Abstract
Arabidopsis thaliana endosperm, a transient tissue that nourishes the embryo, exhibits extensive localized DNA demethylation on maternally inherited chromosomes. Demethylation mediates parent-of-origin–specific (imprinted) gene expression but is apparently unnecessary for the extensive accumulation of maternally biased small RNA (sRNA) molecules detected in seeds. Endosperm DNA in the distantly related monocots rice and maize is likewise locally hypomethylated, but whether this hypomethylation is generally parent-of-origin specific is unknown. Imprinted expression of sRNA also remains uninvestigated in monocot seeds. Here, we report high-coverage sequencing of the Kitaake rice cultivar that enabled us to show that localized hypomethylation in rice endosperm occurs solely on the maternal genome, preferring regions of high DNA accessibility. Maternally expressed imprinted genes are enriched for hypomethylation at putative promoter regions and transcriptional termini and paternally expressed genes at promoters and gene bodies, mirroring our recent results in A. thaliana. However, unlike in A. thaliana, rice endosperm sRNA populations are dominated by specific strong sRNA-producing loci, and imprinted 24-nt sRNAs are expressed from both parental genomes and correlate with hypomethylation. Overlaps between imprinted sRNA loci and imprinted genes expressed from opposite alleles suggest that sRNAs may regulate genomic imprinting. Whereas sRNAs in seedling tissues primarily originate from small class II (cut-and-paste) transposable elements, those in endosperm are more uniformly derived, including sequences from other transposon classes, as well as genic and intergenic regions. Our data indicate that the endosperm exhibits a unique pattern of sRNA expression and suggest that localized hypomethylation of maternal endosperm DNA is conserved in flowering plants.
Keywords: DNA methylation, chromatin, RNA interference, gene imprinting
DNA methylation is a covalent modification of cytosine observed across the tree of life (1). In plants, methylation is mediated by distinct enzymatic systems in the CG, CHG, and CHH contexts (where H is A, C, or T), and regulates gene expression and transposon repression (2). The establishment of plant DNA methylation in all sequence contexts and the maintenance of CHH methylation is mediated by a specialized branch of the RNA interference pathway that generates nuclear-targeted 24-nt small RNA (sRNA) molecules (3, 4). Plants also possess DEMETER (DME) family DNA glycosylases that actively demethylate DNA by excising 5-methylcytosine in all sequence contexts (5, 6), with a preference for relatively euchromatic transposable elements (TEs) (7).
Homologs of DME are expressed in various tissues of the model dicot Arabidopsis thaliana throughout development (8), but DME expression is most prominent in the so-called gamete companion cells: the central and vegetative cells (9, 10). The vegetative cell forms the pollen tube that delivers two sperm cells, one of which fuses with the haploid egg cell to form the embryo, whereas the other fuses with the adjacent diploid central cell to form the nutritive triploid endosperm. Together with a maternally derived seed coat, the endosperm and embryo form a seed. DME activity in the central cell is essential for the specific hypomethylation seen in maternally inherited endosperm chromosomes (7), which establishes parent-of-origin–specific (imprinted) gene expression patterns that are crucial for seed development (9, 11).
sRNAs in the A. thaliana seed also show biased parental origin, with the majority of identified 24-nt sRNAs derived from maternal sequences (12). These sRNAs apparently accumulate in the endosperm and mediate gene expression (12, 13). However, accumulation of maternally derived sRNA is unaffected by mutations in any of the DNA- and histone-modifying enzymes that are known to regulate imprinted gene expression, including DME (14), leaving unresolved the mechanism by which they are generated.
Rice, a monocot that diverged from A. thaliana roughly 150 million years ago (15), possesses marked decreases in CG methylation at specific sites throughout the genome that correlate with endosperm-specific gene expression (16). However, we currently do not know whether the localized endosperm CG hypomethylation in rice is maternal specific, whether it correlates with imprinted gene expression, and whether sRNAs in the rice seed are generally maternally biased. Here, we show that hypomethylation in rice endosperm occurs specifically on maternally inherited chromosomes, is preferentially associated with imprinted genes, and is enriched within regions of open chromatin, suggesting that DME family–mediated demethylation is a conserved feature of flowering plant reproduction. We also show that, unlike in A. thaliana, imprinted 24-nt sRNAs in the endosperm originate from both parental genomes and are associated with demethylated regions, closely resembling the behavior of protein coding genes and suggesting that sRNA accumulation may be regulated differently in the persistent endosperm of rice and the more ephemeral endosperm of A. thaliana.
Results
Resequencing of the Kitaake Cultivar.
To examine parent-of-origin–specific DNA methylation and sRNA expression in rice endosperm, we sequenced the genome of the Kitaake cultivar of japonica rice, achieving 79-fold depth of coverage. Kitaake is a photoperiod-insensitive relative of Nipponbare with a rapid life cycle compared with other rice varieties (∼9 wk from seed to seed) that facilitates genetic experiments. Using the reference MSU version 6.0 genome sequence of the closely related (17) Nipponbare rice cultivar (18, 19), we mapped Kitaake reads, which covered 97% of the Nipponbare genome. We identified 169,819 SNPs between Nipponbare and Kitaake (Dataset S1), allowing us to resolve the parental origin of sequence reads from F1 hybrids of these cultivars.
Maternal Rice Endosperm Genome Shows Strong Site-Specific DNA Hypomethylation.
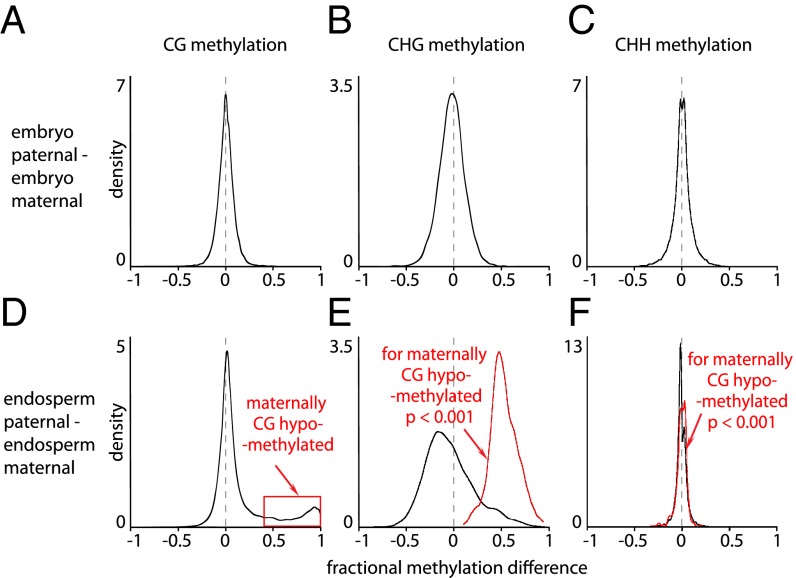
We performed bisulfite sequencing of endosperm and embryo harvested from F1 seeds of Nipponbare and Kitaake reciprocal crosses (Table S1). In both reciprocal crosses, maternal and paternal genomes in 7- to 8-d-old rice embryos display similar methylation patterns in all sequence contexts (the average of reciprocal crosses is shown in Figs. 1 A–F and 2 A–C; results for individual crosses, which are very similar, are shown in Figs. S1 and S2). However, the maternal genome of 7- to 8-d-old rice endosperm is slightly globally hypomethylated and strongly hypomethylated at specific sites in the CG context with respect to the paternal endosperm genome (Figs. 1 A and B and 2D; consistent individual cross data shown in Figs. S1 A and B and S2 A and B), which has similar methylation patterns to both parental complements of the embryo genome (Fig. 1 A and B; Fig. S3A). Thus, the CG hypomethylation of the maternal endosperm genome apparently fully accounts for the localized CG methylation differences we previously reported between rice endosperm and embryo (Fig. S3B) (16), and mirrors the site-specific, DME-mediated CG demethylation of the maternal genome in A. thaliana (7).
Fig. 1.
Maternal and paternal patterns of DNA methylation in rice embryo and endosperm. Genes (A, C, and E) and TEs (B, D, and F) were aligned at the 5′ end (Left) or the 3′ end (Right). Methylation levels within each 100-bp interval for maternal and paternal genomes were averaged between reciprocal crosses and then plotted from 2 kb away from the annotated gene or TE (negative numbers) to 4 kb into the annotated region (positive numbers). The dashed lines at zero represent the points of alignment. CG methylation is shown in A and B, CHG in C and D, and CHH in E and F.
Fig. 2.
Kernel density plots showing the frequency distribution of DNA methylation differences between maternal and paternal complements in 50-bp windows across the genome for embryo (A–C) and endosperm (D–F). Red traces in E and F represent CHG (E) and CHH (F) methylation differences in windows that show fractional CG hypomethylation of the maternal endosperm genome >0.4 (red box in D); p, p-value of a two-sample Kolmogorov-Smirnov test. A shift of the main peak with respect to zero represents a global difference between maternal and paternal genomes. Shoulders at the left and right represent local hypomethylation of the paternal and maternal genomes, respectively.
CHG and CHH methylation of both maternal and paternal genomes is lower in the endosperm than in the embryo (Fig. 1 C–F; Figs. S1 C–F, S2 C–F, and S3 C–F), consistent with our earlier report (16). The maternal endosperm genome is modestly globally hypermethylated in the CHG context compared with the paternal genome (black trace in Fig. 2E), but loci with strong maternal CG hypomethylation are also maternally hypomethylated in non-CG contexts (red traces in Fig. 2 E and F), as they are in A. thaliana (7). This result indicates that the localized demethylation affects all sequence contexts, strongly implicating a DME family glycosylase.
Endosperm Hypomethylated Sites Are Enriched Within Regions of Open Chromatin.
In A. thaliana, DME-mediated demethylation preferentially occurs in short TEs with euchromatic chromatin features and is rare in gene bodies and long heterochromatic TEs (7). To examine the distribution of demethylated sites in rice, we identified 27,669 regions that are significantly hypomethylated at CG sites in the endosperm compared with the embryo (Dataset S2) and used them as a proxy for maternally hypomethylated sequences. These differentially methylated regions (DMRs) span 5.5% of the genome and range in size from 50 bp to 12.95 kb, with a median size of 500 bp and a mean size of 737 bp.
Rice DMRs preferentially occur in genes and intergenic sequences rather than TEs (Fig. 3A). Among repetitive sequences, DMRs are less frequent at large class I long terminal repeat (LTR) TEs and are more abundant in class I non-LTR TEs, short TEs of various types, and TEs that occur within gene bodies (Fig. 3 B and C). Although distinct from the distribution of A. thaliana DMRs (7), the depletion of rice DMRs from long TEs suggests that DME family enzymes preferentially demethylate more accessible euchromatic sequences in rice, as well as in A. thaliana. To test this hypothesis, we compared our methylation results with previously published DNase I hypersensitivity data (20). Indeed, DNA accessibility is correlated with endosperm hypomethylation (Fig. 3D); genic and intergenic regions are generally more accessible than TEs, and class I non-LTR and shorter TEs are more accessible than longer class I LTR and class II TEs (Fig. 3D).
Fig. 3.
Genomic distribution of DMRs between embryo and endosperm. (A–C) 50-bp windows across the genome were assigned to either introns, exons, intergenic regions (no gene or repeat annotation), repeat regions (mostly TEs), or an edge category (the boundaries of gene bodies and repeats), and the number of windows that constitute the embryo methylome and the number that overlapped defined DMRs were counted. Further resolution of repeat windows (black bar in A) is shown in B. Further resolution of Class I non-LTR sequences (pink bar in B) is shown in C. (D) The mean CG methylation difference between embryo and endosperm was calculated for 50-bp windows of varying degrees of DNase I hypersensitivity in two tissues. The mean DNase I hypersensitivity of some sequence elements categorized in A–C is indicated.
Differentially Methylated Regions Are Enriched Around and Within Imprinted Genes.
Because of the strong association between imprinted gene expression and endosperm hypomethylation (9, 11), we examined DNA methylation at genes known to be imprinted in rice endosperm (21). Both maternally and paternally expressed genes are preferentially demethylated compared with other genes (Fig. 4A) and tend to have DMRs in their promoter regions (Fig. 4B). Maternally expressed genes are specifically enriched for DMRs that span the transcription start site (TSS), transcription termination site (TTS), and 3′ region, whereas paternally expressed genes are specifically enriched for DMRs in their bodies (Fig. 4 B and C; Fig. S4). Methylation of the TSS and TTS is correlated with gene silencing (22), consistent with the specific activation of maternally expressed endosperm genes by DNA demethylation. The rice DMR distribution, including the enrichment of DMRs in the bodies of paternally expressed genes, is very similar to that in A. thaliana (7), further reinforcing the mechanistic similarities between monocots and dicots.
Fig. 4.
Enrichment of DMRs between embryo and endosperm at imprinted genes. (A) Kernel density plots of the differences between embryo and endosperm CG methylation in 50-bp windows across the bodies of all annotated genes (black trace) and imprinted subsets (red and blue traces); p, p-value of a two-sample Kolmogorov-Smirnov test. (B) Genes were aligned at the TSS and TTS, and the proportion of genes with a DMR present is plotted for each 100-bp interval within 3 kb of the alignment sites (dashed lines). Within specific genic regions (boxed in gray), maternally (red) and paternally (blue) expressed genes are enriched for DMRs compared with the genome average (green), with significance evaluated using Fisher’s exact test. (C) Examples of maternally (red) and paternally (blue) expressed genes enriched in DMRs. Green bars represent embryo CG methylation, orange bars represent endosperm CG methylation, and red and blue bars represent CG methylation of the maternal and paternal genomes, respectively. Identified DMRs are underlined in red.
Endosperm Exhibits a Unique sRNA Expression Pattern.
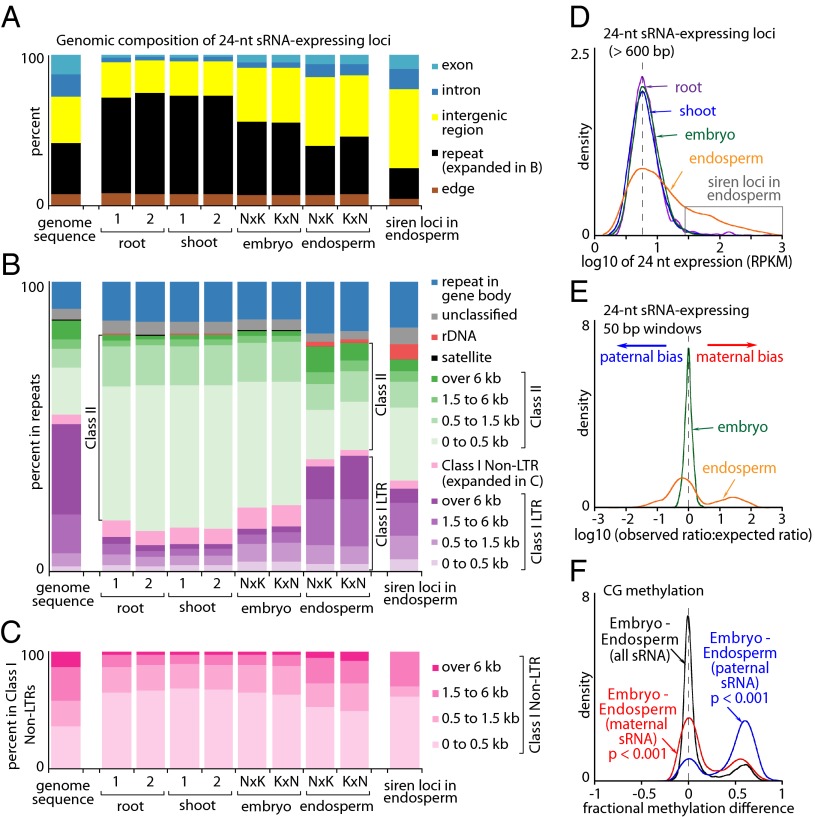
To examine sRNA expression in rice seeds, we sequenced endosperm and embryo sRNA libraries from Nipponbare and Kitaake reciprocal crosses and also libraries from several control tissues (Table S2). Unlike in embryo, seedling root, and seedling shoot, 24-nt sRNAs in the endosperm predominantly map to genic and intergenic regions rather than TEs (Fig. 5A), although embryos show a somewhat intermediate pattern (Fig. 5A). Furthermore, the endosperm 24-nt sRNA reads that do map to TEs indicate a different pattern of TE-associated sRNA expression compared with other tissues (Fig. 5 B and C). The majority of TE-associated sRNA production in embryo, shoot, and root occurs within class II TEs shorter than 500 bp (Fig. 5B). This group is largely comprised of miniature inverted-repeat TEs (MITEs), which preferentially occur near genes in euchromatic regions and are the predominant target of CHH methylation in rice tissues (16, 23). In contrast, 24-nt sRNA production in endosperm is much more evenly distributed across different TE classes and sizes (Fig. 5B), although class I TEs >6 kb are still underrepresented compared with their genomic abundance (Fig. 5 B and C).
Fig. 5.
sRNA (24-nt) expression in the endosperm, embryo, seedling root, and seedling shoot. (A–C) Relative abundance of 24-nt sRNA expression across the genome, subdivided as in Fig. 3 A–C. (D) Kernel density plot of 24-nt sRNA expression intensity in endosperm, embryo, root, and shoot at loci longer than 600 bp (length threshold imposed to exclude microRNA loci). (E) Kernel density plots depicting the prevalence and direction of 24-nt sRNA parental bias. Deviations from zero (dashed line) indicate either maternal bias (deviation to the right) or paternal bias (deviation to the left). (F) Kernel density plots of the differences between embryo and endosperm CG methylation measured in 50-bp windows across all sRNA-producing loci (black trace) and imprinted subsets (red and blue traces); p, p-value of a two-sample Kolmogorov-Smirnov test.
The endosperm is also distinguished by strong expression of 24-nt sRNA from a modest number of loci (Fig. 5D). We termed the genomic regions of high sRNA production in the endosperm small-interfering RNA in endosperm (siren) loci. Siren loci are dispersed intermittently across all 12 chromosomes, account for 74% of 24-nt sRNA in Nip × Kit endosperm and 64% of 24-nt sRNA in Kit × Nip endosperm despite cumulatively spanning only 0.44% of the genome, and have an even stronger than average tendency to occur in genes, intergenic regions, and small TEs (rightmost bars in Fig. 5 A–C).
Imprinting of Endosperm 24-nt sRNA Shares Similarities with That of Genes.
The stringency associated with accurately mapping short reads to parental genomes, along with the sparse distribution of SNPs within regions of sRNA production, resulted in a very small fraction of the total reads from F1 seeds being informative for detection of parental biases (Table S2). For example, of the 12,991 endosperm sRNA-producing loci we identified, only 125 possessed a total of at least 15 parentally sorted reads in each of the reciprocal crosses (Dataset S3). Nevertheless, we were able to assay the general trends of 24-nt sRNA parental expression in embryo and endosperm. We did not detect significant parental bias in the embryo (Fig. 5E), but did find large deviations from the expected 2:1 maternal:paternal ratio in the endosperm (Fig. 5E). Of the 125 informative loci, 15 had a significant maternal bias (p < 0.001; Fisher’s exact test), and 16 had a paternal bias in both reciprocal crosses. Only four loci were unambiguously biallelic (SI Materials and Methods). Like imprinted genes, imprinted 24-nt sRNA-producing loci are enriched in DMRs (Fig. 5F). These results are in contrast to the strong tendency toward maternal sRNA expression described in A. thaliana seeds (12) that is unaffected by DNA methylation (14).
Of the 31 imprinted sRNA loci, four overlap known imprinted genes, with sRNA and mRNA expression always occurring from opposite parental alleles (Fig. 6; Table 1). Of the remaining 27 imprinted sRNA loci, 18 overlap genes of unknown imprinted status (Dataset S3), suggesting that a substantial proportion of imprinted sRNA may correspond to imprinted genes. Notably, the maternally expressed gene Os02g29230 (Fig. 6, middle column) that overlaps DMRs and paternally expressed imprinted sRNAs encodes a DME homolog named ROS1B (24), suggesting the possibility of a complex, multilayered regulatory relationship between DNA demethylation and sRNA pathways.
Fig. 6.
Snapshots of 24-nt sRNA abundance in reads per million (RPM) and CG methylation in rice embryo and endosperm, around paternally expressed (blue) and maternally expressed (red) imprinted genes. At positions where SNPs resolved reads between parental genomes, 24-nt sRNA abundance and DNA methylation of the maternal and paternal genome are represented by red bars and dark blue bars, respectively. Siren loci identified in our analysis are underlined in orange; DMRs identified between embryo and endosperm are underlined in red. Note the different scales for endosperm sRNA compared with the other tissues.
Table 1.
Regions of overlap between imprinted 24-nt small RNA-expressing loci and imprinted genes
| Chr. | sRNA locus start | sRNA locus end | sRNA locus expression bias | Gene locus ID | Gene locus start | Gene locus end | Gene locus expression bias | Functional annotation of gene |
| 1 | 40,538,301 | 40,544,848 | Maternal | Os01g70060 | 40,539,921 | 40,553,708 | Paternal | RNA Pol-II binding domain |
| 2 | 17,344,451 | 17,357,050 | Paternal | Os02g29230 | 17,348,970 | 17,357,069 | Maternal | DEMETER homolog |
| 6 | 19,595,101 | 19,605,500 | Paternal | Os06g33690 | 19,603,058 | 19,605,119 | Maternal | CAPIP1 |
| 7 | 4,701,351 | 4,709,900 | Paternal | Os07g09020 | 4,701,864 | 4,711,518 | Maternal | AGO14 |
Chr., chromosome.
Discussion
Our results indicate that strong site-specific CG endosperm hypomethylation is a conserved feature of flowering plants. As in A. thaliana (7), rice CG hypomethylation occurs at discrete sites with open, euchromatic features [as evidenced by DNase I sensitivity (20); Fig. 3D] on the maternal but not paternal chromosomes and is associated with CHG and CHH hypomethylation at the same sites. These results strongly implicate the activity of a DME family glycosylase in the rice central cell. The rice DME homolog ROS1A (Os01g11900) has an expression pattern and mutant phenotype in reproductive tissues similar to A. thaliana DME (24), and is a good candidate for a functional rice DME analog. Conserved action of DME-like enzymes in monocots is also supported by the reports of localized DNA hypomethylation in the maize central cell (25) and on the maternal genome in the endosperm (25, 26). Furthermore, A. thaliana and rice show similar patterns of DMR enrichment in and around maternally and paternally expressed genes, suggesting that the mechanisms that regulate imprinted expression are conserved despite the lack of overlap between known imprinted genes in A. thaliana, rice, and maize (21, 26–28).
We also show that rice sRNA-producing loci behave like genes with respect to imprinting, in terms of the direction of imprinting biases and correlation with sites of CG hypomethylation. Paternal expression of endosperm small RNAs associated with maternal DMRs may be regulated by the same epigenetic mechanisms that have been proposed to explain paternal mRNA expression linked to maternal DMRs (11). Taken together with the likely conservation of imprinting mechanisms in flowering plants, this suggests that conventionally imprinted sRNA loci may also exist in A. thaliana but have not been detected because of the large amount of maternally encoded sRNA that is not regulated by known imprinting mechanisms (14). It is possible that this accumulation indeed reflects a unique imprinting pathway that does not operate in rice. Alternatively, the maternal bias seen in A. thaliana may have been due to deposition of sRNAs from surrounding maternal tissue such as the seed coat, because the ratio of seed coat tissue to endosperm is greater in A. thaliana seeds than in the much larger rice seeds used in our experiments.
We and others have proposed that a major function of the demethylation that occurs in central and vegetative cells is to produce mobile sRNAs that are transported into the egg and sperm cells to reinforce the methylation and silencing of TEs (7, 10, 11, 29, 30). Generation of the sRNA signal may be facilitated by the relaxation of chromatin in the companion cells (10, 30) and is supported by unusual sRNA patterns in A. thaliana pollen (30). Our finding that the sRNA population of rice endosperm spans a wider range of genomic sequences, including expression from many TE types that are underrepresented in other tissues, is consistent with this idea. Because the sRNA patterns in the relatively mature rice embryos we analyzed are distinct from those in endosperm, our data also strongly argue that the bulk of the hypothesized sRNA exchange must occur during early development, either between the gametes and their companion cells before fertilization and/or between the nascent embryo and endosperm.
Materials and Methods
Illumina Library Construction.
Bisulfite sequencing libraries for Illumina sequencing were constructed essentially as described previously (31). sRNA libraries were constructed as described (32). Because our analyses evaluated DNA methylation and sRNA patterns that were consistent between a set of reciprocal crosses, we did not perform biological replication for each cross.
Allele-Specific Mapping of Reads.
Sequencing reads were sorted to the Nipponbare and Kitaake genomes as described (28). DNA methylation of cytosines within sorted reads was calculated as described (22, 31).
Definition of DMRs.
CG fractional methylation in 50-bp windows (each window averages methylation on both strands) was compared between embryo and endosperm. Windows with embryo fractional methylation at least 0.2 greater than that in endosperm and a Fisher’s exact test p < 0.05 were merged if they occurred within 300 bp. Merged DMRs were retained if the fractional methylation in embryo across the DMR was at least 0.3 greater than that in endosperm and the Fisher’s exact test was p < 10−5.
Identification of Imprinted sRNA-Producing Loci.
Loci of 24-nt sRNA expression were defined by blocking together 50-bp windows with more than five read counts in libraries from the same tissue type when the 50-bp windows were within 500 bp of each other. The number of maternally sorted and paternally sorted endosperm 24-nt reads that mapped within these loci was recorded, and the significance of bias was calculated using Fisher’s exact test.
Supplementary Material
Acknowledgments
We thank W. Schackwitz, M. Joel, and the Joint Genome Institute (JGI) sequencing team for generating Kitaake genome sequence and initial analysis; L. Bartley and E. Marvinney for rice genomic DNA preparation; J. Huff and A. Zemach for suggestions on data analysis and interpretation; A. Zemach, Y. Kim, and T.-F. Hsieh for training in experimental techniques; M. Couvillon for the sRNA sequencing protocol; J. Zhai for pointing out that an imprinted rice sRNA locus overlaps a DME homolog; and Y. Wu and J. Jiang for DNase I hypersensitivity data. This work was partially funded by National Science Foundation Grant IOS-1025890 (to R.L.F. and D.Z.), National Institutes of Health Grant GM69415 (to R.L.F.), a Young Investigator grant from the Arnold and Mabel Beckman Foundation (to D.Z.), and a Fulbright scholarship (to J.A.R.). The JGI is supported by the Office of Science of the US Department of Energy (DOE) under Contract DE-AC02-05CH1123. This work was also supported by the US DOE Office of Biological and Environmental Research through Contract DE-AC02-05CH11231 with Lawrence Berkeley National Laboratory.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE44898), and the Sequence Read Archive (SRA), http://www.ncbi.nlm.nih.gov/sra (accession no. SRX037797).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306164110/-/DCSupplemental.
References
- 1.Zemach A, Zilberman D. Evolution of eukaryotic DNA methylation and the pursuit of safer sex. Curr Biol. 2010;20(17):R780–R785. doi: 10.1016/j.cub.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 2.He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21(3):442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H, Zhu JK. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14(2):142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morales-Ruiz T, et al. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA. 2006;103(18):6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penterman J, et al. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104(16):6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibarra CA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337(6100):1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110(1):33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 10.Schoft VK, et al. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc Natl Acad Sci USA. 2011;108(19):8042–8047. doi: 10.1073/pnas.1105117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer MJ, Fischer RL. Genome demethylation and imprinting in the endosperm. Curr Opin Plant Biol. 2011;14(2):162–167. doi: 10.1016/j.pbi.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mosher RA, et al. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;460(7252):283–286. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- 13.Lu J, Zhang C, Baulcombe DC, Chen ZJ. Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc Natl Acad Sci USA. 2012;109(14):5529–5534. doi: 10.1073/pnas.1203094109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosher RA, et al. An atypical epigenetic mechanism affects uniparental expression of Pol IV-dependent siRNAs. PLoS ONE. 2011;6(10):e25756. doi: 10.1371/journal.pone.0025756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol. 2004;58(4):424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- 16.Zemach A, et al. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA. 2010;107(43):18729–18734. doi: 10.1073/pnas.1009695107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nasu S, et al. Search for and analysis of single nucleotide polymorphisms (SNPs) in rice (Oryza sativa, Oryza rufipogon) and establishment of SNP markers. DNA Res. 2002;9(5):163–171. doi: 10.1093/dnares/9.5.163. [DOI] [PubMed] [Google Scholar]
- 18.Ouyang S, et al. The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res. 2007;35(Database issue):D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Project IRGS. International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436(7052):793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, et al. High-resolution mapping of open chromatin in the rice genome. Genome Res. 2012;22(1):151–162. doi: 10.1101/gr.131342.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo M, et al. A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 2011;7(6):e1002125. doi: 10.1371/journal.pgen.1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 23.Lu C, et al. Miniature inverted-repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol. 2012;29(3):1005–1017. doi: 10.1093/molbev/msr282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ono A, et al. A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J. 2012;71(4):564–574. doi: 10.1111/j.1365-313X.2012.05009.x. [DOI] [PubMed] [Google Scholar]
- 25.Gutiérrez-Marcos JF, et al. Epigenetic asymmetry of imprinted genes in plant gametes. Nat Genet. 2006;38(8):876–878. doi: 10.1038/ng1828. [DOI] [PubMed] [Google Scholar]
- 26.Waters AJ, et al. Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell. 2011;23(12):4221–4233. doi: 10.1105/tpc.111.092668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang M, et al. Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc Natl Acad Sci USA. 2011;108(50):20042–20047. doi: 10.1073/pnas.1112186108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh T-F, et al. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci USA. 2011;108(5):1755–1762. doi: 10.1073/pnas.1019273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutierrez-Marcos JF, Dickinson HG. Epigenetic reprogramming in plant reproductive lineages. Plant Cell Physiol. 2012;53(5):817–823. doi: 10.1093/pcp/pcs052. [DOI] [PubMed] [Google Scholar]
- 30.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136(3):461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh T-F, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324(5933):1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Couvillion MT, et al. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev. 2009;23(17):2016–2032. doi: 10.1101/gad.1821209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.