Abstract
Bacterial capsules are surface layers made of long-chain polysaccharides. They are anchored to the outer membrane of many Gram-negative bacteria, including pathogens such as Escherichia coli, Neisseria meningitidis, Haemophilus influenzae, and Pasteurella multocida. Capsules protect pathogens from host defenses including complement-mediated killing and phagocytosis and therefore represent a major virulence factor. Capsular polysaccharides are synthesized by enzymes located in the inner (cytoplasmic) membrane and are then translocated to the cell surface. Whereas the enzymes that synthesize the polysaccharides have been studied in detail, the structure and biosynthesis of the anchoring elements have not been definitively resolved. Here we determine the structure of the glycolipid attached to the reducing terminus of the polysialic acid capsular polysaccharides from E. coli K1 and N. meningitidis group B and the heparosan-like capsular polysaccharide from E. coli K5. All possess the same unique glycolipid terminus consisting of a lyso-phosphatidylglycerol moiety with a β-linked poly-(3-deoxy-d-manno-oct-2-ulosonic acid) (poly-Kdo) linker attached to the reducing terminus of the capsular polysaccharide.
Keywords: polysaccharide export, capsule assembly
Gram-negative bacteria exploit a variety of virulence factors to colonize particular niches and evade host defenses. One common virulence factor is the capsule, which consists of long-chain capsular polysaccharides (CPSs) anchored in the outer membrane. CPSs are often important for preventing phagocytosis and complement-mediated killing and thus represent an attractive therapeutic target (1, 2).
In Gram-negative bacteria, CPSs are synthesized and translocated to the cell surface by one of two mechanisms; both have been well-studied in Escherichia coli (reviewed in refs. 3 and 4). E. coli isolates causing urinary tract infections, septicemia, and meningitis possess capsules that are assembled via the ATP-binding cassette (ABC) transporter–dependent pathway, which is the focus of this study. Similar assembly systems are found in Neisseria meningitidis, Haemophilus influenzae, and a variety of human and animal pathogens (3, 5). In a landmark paper, Silver and colleagues reported the cloning of the genetic locus for assembly of the E. coli serotype K1 capsule and expression of the K1 antigen in a heterologous host (6). This was the first full polysaccharide gene cluster cloned, and it opened up biochemical and molecular genetic strategies to investigate these and other bacterial glycans. Since then, the E. coli K1 and K5 systems have been influential prototypes for studying CPS assembly via ABC transporter-dependent pathways (3, 4). E. coli K1 CPS consists of polysialic acid (PSA), a homopolymer of α-(2→8)-linked sialic acid (NeuAc), and K5 is composed of a heparosan-like glycan containing glucuronic acid (GlcA) and N-acetylglucosamine (GlcNAc) (7, 8). Identical CPS structures can also be found in other bacteria. For example, α-(2→8)-linked PSA is also produced by N. meningitidis serogroup B and Pasteurella haemolytica serogroup A2 (9, 10), whereas P. multocida type D produces a nonsulfated heparosan CPS polymer (11).
Biosynthesis of these CPSs occurs at the cytoplasmic (inner) membrane, before its export to the periplasm by the system-defining ABC transporter (comprising proteins KpsM and KpsT in E. coli nomenclature) (3, 4). Translocation of CPS from the periplasm to the cell surface requires the periplasmic and outer-membrane proteins KpsE and KpsD. Together, KpsMTED are predicted to form a transenvelope complex (3, 4, 12, 13). KpsMTED functions are not confined to a given CPS repeat-unit structure, and one possible explanation of their broad substrate specificity is the presence of a conserved lipid terminus that may be recognized by the ABC transporter (3, 5, 14, 15). This lipid has been implicated in anchoring CPSs to the outer membrane (16). Mass spectrometry analysis of acid-hydrolyzed PSA from E. coli K1 and K92, as well as N. meningitidis group B, identified dipalmitoylglycerol as a component (17–20). However, direct covalent linkage between the CPS and this lipid has not been established. As an added complication, experiments with E. coli K5 CPS suggested that a 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) residue may exist at the reducing end of the polymer (19, 21), but no definitive chemical structures have been reported. All E. coli wild-type strains require cytidine-5′-monophosphate (CMP)-Kdo as a precursor for the biosynthesis of lipopolysaccharide, which is essential for viability (22), but the genetic loci encoding ABC transporter-dependent CPS assembly pathways in E. coli contain additional copies of genes encoding two of the four enzymes in the CMP-Kdo biosynthesis pathway (3). Although the correlation between the duplicated genes and the proposed terminal Kdo residue has been noted, it does not represent a unifying feature for all bacteria containing these CPS assembly systems because other examples (e.g., N. meningitidis) lack the duplication. This has raised the possibility of subtle variations in reducing terminal structures on different CPSs assembled through a common strategy.
To fully understand the assembly pathway, it is critical to know the precise nature of the lipid terminus and its linkage to the CPS glycan. To that end, we have determined the structure of the reducing terminus of CPSs from E. coli K1 and K5 and N. meningitidis group B, to ask whether they possess an identical lipid terminus. The analysis revealed a unique glycolipid terminus conserved in all three bacteria.
Results
Identification of a Conserved Lipid Terminus.
Structural characterization of a lipid terminus and its linkage region is not feasible with heterogeneous preparations containing high-molecular-mass CPS glycans. As a result, prior studies have investigated material released from CPS preparations treated with acid. Although acid hydrolysates yield information on individual components, they provide no insight into the linkage. Therefore, we developed a strategy that generated highly purified CPS and then reduced the contribution of the CPS with specific endo-acting CPS depolymerases. These glycanases are tail-spike proteins from E. coli K1 and K5 CPS-specific bacteriophages (23, 24). They rapidly depolymerize purified CPS (Fig. S1), but leave the terminal lipid (and any linker domain) intact and connected to the first few residues of the CPS repeat unit. The hydrophobic products from these enzyme digests were purified and analyzed by mass spectrometry (MS).
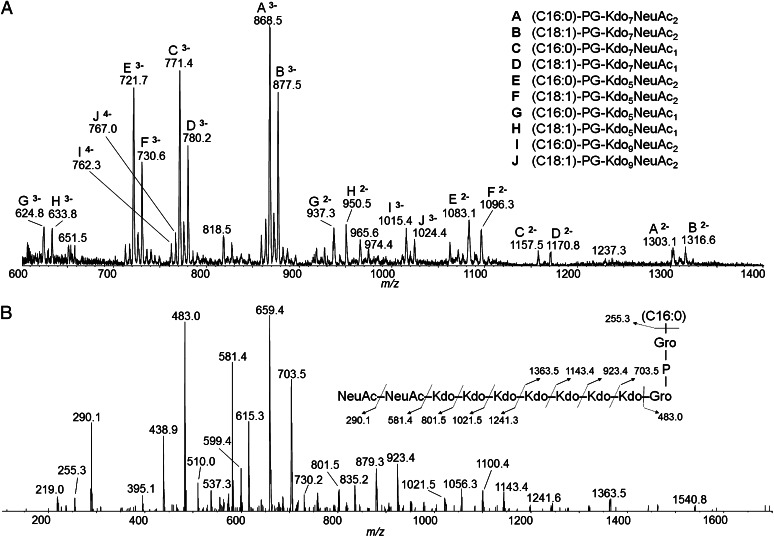
The liquid chromatography (LC)-MS spectrum of the E. coli K1 terminus showed six major species and several minor ones (Fig. 1A). Lyso-phosphatidylglycerol (lyso-PG) was detected together with an unanticipated poly-Kdo linker. As expected, each species contained one or more NeuAc residues from the CPS. Ions A/B and C/D correspond to the calculated masses of lyso-phosphatidylglycerol linked to seven Kdo sugars and either two or one NeuAc residue(s), respectively. MS/MS was performed on these ions to confirm the assignments (Fig. 1B and Fig. S2). The spectrum for ion A revealed characteristic ions corresponding to Kdo and NeuAc, in addition to a major ion at m/z 483, corresponding to the mass of lyso-PG containing palmitate as the acyl chain. MS/MS/MS of the m/z 483 ion confirmed that it is indeed palmitoyl-phosphatidylglycerol, based on the characteristic fragment ions: glycerol2-PO4 (m/z 227) and palmitate (m/z 255) (Fig. S3). Also detected in the MS/MS spectrum of ion A were ions corresponding to lyso-PG linked to multiple Kdo residues, as well as multiple Kdo residues linked to NeuAc, identifying a poly-Kdo linker between the PSA glycan and the lipid moiety. The difference between ions A and B lies in the identity of the acyl chain; ion A contains C16:0, whereas B contains C18:1 (Fig. 1B and Fig. S2). The same is true for ions C and D. In addition to the molecular species containing seven Kdo residues, the spectrum also contained ions corresponding to lyso-PG-Kdo5-NeuAcn (labeled E, F, G, and H) and lyso-PG-Kdo9-NeuAcn (I and J), where n is one or two NeuAc residues (Fig. 1A). In terms of both intensity and number of ions found, the predominant species contained five and seven Kdo residues, whereas nine was less abundant. Interestingly, there were no ions present corresponding to two, four, six, or eight Kdo residues. These experiments were repeated with two additional independent batches of E. coli K1 CPS. The number of Kdo residues was consistent, but one batch contained both lyso-PG and the diacyl-PG anticipated from previous reports (18, 19) (Fig. S4).
Fig. 1.
Identification of the terminal glycolipid structure isolated from E. coli K1 CPS by mass spectrometry. (A) LC-MS data for E. coli K1 CPS terminus. All indicated ions correspond to lyso-phosphatidylglycerol attached to a poly-Kdo linker and one or more NeuAc residues. The composition of the acyl chain is in parentheses and the number of Kdo and NeuAc residues in each ion is indicated. The charge of the ion is shown next to the letter identifying it. (B) LC-MS/MS data for the m/z 868.5 ion. Glycerol and phosphate are designated Gro and P, respectively.
To determine whether the terminal structure is conserved in different genera, CPS was purified from N. meningitidis 992B and the lipid terminus was analyzed by LC-MS (Table 1 and Fig. S5A). The overall features of the terminus were the same, but subtle differences were seen. For example, the acyl chain was more homogeneous (mostly C16:0), and the Kdo5 species was predominant in the spectrum. In addition, endosialidase digestion was not as effective in comparison with K1 CPS (i.e., all ions contained at least two NeuAc residues). E. coli K5 CPS was investigated to test whether the linker was specific to PSA CPS or a more general feature of CPSs synthesized via the ABC transporter-dependent pathway (Table 1 and Fig. S5B). LC-MS revealed the terminal structure was also conserved in this strain but the predominant ions corresponded to a Kdo6 species, with minor ions corresponding to a Kdo8 species. No ions corresponding to an odd number of Kdo residues were found.
Table 1.
Identification of ions in the LC-MS spectra for the terminal glycolipids from N. meningitidis group B, E. coli K5, and E. coli K1 ΔkpsT CPSs
| Linker | Expected m/z | Observed m/z | Species |
| N. meningitidis serogroup B | |||
| Kdo5 | 721.73− | 721.43− | l-PG(C16:0)-Kdo5-NeuAc2 |
| 1,083.02− | 1,082.62− | ||
| 730.33− | 730.73− | l-PG(C18:1)-Kdo5-NeuAc2 | |
| 1,096.02− | 1,096.42− | ||
| 818.73− | 818.43− | l-PG(C16:0)-Kdo5-NeuAc3 | |
| 915.83− | 915.43− | l-PG(C16:0)-Kdo5-NeuAc4 | |
| Kdo7 | 868.43− | 867.93− | l-PG(C16:0)-Kdo7-NeuAc2 |
| E. coli K5 | |||
| Kdo6 | 853.83− | 853.23− | l-PG(C16:0)-Kdo6-(GlcNAc-GlcA)2 |
| 862.43− | 862.13− | l-PG(C18:1)-Kdo6-(GlcNAc-GlcA)2 | |
| 980.23− | 979.63− | l-PG(C16:0)-Kdo6-(GlcNAc-GlcA)3 | |
| 988.93− | 988.93− | l-PG(C18:1)-Kdo6-(GlcNAc-GlcA)3 | |
| 829.54− | 829.34− | l-PG(C16:0)-Kdo6-(GlcNAc-GlcA)4 | |
| 1,106.63− | 1,106.03− | ||
| 1,115.33− | 1,115.23− | l-PG(C18:1)-Kdo6-(GlcNAc-GlcA)4 | |
| Kdo8 | 1,000.53− | 1,000.23− | l-PG(C16:0)-Kdo8-(GlcNAc-GlcA)2 |
| 1,127.03− | 1,126.73− | l-PG(C16:0)-Kdo8-(GlcNAc-GlcA)3 | |
| 1,135.73− | 1,135.73− | l-PG(C18:1)-Kdo8-(GlcNAc-GlcA)3 | |
| 939.64− | 939.64− | l-PG(C16:0)-Kdo8-(GlcNAc-GlcA)4 | |
| 1,253.43− | 1,252.93− | ||
| E. coli K1 ΔkpsT | |||
| Kdo5 | 937.42− | 936.62− | l-PG(C16:0)-Kdo5-NeuAc1 |
| 950.42− | 950.22− | l-PG(C18:1)-Kdo5-NeuAc1 | |
| 721.73− | 721.63− | l-PG(C16:0)-Kdo5-NeuAc2 | |
| 1,083.02− | 1,083.02− | ||
| 730.33− | 730.73− | l-PG(C18:1)-Kdo5-NeuAc2 | |
| 1,096.02− | 1,095.92− | ||
| 818.73− | 818.93− | l-PG(C16:0)-Kdo5-NeuAc3 | |
| Kdo7 | 771.43− | 771.53− | l-PG(C16:0)-Kdo7-NeuAc1 |
| 1,157.52− | 1,158.02− | ||
| 1,170.52− | 1,171.12− | l-PG(C18:1)-Kdo7-NeuAc1 | |
| 868.43− | 868.03− | l-PG(C16:0)-Kdo7-NeuAc2 | |
| 877.13− | 877.33− | l-PG(C18:1)-Kdo7-NeuAc2 | |
| Kdo9 | 1,015.23− | 1,015.33− | l-PG(C16:0)-Kdo9-NeuAc2 |
Structural Analysis of the K1 and K5 Glycolipid Termini.
Composition analysis of trimethylsilyated (TMS)-derivatized hydrophobic moieties in the E. coli K1 sample confirmed the presence of C16:0 fatty acids, glycerol, Kdo, and NeuAc. To identify the linkage of the Kdo residues, methylation analysis of the K1 hydrophobic moiety was performed using the Ciucanu and Kerek method with modifications to preserve the linkages of NeuAc and Kdo (25–27). Roughly equal amounts of 1,2,4,6-tetra-O-acetyl-3-deoxy-5,7,8-tri-O-methyl-octitol and 1,2,6,7-tetra-O-acetyl-3-deoxy-4,5,8-tri-O-methyl-octitol were observed (ratio of 1:1.38), suggestive of an alternating polymer of 2,4- and 2,7-linked Kdo (Fig. S6). A small amount of terminal Kdo was also detected, indicative of some hydrolysis in the sample. The presence of characteristic fragment ions in electron-impact MS confirmed the assignment of 4-, 7-, and terminal Kdo species, and chemical-ionization MS confirmed that all molecular ions were consistent with monolinked Kdo.
To determine the anomeric configuration of the Kdo residues, the K1 and K5 termini were further analyzed by NMR spectroscopy. In the E. coli K1 CPS spectrum, the NeuAc signals are distinct from the Kdo, with the H3 protons from NeuAc at 1.72 (H3 axial) and 2.76 ppm (H3 equatorial) (28). Based on the region of the 1H-1H gradient correlation spectroscopy (gCOSY) and gradient total correlation spectroscopy (gTOCSY) spectra from 1.81 to 1.92 ppm, there are two species in solution that contain signals consistent with Kdo methylene protons (Table 2 and Fig. S7). Both have a difference of >0.5 ppm between the axial and equatorial H3 protons, consistent with β-linkages (29). Unfortunately, the ratio between the major and minor signals could not be determined, due to spectral overlap inherent in the heterogeneous samples. This also prevented assignment of linkages via NMR. Analysis of the E. coli K5 glycolipid indicated that there are at least five species in solution that contain signals consistent with Kdo methylene protons, and these are also consistent with β-linkages (Fig. S8).
Table 2.
1H chemical shifts (ppm) of the two Kdo species in the terminal glycolipid from E. coli K1 CPS
Examination of the Glycolipid Terminus in CPS from Mutants with Defects in the Biosynthesis and Export Pathways.
To provide insight into the structure of the glycolipid terminus pre-export, we examined intracellular CPS accumulated in an E. coli K1 ΔkpsT mutant. KpsT is the polypeptide containing the nucleotide-binding domain for the ABC transporter, and ΔkpsT mutants produce CPS which remains trapped inside the cell (30). The glycolipid terminus of CPS from the ΔkpsT mutant is also monoacylated (Table 1).
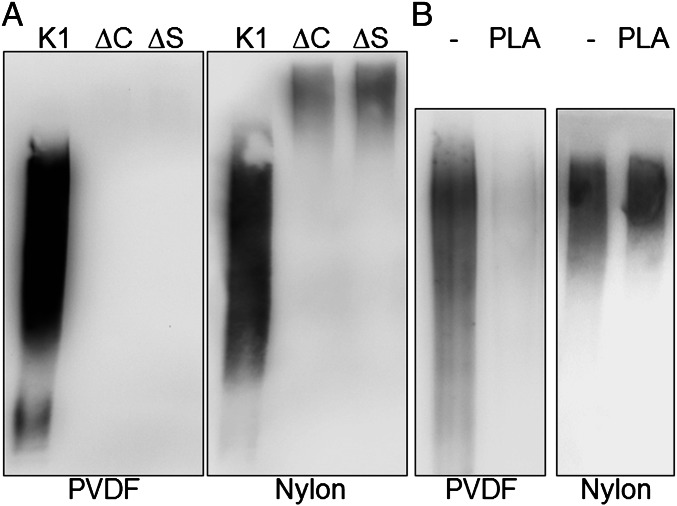
KpsC and KpsS are highly conserved proteins in ABC transporter-dependent CPS assembly systems. They have been implicated in lipidation of CPS in E. coli, but their exact function is unknown (21, 31). To determine whether they were required for synthesizing the glycolipid terminus, ΔkpsC and ΔkpsS mutants were constructed in E. coli K1. As observed previously (32), the mutants are resistant to K1-specific bacteriophage due to a CPS export defect, and each mutant could be complemented in trans, to restore surface CPS (and bacteriophage sensitivity), indicating that the deletions are nonpolar (Fig. S9). CPS from these mutants was first analyzed by immunoblotting with whole-cell extracts and probing with a PSA-specific monoclonal antibody. Initial analyses with immunoblots on polyvinylidine difluoride (PVDF) membranes yielded a strong signal for the parent CPS but no signal for either mutant (Fig. 2A), differing from the established phenotype (33). In contrast, signal was obtained for all samples when transferred to a positively charged nylon membrane, although the apparent molecular masses of the CPSs from the ΔkpsC and ΔkpsS mutants were significantly higher than that of the parent (Fig. 2A). The higher apparent molecular masses are consistent with previous reports for the corresponding mutants (lipA and lipB) in N. meningitidis (31). Similar effects have also been seen in E. coli strains with mutations affecting later stages of CPS export (kpsE) (32). The reason for this is unknown, but it could reflect unregulated chain extension when synthesis and export are uncoupled. When purified CPS from the ΔkpsC mutant was processed, no lipid moiety was isolated, suggesting the difference in behavior on PVDF immunoblots reflected a change in physical properties (i.e., a decrease in hydrophobicity of CPS in the mutants). The same behavior was obtained when parental K1 CPS was pretreated with commercial phospholipase A (PLA), the enzyme that catalyzes the removal of acyl chains from phospholipids (Fig. 2B). Removal of the acyl chain is apparently sufficient to abrogate binding to PVDF, consistent with the conclusion that PSA from ΔkpsC and ΔkpsS mutants lacks the lipid terminus.
Fig. 2.
Determination of the lipidation state of E. coli K1 CPS in ∆kpsC and ∆kpsS mutants. (A) Immunoblots of whole-cell lysates on PVDF or positively charged nylon membranes. These were probed with the monoclonal α-PSA mAb 2-2B. Whereas CPS from the parent (K1) binds to both membranes, CPSs from the mutants (∆C and ∆S) do not bind to PVDF. (B) PLA treatment of purified K1 CPS is sufficient to eliminate binding to PVDF.
Discussion
We have demonstrated that the E. coli K1 and K5 and N. meningitidis group B CPSs are attached to a (predominantly) lyso-phosphatidylglycerol moiety via a unique poly-Kdo linker. Central to this discovery is the development of a technical approach to isolate the intact glycolipid without resorting to treatment with extreme pH or complicated chemical derivatization. The lipid moiety was identical in all three bacteria, but considerable variation was observed in the number of Kdo residues both within and between species.
PG is the second-most common glycerophospholipid in E. coli, accounting for ∼25% and 10% of the inner- and outer-membrane phospholipids, respectively (34). The biosynthesis of E. coli glycerophospholipids proceeds through cytidine-5′-diphosphate (CDP)-diacylglycerol, and lyso-phospholipids, which are comparatively rare, are made from the diacylated species through the action of PLA (35). E. coli possesses two PLA activities. The known outer-membrane PLA enzyme (35) was ruled out by the identification of lyso-PG in the terminal glycolipid of CPS isolated from the corresponding pldA mutant (Fig. S10). Detection of lyso-PG at the terminus of intracellular CPS in the kpsT mutant suggests removal of an acyl chain before export, implicating a currently unidentified cytoplasmic PLA. Earlier studies predicted that the terminus of PSA CPS was diacylated, but the parent ion for the intact PG structure was not detected (17). A subsequent report clearly shows a diacylated phospholipid in N. meningitidis group B CPS (18). The varying results could be explained by the use of different isolates with variation in PLA activities in these studies. Collectively, the data suggest that lyso-PG predominates, but CPS export can apparently occur with either lyso- or diacyl-PG at the terminus.
The lipid terminus has been implicated in mediating attachment of CPS to the cell surface to form the capsule (16, 19), but the reported acid lability of the linkage between the lipid and the CPS (19) may lead to release of material into the culture during growth under laboratory conditions, making it difficult to determine the proportion of CPS covalently linked to the cell surface in vivo. The presence of a terminal lipid does not preclude other factors from participating in surface association of CPSs. For example, one report has suggested that E. coli K1 CPS associates by ionic interactions with the lipopolysaccharide (LPS) core region (36). Sequential truncations of the LPS core structure were used to define the minimal requirement for CPS binding. However, the effect of these truncations on outer-membrane integrity was not examined, so it is currently not possible to rule out a potentially indirect influence on CPS association.
Several reports have suggested that Kdo biosynthesis plays a role in formation of CPS synthesized in the ABC transporter-dependent pathway, but the molecular basis has not been established (21, 33, 37). Establishing the requirement in E. coli is complicated by CMP-Kdo being essential for LPS biosynthesis and viability in wild-type bacteria (38), and by the duplication of genes encoding enzymes involved in CMP-Kdo biosynthesis in the kps locus. This limitation does not exist in N. meningitidis (37), where defects in the CMP-Kdo biosynthesis pathway lead to significant reductions in the amount of PSA CPS (37). However, it was not determined whether the residual CPS was lipidated, nor whether it was translocated to the cell surface.
Unraveling the initial steps in biosynthesis of E. coli CPSs in ABC transporter-dependent pathways has been complicated by conflicting reports concerning the involvement of undecaprenol-linked intermediates. Undecaprenyl monophospho-NeuAc has been proposed as the acceptor for the glycosyltransferases that synthesize the CPS glycan in the K1 system (39), invoking an elaborate two-step process involving transfer of such intermediates to a phospholipid (21, 37). Presumably such a process would require a dedicated “ligase” enzyme, and there is no obvious candidate for this. Furthermore, studies in the E. coli K5 system could identify no involvement of undecaprenol-linked intermediates (21). It has therefore been proposed that these CPSs are polymerized on a phospholipid-based acceptor (40), akin to the Streptococcus pneumoniae type 3 CPS (41). This hypothesis is now complicated by the existence of the poly-Kdo linker. kpsC and kpsS mutants accumulate large aggregates of intracellular CPS (20, 29), but there have been conflicting reports in the past as to whether the CPS is lipidated (18, 31, 33); here we provide evidence that these CPSs are not lipidated in E. coli. Notably, the intracellular aggregates from these mutants lack the characteristic membrane association seen in the aggregates of lipidated CPSs produced by export-deficient mutants (e.g., kpsT) (33). Experiments to reconstruct capsule synthesis in an E. coli background that lacks any capsule biosynthesis genes have shown that kpsC and kpsS are essential for de novo synthesis of CPS in an in vitro (membrane preparation) system (40). The difference in the in vivo and in vitro outcomes is surprising. One explanation is that lyso-PG-Kdon represents the natural acceptor on which these CPSs are polymerized and, in its absence, aberrant initiation may occur in vivo using an unknown acceptor, which may not be membrane-associated. If such a scenario occurs, then KpsC and KpsS could provide a scaffold to allow proper assembly of lyso-PG-Kdon by other (unknown) components. Both proteins have been reported to interact with proteins involved in biosynthesis and export of the final product (12, 42). Alternatively, KpsC and KpsS could be CMP-Kdo–dependent glycosyltransferases.
Kdo is a component of the conserved region of LPS molecules, which are essential for outer-membrane integrity and cell viability in Gram-negative bacteria. There is some variability across bacterial species in the number and linkages of Kdo residues in this part of LPS, but all involve α-linked residues (22). Kdo can also be part of the O antigen repeat unit in some species of bacteria, such as Providencia alcalifaciens O36, and in several CPS antigens, where it can be either α- or β-linked (43–45). Sinorhizobium meliloti strain 1021 rkpZ mutants even produce a “low-molecular-mass” CPS composed of 8–15 β-(2→7)–linked Kdo residues (29), but the glycosyltransferases responsible for synthesizing these structures have not been determined. All Kdo transferases characterized to date are involved in formation of the LPS inner core (Carbohydrate-Active EnZymes database families GT30 and GT73) from CMP-β-Kdo and, as such, are inverting enzymes (46). It is not surprising then that KpsC and KpsS proteins have no significant homology to any known Kdo transferase, because the poly-Kdo linker for these CPSs contains β-linkages.
In summary, we have shown that the reducing termini of representative CPSs from E. coli and N. meningitidis contain lyso-phosphatidylglycerol-poly-Kdo, and that KpsC and KpsS are required for its synthesis/addition. These findings begin to explain various contradictory genetic and biochemical information and address some open questions for arguably the most influential CPS assembly system in microbial glycobiology. We predict that this structure represents a unifying feature of all capsules assembled in an ABC transporter-dependent process.
Materials and Methods
Bacterial Strains.
E. coli EV36 (serotype K1) and N. meningitidis 992B (serogroup B) NRCC4726 were gifts from E. Vimr (University of Illinois at Champaign-Urbana, Urbana, IL) and W. Wakarchuk (Ryerson University, Toronto), respectively. E. coli Bi8337/41 (O10:K5:H4) was obtained from the Statens Serum Institut. E. coli isolates were grown in 1 L LB at 37 °C for 5 h before harvesting. N. meningitidis was grown on GC plates supplemented with 1% glucose at 37 °C in 5% (vol/vol) CO2 for 18 h. E. coli K1ΔkpsC, ΔkpsS, and ΔpldA mutants were made using the λ-red recombinase procedure (47, 48). The complete coding sequences of both kpsC and kpsS were replaced with the Kan cassette, whereas only the internal 54–821 bp of pldA was replaced (SI Materials and Methods). Mutants were confirmed by sequencing.
Biochemical Experiments.
Bacteriophage K1F-sensitivity assays and immunoblotting were performed as previously described (28). The PSA-specific monoclonal antibody 2-2B used in immunoblotting was a gift from M. Apicella (University of Iowa, Iowa City, IA) and is available through EMD Millipore. Phospholipase A reactions were performed in 25 mM Tris⋅HCl (pH 8) containing 0.4 mg/mL bovine phospholipase A2 (Sigma-Aldrich) at 37 °C for 16 h. The expression and purification of maltose binding protein (MalE)-K1 endoneuraminidase has been described previously (23). The gene encoding His6-K5 lyase was ordered from GeneArt (Invitrogen Canada) and cloned into pCW ori+ (49), and the enzyme was expressed and purified as described elsewhere (24).
Isolation of the Glycolipid Terminus.
CPS was purified using a modified Cetavlon-based method (18), followed by gel filtration (see SI Materials and Methods for additional details). Purified CPS was digested in 4 mM NH4OAc at 37 °C for 3–5 h and hydrophobic material was isolated from enzyme digests using a SepPak C8 cartridge (Waters); the bound glycolipid was eluted with 70% acetonitrile and dried.
Mass Spectrometry.
The dried glycolipid was resuspended in 50% ethanol and analyzed by LC-MS/MS using an UltiMate 3000 HPLC system (Thermo Scientific) linked to a 4000 QTRAP mass spectrometer (AB SCIEX). Chromatographic separation was performed on a Kappa BioBasic C18 column (Thermo Scientific) with dimensions of 150 × 0.32 mm. Mobile phase A consisted of 10 mM ammonium acetate in water. Mobile phase B consisted of 10 mM ammonium acetate in methanol. Gradient elution was performed at a flow rate of 8 µL/min. The initial composition of the gradient was 50% B for 3 min before a gradient raised the composition to 95% B over 1 min. The gradient was held at 95% B for 26 min. At 30 min, the gradient was returned to 50% B to reequilibrate the column for 10 min.
Composition Analysis.
The glycosyl composition of the two samples was determined at the Complex Carbohydrate Research Center (CCRC), University of Georgia, by preparing the TMS-methyl glycosides with GC-MS (electron-impact) analysis. Samples were dissolved in 1 M methanolic HCl using brief sonication. The sample was hydrolyzed for 16 h at 80 °C, and then subjected to N-acetylation and trimethylsilylation (Tri-Sil Reagent; Pierce). The resulting TMS-methyl glycosides were analyzed by GC-MS using a 30-m DB-1 capillary column equipped with a mass selective detector. For separation of the TMS-methyl glycoside derivatives, the DB-1 column was programmed from 80 °C to 260 °C over 50 min.
Methylation Analysis.
Linkage (methylation analysis) was also performed at the CCRC. Dried glycolipid was solubilized in dry DMSO with sonication/vortexing before methylation. Methylation reactions were performed using the sodium hydroxide-methyl iodide procedure (25) with modifications (26, 27). As used here, these modifications consist of adding the sodium hydroxide slurry in DMSO to the lyophilized LPS samples and reacting for a brief period (6–7 min) before the addition of excess methyl iodide. Following a 1-h exposure to methyl iodide, the samples were then worked up using modifications designed to preserve the labile Kdo- and NeuAc-ketosidic linkages in the following sequence of steps: (i) Reduction of the Kdo and NeuNAc carboxymethyl groups was performed with lithium triethylborodeuteride (Super-Deuteride; Sigma-Aldrich) in THF (2 h at room temperature); (ii) mild hydrolysis (0.1 M trifluoroacetic acid, 100 °C, 30 min) was used to cleave the Kdo/NeuAc-ketosidic linkages; (iii) reduction of Kdo/NeuAc residues at the C-2 carbonyl group was performed using NaBD4 in water/ethanol; (iv) normal hydrolysis conditions (2 M trifluoroacetic acid, 105 °C, 1 h) were used to cleave the remaining uronosyl and neutral sugar linkages (in the case of sample K5); (v) the newly formed aldehydo-sugars were reduced with NaBD4 in 50 mM NH4OH; and (vi) acetylation of the resulting partially methylated alditols was performed in acetonitrile-pyridine-acetic anhydride containing 4-N,N′-dimethylaminopyridine as catalyst for 3 h at room temperature as described (27). The resulting permethylated alditol acetate derivatives were analyzed by GC-MS (electron-impact) using a 30-m SP-2330 capillary column (Supelco) programmed from 80 to 250 °C. Chemical ionization GC-MS was performed on a 30-m DB-1 capillary column, and samples were also run on an equivalent DB-1 column in the electron-impact mode for comparison.
NMR Spectroscopy.
Spectra were acquired in D2O at 27 °C on an Agilent VNMRS 700-MHz spectrometer equipped with a cryoprobe. The spectra were referenced to an external standard of acetone (2.22 ppm for 1H, 31.07 for 13C). 1D-1H and 1H-1H gCOSY spectra were obtained for both samples, and a 1H-1H gTOCSY spectrum was acquired for the K1 sample. For the 1D, gCOSY, and gTOCSY spectra, the intensity of the residual deuterated water peak was decreased using a presaturation pulse sequence, irradiating at 4.76 ppm. The spectral window for the 1D spectra was 8,446 Hz (from 10.8 to –1.3 ppm), and a Gaussian function was applied interactively to improve the signal-to-noise ratio. Sine-bell functions were applied interactively to all of the 2D spectra to improve signal to noise. The spectral windows for the gCOSY and gTOCSY of the K1glycolipid were 4,562 Hz (from 6.5 to 0 ppm) in both dimensions, and the spectra were acquired with 600 increments in F1 and 16 transients in F2. The spectral window for the gCOSY of the K5 capsule anchor was 5,952 Hz (from 8.0 to 0.5 ppm) in both dimensions, and the spectrum was acquired with 634 increments in F1 and 32 transients in F2.
Supplementary Material
Acknowledgments
We thank Drs. John Kelly, Warren Wakarchuk, and Willie Vann for invaluable discussion. We also thank Drs. L. S. Forsberg and R. W. Carlson (Complex Carbohydrate Research Center) for performing the methylation analysis. This work was supported by funding from the Canadian Institutes of Health Research (FRN-9623 to C.W.) and the Alberta Glycomics Centre (to T.L.L.). C.W. holds a Canada Research Chair, and L.M.W. gratefully acknowledges a Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council. The work at the CCRC was supported by Department of Energy Grant DE-FG-02-93ER20097.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222317110/-/DCSupplemental.
References
- 1.Corbett D, Hudson T, Roberts IS. In: Prokaryotic Cell Wall Compounds. Konig H, Claus H, Varma A, editors. Heidelberg: Springer; 2010. pp. 111–132. [Google Scholar]
- 2.Goller CC, Seed PC. High-throughput identification of chemical inhibitors of E. coli group 2 capsule biogenesis as anti-virulence agents. PLoS One. 2010;5(7):e11642. doi: 10.1371/journal.pone.0011642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 4.Vimr ER, Steenbergen SM. Early molecular-recognition events in the synthesis and export of group 2 capsular polysaccharides. Microbiology. 2009;155(Pt 1):9–15. doi: 10.1099/mic.0.023564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuthbertson L, Kos V, Whitfield C. ABC transporters involved in export of cell surface glycoconjugates. Microbiol Mol Biol Rev. 2010;74(3):341–362. doi: 10.1128/MMBR.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silver RP, et al. Molecular cloning of the K1 capsular polysaccharide genes of E. coli. Nature. 1981;289(5799):696–698. doi: 10.1038/289696b0. [DOI] [PubMed] [Google Scholar]
- 7.McGuire EJ, Binkley SB. The structure and chemistry of colominic acid. Biochemistry. 1964;3:247–251. doi: 10.1021/bi00890a017. [DOI] [PubMed] [Google Scholar]
- 8.Vann WF, Schmidt MA, Jann B, Jann K. The structure of the capsular polysaccharide (K5 antigen) of urinary-tract-infective Escherichia coli 010:K5:H4. A polymer similar to desulfo-heparin. Eur J Biochem. 1981;116(2):359–364. doi: 10.1111/j.1432-1033.1981.tb05343.x. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharjee AK, Jennings HJ, Kenny CP, Martin A, Smith IC. Structural determination of the sialic acid polysaccharide antigens of Neisseria meningitidis serogroups B and C with carbon 13 nuclear magnetic resonance. J Biol Chem. 1975;250(5):1926–1932. [PubMed] [Google Scholar]
- 10.Aalto J, Pelkonen S, Kalimo H, Finne J. Mutant bacteriophage with non-catalytic endosialidase binds to both bacterial and eukaryotic polysialic acid and can be used as probe for its detection. Glycoconj J. 2001;18(10):751–758. doi: 10.1023/a:1021147316647. [DOI] [PubMed] [Google Scholar]
- 11.DeAngelis PL, Gunay NS, Toida T, Mao WJ, Linhardt RJ. Identification of the capsular polysaccharides of type D and F Pasteurella multocida as unmodified heparin and chondroitin, respectively. Carbohydr Res. 2002;337(17):1547–1552. doi: 10.1016/s0008-6215(02)00219-7. [DOI] [PubMed] [Google Scholar]
- 12.Rigg GP, Barrett B, Roberts IS. The localization of KpsC, S and T, and KfiA, C and D proteins involved in the biosynthesis of the Escherichia coli K5 capsular polysaccharide: Evidence for a membrane-bound complex. Microbiology. 1998;144(Pt 10):2905–2914. doi: 10.1099/00221287-144-10-2905. [DOI] [PubMed] [Google Scholar]
- 13.McNulty C, et al. The cell surface expression of group 2 capsular polysaccharides in Escherichia coli: The role of KpsD, RhsA and a multi-protein complex at the pole of the cell. Mol Microbiol. 2006;59(3):907–922. doi: 10.1111/j.1365-2958.2005.05010.x. [DOI] [PubMed] [Google Scholar]
- 14.Silver RP, Prior K, Nsahlai C, Wright LF. ABC transporters and the export of capsular polysaccharides from Gram-negative bacteria. Res Microbiol. 2001;152(3-4):357–364. doi: 10.1016/s0923-2508(01)01207-4. [DOI] [PubMed] [Google Scholar]
- 15.Roberts IS, Mountford R, Hodge R, Jann KB, Boulnois GJ. Common organization of gene clusters for production of different capsular polysaccharides (K antigens) in Escherichia coli. J Bacteriol. 1988;170(3):1305–1310. doi: 10.1128/jb.170.3.1305-1310.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jann B, Jann K. In: Bacterial Capsules. Jann K, Jann B, editors. Berlin: Springer; 1990. pp. 19–42. [Google Scholar]
- 17.Gotschlich EC, Fraser BA, Nishimura O, Robbins JB, Liu TY. Lipid on capsular polysaccharides of Gram-negative bacteria. J Biol Chem. 1981;256(17):8915–8921. [PubMed] [Google Scholar]
- 18.Tzeng YL, et al. Translocation and surface expression of lipidated serogroup B capsular polysaccharide in Neisseria meningitidis. Infect Immun. 2005;73(3):1491–1505. doi: 10.1128/IAI.73.3.1491-1505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer W, Schmidt MA, Jann B, Jann K. Structure of the Escherichia coli K2 capsular antigen. Stereochemical configuration of the glycerophosphate and distribution of galactopyranosyl and galactofuranosyl residues. Biochemistry. 1982;21(6):1279–1284. doi: 10.1021/bi00535a027. [DOI] [PubMed] [Google Scholar]
- 20.Corcoran AT, Annuk H, Moran AP. The structure of the lipid anchor of Campylobacter jejuni polysaccharide. FEMS Microbiol Lett. 2006;257(2):228–235. doi: 10.1111/j.1574-6968.2006.00177.x. [DOI] [PubMed] [Google Scholar]
- 21.Finke A, Bronner D, Nikolaev AV, Jann B, Jann K. Biosynthesis of the Escherichia coli K5 polysaccharide, a representative of group II capsular polysaccharides: Polymerization in vitro and characterization of the product. J Bacteriol. 1991;173(13):4088–4094. doi: 10.1128/jb.173.13.4088-4094.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morley TJ, Willis LM, Whitfield C, Wakarchuk WW, Withers SG. A new sialidase mechanism: Bacteriophage K1F endo-sialidase is an inverting glycosidase. J Biol Chem. 2009;284(26):17404–17410. doi: 10.1074/jbc.M109.003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JE, et al. The K5 lyase KflA combines a viral tail spike structure with a bacterial polysaccharide lyase mechanism. J Biol Chem. 2010;285(31):23963–23969. doi: 10.1074/jbc.M110.127571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131(2):209–217. [Google Scholar]
- 26.Cointe D, Leroy Y, Chirat F. Determination of the sialylation level and of the ratio alpha-(2—>3)/alpha-(2—>6) sialyl linkages of N-glycans by methylation and GC/MS analysis. Carbohydr Res. 1998;311(1-2):51–59. doi: 10.1016/s0008-6215(98)00196-7. [DOI] [PubMed] [Google Scholar]
- 27.Anumula KR, Taylor PB. A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal Biochem. 1992;203(1):101–108. doi: 10.1016/0003-2697(92)90048-c. [DOI] [PubMed] [Google Scholar]
- 28.Vliegenthart JF, Dorland L, van Halbeek H, Haverkamp J. NMR spectroscopy of sialic acids. In: Schauer R, editor. Sialic Acids: Chemistry, Metabolism and Function. Vol 10. New York: Springer; 1982. pp. 127–172. [Google Scholar]
- 29.Fraysse N, et al. Sinorhizobium meliloti strain 1021 produces a low-molecular-mass capsular polysaccharide that is a homopolymer of 3-deoxy-D-manno-oct-2-ulosonic acid harboring a phospholipid anchor. Glycobiology. 2005;15(1):101–108. doi: 10.1093/glycob/cwh142. [DOI] [PubMed] [Google Scholar]
- 30.Pavelka MS, Jr, Hayes SF, Silver RP. Characterization of KpsT, the ATP-binding component of the ABC-transporter involved with the export of capsular polysialic acid in Escherichia coli K1. J Biol Chem. 1994;269(31):20149–20158. [PubMed] [Google Scholar]
- 31.Frosch M, Müller A. Phospholipid substitution of capsular polysaccharides and mechanisms of capsule formation in Neisseria meningitidis. Mol Microbiol. 1993;8(3):483–493. doi: 10.1111/j.1365-2958.1993.tb01592.x. [DOI] [PubMed] [Google Scholar]
- 32.Larue K, Ford RC, Willis LM, Whitfield C. Functional and structural characterization of polysaccharide co-polymerase proteins required for polymer export in ATP-binding cassette transporter-dependent capsule biosynthesis pathways. J Biol Chem. 2011;286(19):16658–16668. doi: 10.1074/jbc.M111.228221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cieslewicz M, Vimr E. Thermoregulation of kpsF, the first region 1 gene in the kps locus for polysialic acid biosynthesis in Escherichia coli K1. J Bacteriol. 1996;178(11):3212–3220. doi: 10.1128/jb.178.11.3212-3220.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishinaga M, Kanamoto R, Kito M. Distribution of phospholipid molecular species in outer and cytoplasmic membrane of Escherichia coli. J Biochem. 1979;86(1):161–165. [PubMed] [Google Scholar]
- 35.Raetz CR. Enzymology, genetics, and regulation of membrane phospholipid synthesis in Escherichia coli. Microbiol Rev. 1978;42(3):614–659. doi: 10.1128/mr.42.3.614-659.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiménez N, et al. Effects of lipopolysaccharide biosynthesis mutations on K1 polysaccharide association with the Escherichia coli cell surface. J Bacteriol. 2012;194(13):3356–3367. doi: 10.1128/JB.00329-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzeng YL, et al. KpsF is the arabinose-5-phosphate isomerase required for 3-deoxy-D-manno-octulosonic acid biosynthesis and for both lipooligosaccharide assembly and capsular polysaccharide expression in Neisseria meningitidis. J Biol Chem. 2002;277(27):24103–24113. doi: 10.1074/jbc.M200931200. [DOI] [PubMed] [Google Scholar]
- 38.Meredith TC, Aggarwal P, Mamat U, Lindner B, Woodard RW. Redefining the requisite lipopolysaccharide structure in Escherichia coli. ACS Chem Biol. 2006;1(1):33–42. doi: 10.1021/cb0500015. [DOI] [PubMed] [Google Scholar]
- 39.Troy FA, Vijay IK, Tesche N. Role of undecaprenyl phosphate in synthesis of polymers containing sialic acid in Escherichia coli. J Biol Chem. 1975;250(1):156–163. [PubMed] [Google Scholar]
- 40.Andreishcheva EN, Vann WF. Gene products required for de novo synthesis of polysialic acid in Escherichia coli K1. J Bacteriol. 2006;188(5):1786–1797. doi: 10.1128/JB.188.5.1786-1797.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cartee RT, Forsee WT, Yother J. Initiation and synthesis of the Streptococcus pneumoniae type 3 capsule on a phosphatidylglycerol membrane anchor. J Bacteriol. 2005;187(13):4470–4479. doi: 10.1128/JB.187.13.4470-4479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steenbergen SM, Vimr ER. Biosynthesis of the Escherichia coli K1 group 2 polysialic acid capsule occurs within a protected cytoplasmic compartment. Mol Microbiol. 2008;68(5):1252–1267. doi: 10.1111/j.1365-2958.2008.06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knirel YA, Shevelev SD, Perepelov AV. Higher aldulosonic acids: Components of bacterial glycans. Mendeleev Commun. 2011;21(4):173–182. [Google Scholar]
- 44.Schmidt MA, Jann K. Structure of the 2-keto-3-deoxy-D-manno-octonic-acid-containing capsular polysaccharide (K12 antigen) of the urinary-tract-infective Escherichia coli O4:K12:H−. Eur J Biochem. 1983;131(3):509–517. doi: 10.1111/j.1432-1033.1983.tb07291.x. [DOI] [PubMed] [Google Scholar]
- 45.Altman E, Brisson JR, Gagné SM, Perry MB. Structure of the capsular polysaccharide of Actinobacillus pleuropneumoniae serotype 5b. Eur J Biochem. 1992;204(1):225–230. doi: 10.1111/j.1432-1033.1992.tb16628.x. [DOI] [PubMed] [Google Scholar]
- 46.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Datta S, Costantino N, Court DL. A set of recombineering plasmids for Gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 48.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wakarchuk WW, Campbell RL, Sung WL, Davoodi J, Yaguchi M. Mutational and crystallographic analyses of the active site residues of the Bacillus circulans xylanase. Protein Sci. 1994;3(3):467–475. doi: 10.1002/pro.5560030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.