One selected line, IP637, is similar to wild-type (WT) with respect to brood size and viability, suggesting that expression of BmFeCH::GFP has no major detrimental effect under normal culture conditions.
Control (WT) C. elegans grow on chemically defined axenic basal medium (ABM) if supplemented with hemin (refs. 5 and 20) and this study) but not PPIX (Fig. S3). Worms grown in ABM or PPIX-containing ABM do not die but arrest after one or two molts and fail to reach adulthood. Transgenic IP637 worms were also assayed for growth on axenic medium and were able to grow to adulthood when supplemented with either PPIX or to a lesser extent hemin (Fig. S3). FeCH inhibitor studies were performed with IP637 worms. In the presence of PPIX, their growth was inhibited when NMMP was added to the media. This effect could be partially rescued by addition of hemin to the media (Fig. S3). Taken together, these results indicate that BmFeCH is functionally expressed in C. elegans.
BmFeCH Essentiality.
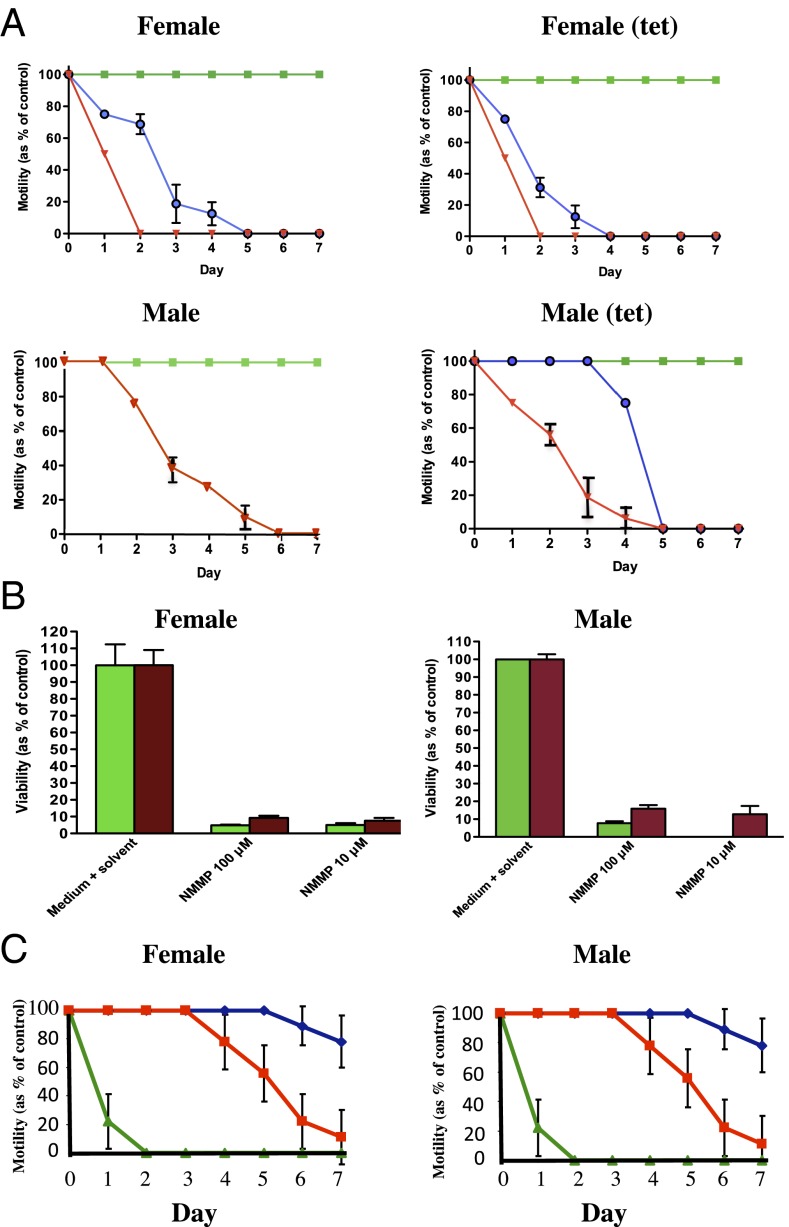
Because B. malayi contains the Wolbachia endosymbiont that encodes a functional FeCH gene and is sensitive to antibiotics (such as the tetracyclines), we were able to assess the relative contributions of wBmFeCH and BmFeCH by comparing the effect of NMMP on adult B. malayi and tetracycline-treated B. malayi adults cleared of their Wolbachia. We measured Wolbachia levels by quantitative PCR (qPCR) and measured worm motility in both groups. Motility in adult B. malayi females and males was significantly reduced by exposure to NMMP relative to untreated controls (Fig. 5A). Viability in both adult B. malayi and tetracycline-treated B. malayi was also significantly decreased in all NMMP treatment groups compared with the untreated controls (Fig. 5B). These results suggest that the inhibition was not Wolbachia specific, as similar results were obtained in antibiotic- and nonantibiotic-treated groups. Presumably, both wBmFeCH and BmFeCH are affected by NMMP, further suggesting the worm’s requirement for heme biosynthesis for viability. The sensitivity of filarial nematode FeCH to NMMP was also demonstrated in studies using adult A. viteae, a species lacking Wolbachia. Male and female worms showed reduced motility by approximately day 4 of exposure to 100 μM NMMP and appeared moribund by day 7 (Fig. 5C).
Fig. 5.
Dynamic effects of the FeCH inhibitor NMMP on motility and viability of adult males and females of B. malayi, tetracycline-treated B. malayi, and A. viteae maintained for 7 d in vitro. (A) Motility of tetracycline-treated B. malayi and untreated control worms cultured in the presence of 100 μM (red), 10 μM (blue) and 0 μM (green) NMMP. [Dose response of control male worms to 10 μM NMMP not performed]. Motility was measured on a scale of 0 to 4 and the data plotted as a percentage of the control worms (untreated with NMMP) at each time point set at 100%. (B) Viability of tetracycline-treated B. malayi (maroon bars) and untreated control worms (green bars) cultured in the presence of 100 μM, 10 μM, or 0 μM NMMP for 7 days. Viability was determined by the MTT assay and the value of the control group was set to 100. Viability of control male worms after exposure to 10 μM NMMP not performed. (C) Motility of A. viteae male and female worms cultured in the presence of 100 μM NMMP (red), 100 μM ivermectin (green), or no inhibitor (blue). Motility was measured on a scale of 0 to 4 and the data plotted as a percentage of control worms (untreated), set at 100%.
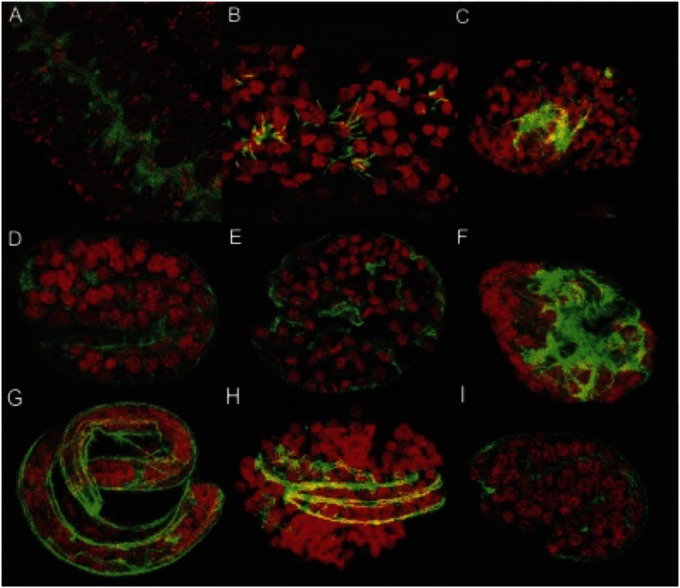
RNA interference (RNAi) was used to investigate the effect of FeCH depletion upon the B. malayi female germ line and during embryonic development. Unlike C. elegans, embryonic development in B. malayi takes place entirely within the uterus, so RNAi in B. malayi potentially affects every developmental stage. We found that FeCH RNAi led to consistent abnormalities in the nuclei in the germ line as well as in embryos. Untreated WT ovaries displayed spheroid nuclei surrounded by numerous Wolbachia, and organized around a central rachis in a syncytium (Fig. 6A) whereas FeCH RNAi-treated females exhibited misshapen germ-line nuclei, often lenticular or polygonal, and also occasionally characterized by an uneven Wolbachia distribution and abnormal rachis (Fig. 6B). The same misshapen nuclei were observed in all developing embryos (n > 500), associated with embryonic elongation and actin cytoskeleton defects (Fig. 6 C and E). The majority of elongated embryos exposed to FeCH heterogeneous short interfering RNA (hsiRNA) eventually ruptured, likely due to cytoskeleton defects compromising overall hypodermal integrity (Fig. 6 F and H). The observation of similar phenotypes when using hsiRNA mixtures derived from different regions of BmFeCH indicates that the defects are specific to FeCH transcript reduction rather than nonspecific off-target effects. By contrast, mature microfilariae did not display any phenotype, perhaps because of reduced FeCH metabolism or insensitivity to RNAi at this stage. Control worms incubated in the absence of RNA (Fig. 6 A, D, and G) or in the presence of hsiRNA derived from a plasmid vector (Fig. 6I) did not show any defects.
Fig. 6.
BmFeCH RNAi and fluorescence-based phenotype analysis in female B. malayi. (A, D, and G) control worms (WT) incubated in the absence of RNAi. (B, E, and H) worms exposed to hsiRNA derived from the 5′ region of BmFeCH. (C and F) B. malayi worms exposed to hsiRNA derived from the 3′ end of BmFeCH. (I) B. malayi exposed to control hsiRNA derived from a cloning vector. A and B show nuclei and Wolbachia in a syncytium around the rachis of the ovary. C–E and I show developing embryos. F–H show elongated embryos. DNA is stained with propidium iodide (red) and actin stained with phalloidin (green).
Discussion
We report a functional LGT in a nematode species parasitic in the Metazoa. LGT is widespread among certain nematodes and may have played a role in the remarkable evolutionary success of this phylum and in the adoption of parasitic lifestyles. Genomes of several plant-parasitic nematodes contain genes acquired by LGT that encode enzymes involved in plant cell wall degradation (2, 3). LGT has previously been noted in animal parasitic filarial nematodes (7–9), but thus far none have been shown to be functional. BmFeCH is functional and essential, as indicated by multiple lines of evidence, including E. coli complementation, heme auxotrophy complementation, ISH experiments showing ubiquitous expression in males and females, inhibitor studies, and the lethal effect of RNAi silencing of BmFeCH in B. malayi embryos.
We suggest that heme biosynthesis in the Nematoda was completely lost (or alternatively, it was never present) but that the last step, catalyzing porphyrin metallation, was acquired by LGT in an ancestor of modern nematodes where it (re)assumed function in the same compartment as in other heme-synthesizing eukaryotes. Although BmFeCH is most closely related to α-proteobacterial FeCH, during its evolution within metazoan organisms, the gene has evolved into a typical eukaryotic gene by acquiring introns and exons. BmFeCH also encodes an additional N-terminal mitochondrial targeting sequence that is not found in α-protobacterial FeCHs but is conserved across filarial FeCHs. The mitochondrial localization, revealed by transgene expression in C. elegans, is consistent with localization in other eukaryotic heme prototrophs, where FeCH is targeted to the inner surface of the inner mitochondrial membrane and is involved in porphyrin metallation (21).
B. malayi transcript analysis revealed the presence of a BmFeCH variant, differing at 4 amino acid positions (E24K, I29M, E32K, and E269D). Both variant proteins are able to complement the E. coli hemH mutant, indicating that the mutations result in functional polymorphisms. Two highly similar FeCH variants can be identified in the emerging genome sequences of various other filarial nematodes (this study and ref. 15).
All analyzed nematode clades also appear to contain genes coding for a highly degenerate FeCH-like protein (this study and ref. 15). However, we note that they lack the key residues of the catalytic domain and failed to complement the E. coli ΔhemH mutant even when PPIX was added to the culture media. Furthermore, C. elegans only has this FeCH-like gene and is unable to use PPIX for growth (5). RNAi experiments and knockout mutants in C. elegans indicate that the nematode FeCH-like protein is not essential for C. elegans viability, at least under standard laboratory conditions (wormbase.org). Studies addressing the function of these FeCH-like proteins are warranted.
It is not obvious why some filarial nematodes have functional FeCH genes when they also have Wolbachia endosymbionts that contain a complete heme biosynthetic pathway. Possibly, the FeCH gene enables nematode survival on PPIX in life-cycle stages where exogenous heme is scarce or Wolbachia titer is low. Because the FeCH gene does not appear to have originated from the Wolbachia endosymbiont and has been retained in filarial species not harboring Wolbachia, it seems unlikely that this gene plays a role in Wolbachia–host interactions. Rather, the presence of LGT FeCH in other nematode clades (such as in Strongyloides spp; clade IV) and the essentiality in filarial nematode embryos point to a model whereby a FeCH may help meet high heme demands at critical stages in the parasite life cycle. However, it is possible that, despite retaining FeCH activity, the protein has assumed an alternative essential cellular function distinct from porphyrin metallation. The finding of FeCH only in filarial nematodes and members of the genus Strongyloides is suggestive of a role in adaptations toward animal parasitism in these groups (16).
FeCH has assumed a critical role in filarial nematode species where it is essential. The bacterial ancestry of BmFeCH results in considerable divergence between these nematode enzymes and their mammalian counterparts. Such divergence lends an opportunity for targeting FeCH using drugs specific to bacterial enzymes for filariasis control. It was suggested that the presence of BmFeCH, along with the FeCH-like protein and the wBmFeCH (from Wolbachia) in the same parasitic organism, might represent a serious challenge to efforts aimed at developing FeCH as a drug target (15). However, our findings indicate that nematode FeCH-like proteins do not function as FeCHs. Conversely, RNAi indicates that BmFeCH is essential for B. malayi and that the FeCH inhibitor NMMP impairs both filarial motility and viability. Accordingly, drug screens targeting filarial FeCHs are warranted.
Materials and Methods
Additional information is provided in SI Materials and Methods.
Filarial Nematode Material.
B. malayi worms were purchased from TRS Labs. O. volvulus worms were generously provided by Sara Lustigman (New York Blood Center, New York, NY). A. viteae worms were a gift from Kenneth Pfarr (University Hospital Bonn, Bonn, Germany) and Peter Fischer (Washington University, St Louis, MO). D. immitis RNA samples were kindly provided by Michael Crawford (Divergence, St. Louis, MO).
FeCH Cloning into E. coli and Expression of FeCH Genes.
Worm RNA extractions were performed using the RNeasy mini kit (Qiagen). Rapid Amplifications of 5′ and 3′ cDNA ends (RACE) were carried out with the SMARTer RACE cDNA amplification kit (Clontech) from RNA preparations of B. malayi, O. volvulus, D. immitis, and A. viteae worms. Subsequent PCR amplification and DNA sequencing between the 3′ and 5′ ends enabled acquisition of complete coding sequences. The newly acquired B. malayi FeCH (BmFeCH) and A. viteae FeCH (AvFeCH) coding sequences were cloned into pET21a+ vector (Novagen) for production of recombinant proteins with C-terminal 6XHis-tags in E. coli using methods established previously (22). For controls and for comparative purposes, we produced recombinant FeCH from human (HsFeCH), Wolbachia from B. malayi (wBmFeCH) and E. coli (EcFeCH) as described previously (22).
Ferrochelatase Enzyme Assays.
Enzyme activities were assayed using purified recombinant N-terminal 6XHis-tagged BmFeCH, AvFeCH, wBmFeCH, and HsFeCH proteins at 37 °C for 10–20 min, essentially as previously described (23). For NMMP (N-methyl mesoporphyrin; Frontier Scientific) inhibitor studies, assays were initiated by the addition of the substrate protoporphyrin IX (PPIX) after a 10-min enzyme preincubation with different concentrations of inhibitor.
Sequence and Phylogenetic Analysis.
For alignments, putative FeCH protein sequences obtained from the different filarial species, along with FeCH orthologs retrieved from National Center for Biotechnology Information GenBank (ncbi.nlm.nih.gov) following BLASTX similarity searches, were aligned using CLUSTALX 1.83 (24). After manual refinement, the sequences in the alignment were further analyzed by TargetP 1.1 (25) for predicting subcellular targeting signals. BmFeCH cDNA sequences were compared with their genomic locus sequence by LFasta (pbil.univ-lyon1.fr/lfasta.php).
For the consensus tree construction, sequences were aligned and curated using the phylogeny.fr web service at phylogeny.fr (26). Sequence alignments were performed with T-Coffee (v6.85) (27). After alignment, the sequences were curated, and ambiguous regions (i.e., containing gaps and/or poorly aligned) were removed with Gblocks (v0.91b) (28). The curated alignment containing 126 sites was analyzed locally using MrBayes (v3.2.1 x64) (29). Trees were visualized with FigTree v1.4.0 (tree.bio.ed.ac.uk), and final figures were created in InkScape (inkscape.org).
E.coli ΔhemH Complementation with Heterologous FeCH Genes.
The E. coli ΔhemH deletion strain VS200 has been described previously (19). The pET21a vectors containing the corresponding human, B. malayi, Wolbachia, and E. coli putative FeCH genes were transformed into the E. coli ΔhemH strain, both with and without RIL plasmid cotransformation. Transformants were selected on 20-µM hemin-containing LB plates with appropriate antibiotics and incubated at 37 °C overnight. Hemin was freshly prepared as a 5-mM stock solution in 50% (vol/vol) ethanol containing 0.02 M NaOH. The selected transgenic clones were further tested on LB plates with no hemin addition.
E.coli-Based FeCH Inhibitor Assays.
The E. coli hemH mutant strain VS200, containing separately HsFeCH, BmFeCH, AvFeCH, wBmFeCH, or EcFeCH genes, was used in inhibitor assays. Transgenic E. coli (grown to 0.6–0.8 OD600 in LB medium) were diluted to 0.01 OD600 before initiating growth assays in the presence or absence of different concentrations of NMMP. The E. coli cells were grown at 30 °C for 3 h before determination of cell growth by measurement of the final OD600 values. The average cell growth ratio (final OD600/0.01) for untreated control was set at 1.0.
Ex Vivo B. malayi and A. viteae Motility and Viability Tests.
B. malayi adults were isolated from jirds. For Wolbachia-depleted B. malayi, jirds were treated at Liverpool School of Tropical Medicine (Liverpool, United Kingdom) with 2.5 mg/mL tetracycline (Sigma) in drinking water for 6 wk (with tetracycline/water prepared fresh daily) plus 2 wk of water alone before recovery of adult worms. The reduction in Wolbachia load following tetracycline treatment was determined by quantitative PCR (qPCR) to determine the copy number of the genes encoding Wolbachia surface protein (wsp) and B. malayi GST (gst) as previously described (30). Following treatment, Wolbachia loads were reduced by 99.24% in female worms and 99.91% in male worms. Worms were cultured in the presence of different concentrations of NMMP for 7 d. Motility was scored daily using a scoring system described previously (31), with 0 being immotile and 4 being highly active and motile and the results for each group presented as a percentage of the control group. Viability was evaluated at day 7 using the MTT assay (32, 33). The reduction of MTT for each group is presented as a percentage of the control group.
Adult male and female A. viteae were incubated with different concentrations of the FeCH inhibitor NMMP for 7 d (three replicates per experiment). The anthelmintic ivermectin (100 μM; Sigma) was used as a positive control. Motility was measured daily (as described above and in ref. 31).
In Situ Hybridization.
Parasite material and slide preparation, RNA probe synthesis, and ISH procedures were performed as described previously (34). For these experiments, 10-μm frozen sections cut from adult B. malayi worms were used. Digoxigenin-labeled RNA probes were synthesized from a 467-bp fragment of BmFeCH that was subcloned by the following primer pair: forward primer, 5′-CGAATGCTTCCATTTTCGAT-3′; reverse primer, 5′-TCGAAAAGAATCGGCAAGTT-3′. Sense and antisense RNA probes were used for ISH, and the sense probe served as a negative control.
B.malayi RNAi.
B. malayi females were soaked in 1 μM BmFeCH hsiRNA for 2 d (changing the medium and RNA every 12 h). The hsiRNA mixtures were prepared from (i) a 514-bp fragment corresponding to nucleotide positions 170–683 of the BmFeCH ORF, and (ii) a 393-bp fragment corresponding to positions 663–1055 of the ORF. Methods for preparing hsiRNA mixtures for use in RNAi and the subsequent fixation and staining protocols have been described previously (35). Two different preparations of hsiRNA derived from the 5′ end of BmFeCH were used in separate experiments whereas hsiRNA derived from the 3′ region was used once. Controls included worms similarly cultured both in the absence of any RNA or in the presence of 1 μM control hsiRNA (Lit28i Polylinker ShortCut siRNA Mix; New England Biolabs).
C.elegans Strains, Constructs, and Transgenesis.
C. elegans N2 strain var. Bristol was obtained from the C. elegans stock-center (Caenorhabditis Genetics Center, University of Minnesota) and cultured by standard methods unless otherwise stated. unc-119(ed3) mutants were used in this work (36). The following transgenic C. elegans strains were also generated and used:
IP633: unc-119(ed3), nbEx172 [pDP#MM016b; psur5NTER-BmFeCH-GFP]
IP634: unc-119(ed3), nbEx173 [pDP#MM016b, psur5BmFeCH-GFP]
IP637: nbIs171 [pDP#MM016b; psur5BmFeCH-GFP].
Integrant line IP637 was generated from strain IP634 using the standard UV light array integration procedure (37)
A general backbone for all C. elegans transgenic constructs was prepared by modifying the GFP expression vector pPD95.75 (gift of Andrew Fire Department of Genetics, Stanford University, Palo Alto, CA). Constructs were injected into unc-119(ed3) worms according to standard protocols (38). Plasmid pDP#MM016b, carrying a unc-119(+) rescue construct (36), was used as a cotransformation marker. For each construct, at least two independent transgenic lines were established and inspected for consistency in expression patterns; a single line was used in further analysis as a reference strain.
C.elegans Microscopy.
Mitochondrial in vivo staining for colocalization experiments was performed by growing worms in the dark on nematode growth medium (NGM) standard plates containing MitoTracker Red CMXRos (2 μg/mL; Molecular Probes). Differential interference contrast (DIC) micrographs were acquired with a Zeiss Axiovert 200MT microscope (Zeiss) equipped with epifluorescence and processed using the Axiovision package release 4.5. Confocal pictures were acquired with a LSM 510 META laser scanning microscope (Zeiss) and processed with the LSM Zeiss package release 4.2.
Axenic Growth of C. elegans.
Axenic growth of C. elegans was by established procedure (39) with some minor modifications. Axenic cultures were produced using the standard sodium hypochlorite treatment (40). Eggs were allowed to hatch overnight in axenic basal medium (ABM) in the absence of a heme source. The concentration of live L1 larvae was adjusted to obtain an average concentration of 15–30 animals per 100 μL. A porphyrin source was added in the form of hemin chloride or PPIX at a final concentration of 10 mg/L, equivalent to 15.3 μM hemin chloride and 17.8 μM PPIX. A control lacking any porphyrin was also included. Worms were cultured in 96-well plates in a 100 μL volume for 11 d at 25 °C. Axenic medium supplemented with hemin supports slow growth of C. elegans, allowing for the completion of only one generation cycle in this period. Twenty-four wells were cultured for a given strain and condition. The number of parental animals reaching adulthood was scored and averaged. NMMP inhibitor assays were performed by supplementing the cultures containing hemin or PPIX with NMMP at 10 μM and assaying growth of one generation of worms as described above. A solvent-control was included in the NMMP test.
Supplementary Material
Acknowledgments
We thank T. A. Dailey and A. Luck for constructive comments on the manuscript and D. Comb, W. Jack, and J. Ellard for their interest and support. This work was funded by New England BioLabs, Inc., NIH Grant DK96501 (to H.A.D.), and by a grant from the Bill and Melinda Gates Foundation (to the Liverpool School of Tropical Medicine as part of the Anti-Wolbachia Consortium).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The FeCH cDNA sequences reported in this paper have been deposited in the GenBank database [accession nos. GQ895739 (Brugia malayi), GQ895740 (B. malayi variant FeCH), GQ895742 (Onchocerca volvulus), GQ895743 (Dirofilaria immitis), and GQ895741 (Acanthocheilonema viteae)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304049110/-/DCSupplemental.
References
- 1.Lambshead PJD. Marine nematode diversity. In: Chen ZX, Chen SY, Dickson DW, editors. Nematology: Advances and Perspectives. Vol 1. Wallingford, UK: CABI Publishing; 2004. pp. 439–468. [Google Scholar]
- 2.Danchin EG, et al. Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proc Natl Acad Sci USA. 2010;107(41):17651–17656. doi: 10.1073/pnas.1008486107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whiteman NK, Gloss AD. Parasitology: Nematode debt to bacteria. Nature. 2010;468(7324):641–642. doi: 10.1038/468641a. [DOI] [PubMed] [Google Scholar]
- 4.Ghedin E, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317(5845):1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao AU, Carta LK, Lesuisse E, Hamza I. Lack of heme synthesis in a free-living eukaryote. Proc Natl Acad Sci USA. 2005;102(12):4270–4275. doi: 10.1073/pnas.0500877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster J, et al. The Wolbachia genome of Brugia malayi: Endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3(4):e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenn K, et al. Phylogenetic relationships of the Wolbachia of nematodes and arthropods. PLoS Pathog. 2006;2(10):e94. doi: 10.1371/journal.ppat.0020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunning Hotopp JC, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317(5845):1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 9.McNulty SN, et al. Endosymbiont DNA in endobacteria-free filarial nematodes indicates ancient horizontal genetic transfer. PLoS ONE. 2010;5(6):e11029. doi: 10.1371/journal.pone.0011029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hotez PJ, Fenwick A, Savioli L, Molyneux DH. Rescuing the bottom billion through control of neglected tropical diseases. Lancet. 2009;373(9674):1570–1575. doi: 10.1016/S0140-6736(09)60233-6. [DOI] [PubMed] [Google Scholar]
- 11.Molyneux DH, Bradley M, Hoerauf A, Kyelem D, Taylor MJ. Mass drug treatment for lymphatic filariasis and onchocerciasis. Trends Parasitol. 2003;19(11):516–522. doi: 10.1016/j.pt.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Osei-Atweneboana MY, et al. Phenotypic evidence of emerging ivermectin resistance in Onchocerca volvulus. PLoS Negl Trop Dis. 2011;5(3):e998. doi: 10.1371/journal.pntd.0000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwab AE, Boakye DA, Kyelem D, Prichard RK. Detection of benzimidazole resistance-associated mutations in the filarial nematode Wuchereria bancrofti and evidence for selection by albendazole and ivermectin combination treatment. Am J Trop Med Hyg. 2005;73(2):234–238. [PubMed] [Google Scholar]
- 14.Dailey HA, Dailey TA. In: Ferrochelatase: The Porphyrin handbook. Kadish KM, Smith KM, Guilard R, editors. Vol 12. New York: Academic; 2003. pp. 93–121. [Google Scholar]
- 15.Elsworth B, Wasmuth J, Blaxter M. NEMBASE4: The nematode transcriptome resource. Int J Parasitol. 2011;41(8):881–894. doi: 10.1016/j.ijpara.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Nagayasu E, et al. Transcriptomic analysis of four developmental stages of Strongyloides venezuelensis. Parasitol Int. 2013;62(1):57–65. doi: 10.1016/j.parint.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Dailey TA, Woodruff JH, Dailey HA. Examination of mitochondrial protein targeting of haem synthetic enzymes: In vivo identification of three functional haem-responsive motifs in 5-aminolaevulinate synthase. Biochem J. 2005;386(Pt 2):381–386. doi: 10.1042/BJ20040570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dailey HA, Fleming JE. Bovine ferrochelatase. Kinetic analysis of inhibition by N-methylprotoporphyrin, manganese, and heme. J Biol Chem. 1983;258(19):11453–11459. [PubMed] [Google Scholar]
- 19.Nakahigashi K, Nishimura K, Miyamoto K, Inokuchi H. Photosensitivity of a protoporphyrin-accumulating, light-sensitive mutant (visA) of Escherichia coli K-12. Proc Natl Acad Sci USA. 1991;88(23):10520–10524. doi: 10.1073/pnas.88.23.10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hieb WF, Stokstad EL, Rothstein M. Heme requirement for reproduction of a free-living nematode. Science. 1970;168(3927):143–144. doi: 10.1126/science.168.3927.143. [DOI] [PubMed] [Google Scholar]
- 21.Sellers VM, Wu CK, Dailey TA, Dailey HA. Human ferrochelatase: Characterization of substrate-iron binding and proton-abstracting residues. Biochemistry. 2001;40(33):9821–9827. doi: 10.1021/bi010012c. [DOI] [PubMed] [Google Scholar]
- 22.Wu B, et al. The heme biosynthetic pathway of the obligate Wolbachia endosymbiont of Brugia malayi as a potential anti-filarial drug target. PLoS Negl Trop Dis. 2009;3(7):e475. doi: 10.1371/journal.pntd.0000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsson U, Billberg A, Sjövall S, Al-Karadaghi S, Hansson M. In vivo and in vitro studies of Bacillus subtilis ferrochelatase mutants suggest substrate channeling in the heme biosynthesis pathway. J Bacteriol. 2002;184(14):4018–4024. doi: 10.1128/JB.184.14.4018-4024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2(4):953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 26.Dereeper A, et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 28.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 29.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 30.McGarry HF, Egerton GL, Taylor MJ. Population dynamics of Wolbachia bacterial endosymbionts in Brugia malayi. Mol Biochem Parasitol. 2004;135(1):57–67. doi: 10.1016/j.molbiopara.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Rao R, Well GJ. In vitro effects of antibiotics on Brugia malayi worm survival and reproduction. J Parasitol. 2002;88(3):605–611. doi: 10.1645/0022-3395(2002)088[0605:IVEOAO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 32.Comley JC, Rees MJ, Turner CH, Jenkins DC. Colorimetric quantitation of filarial viability. Int J Parasitol. 1989;19(1):77–83. doi: 10.1016/0020-7519(89)90024-6. [DOI] [PubMed] [Google Scholar]
- 33.Townson S, Tagboto S, McGarry HF, Egerton GL, Taylor MJ. Onchocerca parasites and Wolbachia endosymbionts: Evaluation of a spectrum of antibiotic types for activity against Onchocerca gutturosa in vitro. Filaria J. 2006;5:4. doi: 10.1186/1475-2883-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang D, Li BW, Fischer PU, Weil GJ. Localization of gender-regulated gene expression in the filarial nematode Brugia malayi. Int J Parasitol. 2008;38(5):503–512. doi: 10.1016/j.ijpara.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Landmann F, Foster JM, Slatko BE, Sullivan W. Efficient in vitro RNA interference and immunofluorescence-based phenotype analysis in a human parasitic nematode, Brugia malayi. Parasit Vectors. 2012;5:16. doi: 10.1186/1756-3305-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141(3):977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitani S, Du H, Hall DH, Driscoll M, Chalfie M. Combinatorial control of touch receptor neuron expression in Caenorhabditis elegans. Development. 1993;119(3):773–783. doi: 10.1242/dev.119.3.773. [DOI] [PubMed] [Google Scholar]
- 38.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10(12):3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houthoofd K, et al. Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans. Exp Gerontol. 2002;37(12):1371–1378. doi: 10.1016/s0531-5565(02)00173-0. [DOI] [PubMed] [Google Scholar]
- 40.Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1988. pp. 587–606. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.