Significance
DNA palindromes were crystallized in complexes with HetR, a transcription factor required for heterocyst differentiation in the nitrogen-fixing cyanobacterium Anabaena. In three complexes, we observed hydrogen bonding of a single glutamate side chain with three successive cytosines in the DNA. The feature of three successive GC pairs in each arm of the palindrome is conserved in other filamentous cyanobacteria. These cyanobacteria contain HetR proteins, each of which contains glutamate in that critical position. This unique interaction between a protein factor and its DNA target is so important that it is invariant across cyanobacteria from environments around the world.
Keywords: heterocyst differentiation, mutagenesis, X-ray crystallography
Abstract
HetR is an essential regulator of heterocyst development in cyanobacteria. Many mutations in HetR render Anabaena incapable of nitrogen fixation. The protein binds to a DNA palindrome upstream of hetP and other genes. We have determined the crystal structures of HetR complexed with palindromic DNA targets, 21, 23, and 29 bp at 2.50-, 3.00-, and 3.25-Å resolution, respectively. The highest-resolution structure shows fine details of specific protein–DNA interactions. The lower-resolution structures with longer DNA duplexes have similar interaction patterns and show how the flap domains interact with DNA in a sequence nonspecific fashion. Fifteen of 15 protein–DNA contacts predicted on the basis of the structure were confirmed by single amino acid mutations that abolished binding in vitro and complementation in vivo. A striking feature of the structure is the association of glutamate 71 from each subunit of the HetR dimer with three successive cytosines in each arm of the palindromic target, a feature that is conserved among all known heterocyst-forming cyanobacteria sequenced to date.
The filamentous cyanobacterium Anabaena differentiates specialized cells, the heterocysts, at regular intervals along each filament when fixed nitrogen is absent from its environment. The intervals separating heterocysts from each other are comprised of 10–20 CO2-fixing, O2-evolving vegetative cells. The precise fraction of differentiated cells and their pattern are determined by interactions among transcription factors, particularly the HetR activator, and several negative regulators, including the PatS peptide RGSGR and the protein HetN, which contains the sequence RGSGR in its interior (1–3).
Long before the negative regulators were discovered, Wolk proposed that the heterocyst pattern was determined by some product of nitrogen fixation that diffused along the filament, from its source in a heterocyst (4). As vegetative cells continue to divide, the distance between two heterocysts increases until the concentration of fixed nitrogen falls below some threshold in a vegetative cell approximately halfway between the heterocysts. That cell then differentiates into a heterocyst. There are some clues to the biochemical mechanisms involved in differentiation. Two proteins, PII and NtcA, respond to the relative levels of glutamine and 2-oxoglutarate (2OG) (5–7). When 2OG is high, signaling nitrogen starvation, PII and NtcA activate transcription of NrrA (8), which in turn activates transcription of another protein, HetR, which also functions as a transcription activator. However, what creates the gradient of HetR, assuming that its activity is the critical variable in establishing the pattern? The answer seems to be a gradient of the PatS peptide RGSGR (9) early in the process, and later, a gradient of the protein HetN, which also contains the sequence RGSGR (9, 10). Both of these “regulators” interact with HetR, preventing it from binding to specific DNA targets, by a mechanism that is still not fully understood. Constitutively expressed HetR is rapidly degraded by cellular protease(s). Therefore, the gradient of HetR is due to a gradient of degradation caused by proteins or peptides produced in the heterocyst and diffused along the filament (10).
HetR alone is not likely to be directly responsible for the activation of transcription of all 1,500 genes that are transcribed differentially in heterocysts. Indeed, only a few genes are known to be activated by HetR. Among these are hetR itself, patS, hetP, and hetZ (11, 12). The latter two potentially encode transcription factors activating other genes, providing the beginning of a cascade that could eventually include all of the genes needed for differentiation. Additionally, it is possible that HetR binds other proteins that extend its range of DNA-binding activity. For all these reasons, it is desirable to determine the 3D structures of the proteins mentioned, as well as their DNA targets and their complexes with other proteins, still to be identified.
HetR is highly conserved among filamentous cyanobacteria (13). We previously cloned and purified HetR from five cyanobacteria, including the thermophile Fischerella, and showed that all of them complement a hetR deletion mutant of Anabaena. We then solved the 3D structure of the Fischerella HetR dimer by X-ray crystallography (14). That structure contains four distinct domains: an extended DNA-binding unit containing helix-turn-helix (HTH) motifs comprised of the two N-terminal sequences; two histidine-rich flaps protruding on either side of the extended structure; and finally a hood comprised of the two C-terminal sequences. The specific functions of the flap and hood domains remain unknown. We modeled the known sequence of a target DNA (the binding site upstream of the hetP gene in Anabaena) into the Fischerella structure and confirmed the outlines of the proposed complex by mutagenesis of three key residues in the HTH domain (14). We also modeled the peptide RGSGR and proposed three locations in HetR as possible binding sites, two of which would directly interfere with the formation of the HetR–DNA complex (14). Here we present the results of further work on the structure of the complex of the Fischerella HetR dimer with several target DNA palindromes based on Anabaena and Fischerella genome sequences upstream of the conjugate hetP genes.
Results and Discussion
Structure of the HetR–DNA Complex.
It has been proposed that in Anabaena, HetR recognizes a 17-bp semipalindromic DNA sequence (12); however, a shorter (15 bp) consensus sequence was reported recently (11), and longer binding sites were proposed based on the crystal structure of the HetR dimer (14). Based on the comparison of HetR binding sites within the hetP promoter of several filamentous cyanobacteria, a consensus sequence recognized by HetR can be derived (5′-tnantngnGGGtcaanCCCanca-3′; Fig. S1; capitals refer to residues conserved in all known palindromes, lower case refer to residues not completely conserved, n can be a, g, c, or t). The most prominent features of this HetR recognition sequence are three consecutive GC base pairs in each arm of the palindrome separated by 5 bp that are not conserved (Fig. S1). The consensus sequence was used to design DNA sequences for structure determination experiments. The target sequence palindromic twofold symmetry is broken by insertion of 1 bp (almost exclusively TA) in the middle. We number this base pair “0,” and the other bases are numbered with a “+” sign to the right and a “−” sign to the left on the top strand and with opposite signs on the bottom strand. This numbering reflects the palindromic symmetry and helps to describe protein interactions with bases and phosphate moieties. In view of the crystal structure of the protein, it was expected that the DNA-binding unit would cover ∼17 bp but that the flap domains extend further out, so a number of DNA fragments (19–31 bp) were designed for cocrystallization experiments. We obtained crystals for seven DNA fragments and determined structures for three of them (sequences shown in Fig. S1). The crystal structure of HetR from Fischerella MV11 in complex with the 21-bp DNA target was determined first, at 2.50-Å resolution, using molecular replacement and HetR [Protein Data Bank (PDB) entry 3QOD] as a search model. Additional structures were determined with longer DNA sequences: 23 bp at 3.00-Å resolution and 29 bp at 3.25-Å resolution. These structures were determined by molecular replacement using the HetR–21-bp DNA complex (PDB entry 4IZZ) as a search model.
DNA Binding.
The HetR-DNA complex structure determined at 2.50 Å is of high quality (Fig. S2), exhibits good crystallographic and geometric statistics (Table S1), and allows detailed visualization of protein–DNA interactions, including many solvent-mediated contacts. The crystals of the complex diffract to higher resolution than the free protein (resolution limit, 3.00 Å; PDB entry 3QOE). On DNA binding, HetR becomes better ordered and may become more resistant to proteases. A similar observation has been previously reported for the pheromone-binding transcription activator TraR from A. tumefaciens (15).
The structure of the HetR protein complexed with DNA is similar to the previously reported structure of free HetR (14). As proposed earlier, HetR interacts with DNA as a dimer, predominantly using its N-terminal domains (residues 1–98; Fig. 1A). This unit has an extended, strongly positively charged surface decorated with Arg, Lys, and His residues (Fig. 1B). It has two symmetrically positioned HTH DNA-binding motifs spaced ∼51 Å apart, covering 19 bp of the DNA target. In general, the HetR structure complexed with the 21-mer DNA superimposes well with the apo form, with an overall rmsd of 2.43 Å for Cα atoms. The DNA-binding unit and hood domains of the dimer are the most similar and superimpose without major conformation change. Larger changes are observed in the orientation of the flap domains (Fig. 1C). The whole HetR dimer becomes more symmetric in the presence of DNA. Overall, the flap orientations are adjusted to provide a more extended interaction with the twofold symmetric DNA duplex. The DNA duplex bends toward the protein and follows the curvature of the protein’s positively charged surface. In the complex of HetR with the 29-bp duplex, the DNA continues along flap domains (Fig. 1A) that have positively charged patches aligned with the DNA phosphate backbone (Fig. 1B).
Fig. 1.
Structure of HetR–DNA complex. (A) Overall structure of the complex of HetR with a 29-bp DNA duplex. The HTH motifs of the DNA-binding unit penetrate the major groove of the DNA. The flap domains interact with the phosphate backbone. The hood domain is presented to the solvent and is available for interactions. Potential peptide RGSGR binding sites identified previously (14) within the DNA-binding unit and on the interface between the HTH and flap domains would interfere with the formation of the protein–DNA complex. DNA curvature is shown as a line through the center of the DNA helix. (B) Charge distribution on the HetR surface interacting with DNA using 21-bp DNA. (C) Conformational changes of the HetR protein on DNA binding. The largest changes are observed within the flap domains. Apo form HetR (in pale) (PDB entry 3QOD) and HetR with the 21-bp DNA bound (in blue and green) are compared. The protein dimers were superimposed: a total of 588 protein residues from HetR-DNA and 574 from Apo-HetR, with an overall rmsd of 2.43 Å with Cα positions. The DNA-binding unit and hood domain of the dimer are well superimposed, but the flap domains of the HetR-DNA complex deviate from that of the apo form, making the dimer more symmetric in the presence of DNA. Overall, the flaps fit better to the extended DNA in both directions away from the twofold axis.
The DNA geometry was analyzed using the program “Curve+” (16). The DNA duplex is distorted, with the average distance between bases being 3.2 Å (32 Å per turn), somewhat shorter than observed for a typical B-DNA (3.4 Å). The 21-bp DNA is bent toward the protein to wrap around the DNA binding unit of HetR. The total bend is 29.3°, with the largest bending (3.6–4.9°) occurring at the A/T track (T-2T-1A0A+1A+2) in the middle. The non-Watson–Crick AA base pair placed at position 0 produces a large opening. The majority of the local and more specific deformations (X-displacement, inclination, buckle, propel and roll) are concentrated on the G-6G-5G-4/C+4C+5C+6 regions. These distortions widen and deepen the major groove at G-5G-4/C+4C+5. Overall, there are three tracks: two comprised of C/G pairs resembling A-form DNA (−6 to −4 and 4–6) containing all base-specific interactions with HetR and one at A/T (−1 to 1) with the central A/A mis-pair maintaining the B-form DNA. (The mis-pair is in our synthetic palindrome; in the genomic counterpart, it is a conventional AT base pair.)
Many HetR–DNA contacts are distributed over the entire length of the 21-bp duplex. These contacts include intricate networks of interactions involving side chains, the main chains, N-terminal portions of α-helices, and solvent on the protein side, as well as bases and phosphate moieties of the DNA duplex (Figs. 2–4; Fig. S3). No contacts of protein with sugar moieties are observed. Both HetR subunits contribute residues to each arm of the palindrome. The HTH motifs, comprised of helices H4 and H5 (and H4′ and H5′) connected by a short loop, are placed symmetrically in the major groove. Helix H5 is positioned perpendicularly to the surface of bases, whereas helix H4 runs parallel, with its N-terminal portion pointing into a phosphate group(P−9/−10). The majority of interactions are contributed by side chains (and main chain) from these two helices (Arg34, His35, Asp40, Lys47, Asn60, Arg62, His69, Glu71, Lys73, Arg74, and Lys76). Additional interactions are provided by side chains and the main chain of the flap domain (Ser179, Glu180, Ala181, and Arg188, and possibly Arg198 and Glu200). The same interactions are observed in both arms of the palindrome except for a few water-mediated contacts.
Fig. 2.
Details of DNA binding. Interaction of Glu61, Lys73, and Arg62 with bases is shown. HTH motif interaction in the major groove: base-specific interaction includes Glu71 (chain A) contacting three consecutive cytosine bases at positions 3, 4, and 5, Lys73 making hydrogen bonds to O6 and N7 of guanine (−5), and Arg62 interacting with adenine at −7 and guanine at −8. Direct interactions of His69 and Ser179 with phosphate are shown. The partial positive charge from the N-terminal end of helix H4 dipole also provides electrostatic interaction with the DNA phosphate backbone.
Fig. 4.
Mutagenesis: in vitro DNA-binding and in vivo complementation experiments. Under each mutation the outcomes of two experiments are listed. DNA binding measured by band shift is highlighted blue and complementation is highlighted pale. +, positive; −, no binding detected and no complementation. Downward arrows indicate weaker complementation based on heterocyst frequency. WT HetR complementation gave ∼8.5% heterocysts at 48 h. Blank fields mean no results.
The interactions can be divided into four classes: direct contacts to bases and phosphates, water-mediated and van der Waals contacts, and α-helix–dipole interactions. Because there is some asymmetry of interaction observed, we first describe the half of the complex where most of the interactions are provided by the A subunit and then indicate differences. Direct interactions with phosphate groups result in “fastening” DNA to protein with a specific register. These direct interactions link Lys76 with P−7/−8, Asn60 with P−8/−9/−10, amides of Leu61 and Arg62 with P−9/−10, Ser179 with P+3/+4 (Figs. 2 and 3), and amides of Glu180 and Ala181 with P+4/+5. From the opposite subunit, His69′ interacts with P+3/+4 (Figs. 2 and 3) and Arg74′ with P0/+1, and Arg188− forms a salt bridge with P−9/−10. Water-mediated interactions involve Arg34, His35, and Asp40 with P+2/+3 (with Asp40 approaching from the minor groove), and from the opposite subunit, Lys47′ and Arg74′ with P0/+1. In total, nine phosphate moieties are involved in binding on both strands of the duplex. The side chain of the Arg188 residue is very flexible and exists in multiple conformations in both subunits. At the current resolution, not all bound water molecules are observed. However, it is known from other high-resolution structures that at certain positions water molecules are bound to DNA bases (17). Therefore, it is likely that some of the longer-distance “contacts” (∼4–5 Å) found in our structures are additional solvent-mediated interactions.
Fig. 3.
Summary of DNA binding by HetR. Schematic diagram of the synthetic 21-bp DNA sequence is shown. HetR residues of chain A are labeled brown and chain B red. Direct hydrogen bonds are shown in green and water-mediated in blue. The residues making specific contact to bases are shaded pale. The phosphate group numbering in the text refers to 3′ phosphate of the nucleotide.
Three α-helices contribute N-terminal helix–dipole interactions with half of the binding site: H4, H3, and H10′ of the opposite subunit. The two-turns-long H4 runs across the major groove, whereas the partly positively charged N terminus of H4 points toward a negatively charged DNA phosphate group at position −8. The amides of Leu61 and Arg62 of H4 make a direct bridging hydrogen bond to P−9/−10 oxygens. Asn60 caps the N terminus of H4 and its O is at the right distance (3.1–3.3 Å) to accept hydrogen bonds from three N-terminal main chain amides at positions 61, 62, and 63 (and it interacts with P−9/−10). Arg62 is involved in a complex network of interactions with Arg188′ from the opposite subunit (both bind to P−9/−10) and through water with guanine at position −8. Ser179′ from H10 of the opposite subunit forms direct hydrogen bonds with P+3/+4, and the amide of Glu180 makes hydrogen bonds with P+4/+5. A very similar interaction pattern is observed at the N terminus of H10 with Ser179 interacting with P+3/+4 and the amides of Glu180 and Ala181 interacting with P+4/+5 (Fig. 2). The interaction of the H3 N terminus is different, as it involves a complex pattern of interacting side chains (Arg34, His35, Asp40, and His69’), water molecules, and P+2/+3 moieties.
The base-specific HetR interactions occur only in the major groove, whereas no interaction with bases in the minor groove is observed. The amino acid sequence of H4 and the N-terminal region of H5 are highly conserved in the HetR family. The longer H5 runs perpendicular to the surface of bases with its N terminus pointing toward DNA, providing additional electrostatic attraction. There are three important interactions in what appear to be sequence-specific contacts with the edges of the bases. A unique base-specific interaction is provided by Glu71 from the loop between H4 and H5 facing the bases. This residue forms 2.7-, 3.1-, and 3.5-Å hydrogen bonds with three amino groups of C-3, C-4, and C-5, respectively (Figs. 2 and 3). A second base-specific interaction involves Lys73 interacting directly with O6 and N7 of G-5 and potentially via water with N7 of G-6. The positively charged amino group of Lys73 is 5.5 Å away from the Glu71 carboxylate providing a suitable electrostatic environment. Arg62 complements these interactions, hydrogen bonding to A-7 and G-8. Glu71, Lys73, and Arg62 are strictly conserved in the HetR family. Thus, the Glu71 and Lys73 contacts require the three consecutive GC base pair sequences in each arm of the palindrome, defining the requirement for the sequence (GGGnnnnnCCC; n can be a, g, c, or t; Fig. S1). Interestingly, somewhat different interactions are observed in the other arm of the semipalindrome. There, Glu71 forms direct hydrogen bonds with the amino groups of C-3 and C-4 and with C-5 through a water molecule; Lys73 interacts with N7 of G-6; and Arg62 interacts via water with G-8. This arm of the palindrome has four additional water-mediated interactions (three to bases and one to phosphate (Arg188 to P−9/−10; Fig. 3).
It appears that the crystal has captured the protein in two states: one in which all possible protein–DNA direct contacts defining specificity have been satisfied (left arm) and one, as it were, just before the completion of the recognition process involving several water-mediated contacts (right arm). Before association, both protein and DNA are fully hydrated, and solvent molecules must be released from interacting surfaces before making specific contacts. Some water-mediated interactions are energetically quite favorable and can be trapped in the crystal structure. This result is consistent with previous observations of solvent present on protein–DNA interfaces (18–26).
How can a glutamic acid side chain define the DNA sequence? Glu and Asp residues are quite often found on protein–DNA interfaces (18, 27, 28). Both hydrogen bond–accepting acidic residues show good base-binding potential because, similar to Arg, Lys, and Asn, they can make two hydrogen bonds with bases and can orient bases in coplanar fashion (29). These residues are often found to specifically bind bases of nucleotides (29). It has been observed that glutamic acid prefers to interact with cytosine and can form a Watson–Crick pseudo-pair with two hydrogen bonds as reported in the structures of the HhaI and HaeIII methyltransferases (PDB entries 1MHT, 4MHT, and 1DCT) (30, 31). Analysis of more than 100 structures of protein–DNA complexes determined at high resolution (available in the PDB) reveals that Asp and Glu have a strong preference for cytosine and a somewhat weaker preference for adenine (32). The N4 of cytosines seems to be a particularly attractive contact. In the crystal structure of the Max transcription factor with its DNA target, Glu22 makes four hydrogen bonds to three bases (two consecutive C bases and an A base), defining the sequence CCA of its DNA target (28). This case is similar to the one we observe in the HetR–DNA structure where Glu71 makes hydrogen bonds to three consecutive cytosines.
The GC base pairs in positions 3 and 4 are absolutely conserved (Fig. S1A). Position 5 is slightly less conserved, but the GC base pair is highly preferred. At position 5, C is sometimes substituted by A or G. Substituting C with A places the NH2 group at a somewhat different position, but this should still accommodate the water-mediated interaction with Glu71. Substituting C with G may be similarly favorable. It has been suggested that the interaction of Glu with A and G shows a positive water enhancement coefficient (32). Interestingly, these substitutions are observed in only one arm of the palindrome. At position 6, the interacting G is often replaced with T or A and sometimes with C (Fig. S1). Lys73 seems to be able to switch between base −5 and −6, allowing for some flexibility. Other positions in the palindrome are clearly less important in terms of sequence conservation. Although there may be a preference for purine in position 8 to interact with Arg62, G and A are seen equally. The design of the HetR DNA-binding unit is quite simple. The strongly positively charged surface attracts the phosphate backbone of the DNA duplex and distorts the duplex to closely follow the protein surface. Two negatively charged glutamate side chains project toward the base edges in the major groove and can be accommodated only by the partly positively charged amino groups of cytosine. Lys73 may provide some additional neutralization charge. This interaction is the most preferred of Glu with bases (32) and is found quite often on protein–DNA interfaces. The uniqueness of the HetR case is that this interaction covers three consecutive cytosines in the sequence. More often, the interaction of Glu with a CA step is observed. Additional specificity is provided by Lys73 interacting directly with G-5 and Arg62 with A-7 and G-8. Conservation of other base pairs in the DNA sequence may be needed only to accommodate DNA distortion (indirect recognition) (33).
Mutagenesis.
Our structures are consistent with previously reported and new mutagenesis data. Mutations of three basic residues from the HTH motif in Anabaena HetR (Arg62 in H4, and Lys72 and Lys73 in H5) to glutamic acid were found to be deficient in binding to a 29-bp palindromic oligonucleotide containing the Anabaena HetR recognition sequence; all of these mutants failed to complement the hetR mutant (14). Two other mutations that reduce heterocyst differentiation substantially have been reported at positions G36 and H69 (34). H69 is in the HTH motif and is involved in binding directly to the oxygen of phosphate P+2/+3. Mutation of this residue should interfere with DNA binding and indeed H69D is completely inactive in complementation (Fig. 4). G36 is in a short 310 helix near the N terminus of H3; its mutation may interfere with the orientation of this helix and with the conformations of Arg34 and His35, which provide solvent-mediated interactions with the phosphate backbone. Another DNA binding–deficient mutation, D17E, results in a Het− phenotype in Anabaena (35). D17 is on the dimer interface and its substitution may affect dimer stability.
With the detailed structure in hand, we made a unique series of mutations in HetR (Fig. 4). A remarkable result involves Glu71 mutation to Arg, Gln, or Asn, which results in no binding and no complementation. This result suggests that substitution of two oxygen atoms in Glu71 and Glu71′ with nitrogen atoms (Glu to Gln mutation) in the 70-kDa HetR dimer is sufficient to abolish DNA binding as measured in our EMSA experiments. The Arg62 side chain, previously mutated to Glu, was changed to Asp and Ala. The Asp replacement gave the same result as Glu (no binding, no complementation), whereas the Ala replacement produced a band shift in EMSA, indicating DNA binding, but did not complement. Lys73 and Lys76 gave the same results: replacement with either Glu or Asp gave no binding and no complementation, but replacement with Ala indicated binding but no complementation. These results also suggest that the role of Lys73 in neutralizing the charge of Glu71 is not essential. Replacement of Glu168 in the flap domain with Arg or Lys reduced binding and complementation, but replacement with Asp had no effect (see more discussion below on the oligomeric state of the complex). Other “control” replacements had little effect on binding or complementation: Pro172 to Glu, Asp, or Ala; Lys139 to Asp or Ala; Gln143 to Ala; and Gln247 to Asp. P172 is in a short stretch between H9 and 310 helix 5, about 70 residues into the flap; K139 and Q143 are likewise well into the flap. Q247 is about 30 residues into the hood domain. Not all mutations in the flap domain are unseen by DNA. We believe that the all-or-none results of complementation by the mutants are most likely due to increased degradation of HetR by protease(s) when it is not bound to DNA. Similarly, the Ala-replacement HetR that binds DNA in vitro but does not complement in vivo can probably be explained by failure to be stabilized against degradation by DNA binding in vivo.
Role of the Flap Domain in DNA Binding.
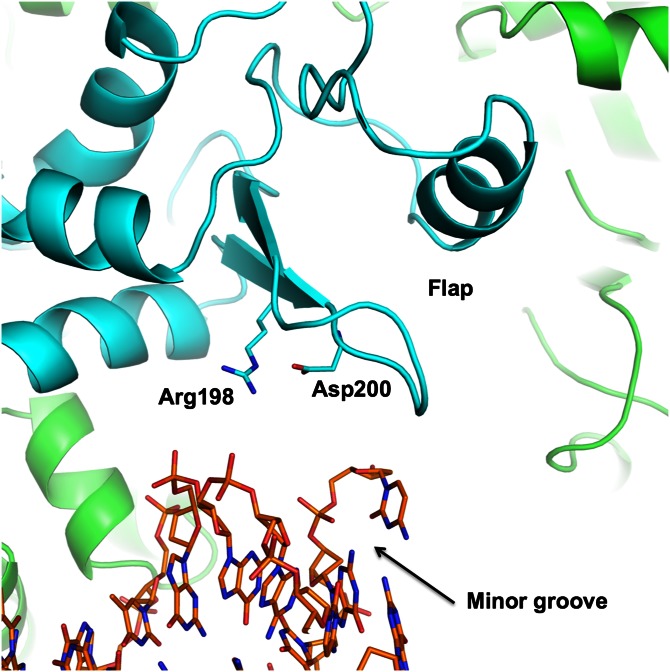
The 21-bp DNA target corresponds approximately to the length of the DNA-binding unit, and the observed interactions cover almost the entire length of the DNA duplex. As mentioned earlier, several residues from the flap domains contribute to the interactions with shorter DNA sequences (Ser179, Glu180, Ala181, and Arg188). To further assess the potential role of the flap domains in DNA binding, we determined structures with 23- and 29-bp DNA duplexes. These longer constructs add one or four more base pairs on each side of the target sequence and extend the length of the DNA by ∼6.4–∼26 Å. The flap surface facing DNA is decorated with positively charged residues (Lys187, Arg188, and Arg198). The DNA duplex follows the surface of the protein, and it approaches the positively charged patches. Arg188 makes direct (18) contact with the oxygen of phosphate P−9/−10. Arg198 is in position to interact with the oxygen of P−10/−11, mediated by water. The β hairpin in the flap domain runs along the minor groove of DNA. Although making no apparent direct contacts in our structure, the Arg198 and Asp200 side chains hydrogen bond with each other and are in position (∼5 Å away) to contact phosphate groups through water-mediated hydrogen bonds, one on the minor groove side (Fig. 5). Unfortunately, at the current resolution of our structure (3.25 Å), water molecules in the minor groove are not visible. Clearly, the DNA can interact with the DNA-binding unit and both flap domains. The flap surface close to the DNA and HTH motifs may also be a good candidate for the binding site of the inhibitory peptide RGSGR, whose binding there would interfere with accommodating DNA along this path.
Fig. 5.
Interaction of flap domain with DNA. The β hairpin in the flap domain runs along the narrow minor groove of DNA. Although making no apparent direct contact, residues Arg198 and Asp200 may contact phosphate groups through water-mediated hydrogen bonds (at the resolution of the structure, water molecules in the minor groove are not visible). Distances to the phosphate groups are ∼5 Å, making possible minor groove-bridging interactions.
Possible HetR Tetramer.
We have previously shown that HetR is a dimer in solution and in the crystal (14). Crystal structures with shorter DNA (23) also show a dimeric arrangement. Surprisingly, the crystal structure of the complex with a 29-bp DNA duplex clearly shows a tetramer (Fig. 6). This tetramer is formed mainly by interaction of the flap domains. The interface is quite extensive (2,346 Å2) and involves salt bridges (Arg120/Asp169′), hydrogen bonds (Asp147-Arg117/Arg101′, Glu112/Glu112′, Gln105/Arg117′, Ser142/Glu229′), and van der Waals interactions Tyr108/Pro113′, Tyr108/Tyr108′), resulting in more hydrophilic than hydrophobic contact area (1,247 vs. 1,104 Å2). Interestingly, as mentioned earlier, Glu168 is on the tetramer interface, although it does not appear to contribute to any direct contact, but it is close to Asp169 and Arg165 and may contribute to the electrostatic potential in this region and stabilize the tetramer. In fact, mutations of Glu168 to Arg or Lys reduce DNA binding and complementation but a mutation to Asp is neutral. In the tetramer, DNA duplexes are parallel to each other and are ∼70 Å apart. One side of this ∼180-kDa assembly is relatively flat with DNA duplexes exposed. The other side contains hood and flap domains forming a large channel that could accommodate an ∼10-kDa protein domain (Fig. 6B). This open channel allows for interaction with HetR, as well as with the minor groove of DNA. This arrangement may explain some limited DNA sequence conservation outside of the HetR recognition sequence. Experimental evidence for a low mobility complex in electrophoresis assays has been reported (36).
Fig. 6.
HetR tetramers observed in the crystal of the complex with the 29-bp DNA duplex. (A) Overall structure of HetR tetramer with two 29-bp DNA duplexes bound. (B) The view into a large channel formed between HetR dimers and DNA in the tetramer. Protein and DNA are shown as solvent-accessible surface. One HetR dimer is in blue and green, the other in light blue and light green, and DNA is red/orange/blue.
Does tetramer formation have functional relevance? HetR recognizes multiple DNA targets and regulates multiple genes in cyanobacteria. For example, there are eight potential binding sites in Scytonema hofmanni 7110 and four in Chlorogloeopsis fritschii 9212 (Fig. S1). By bringing together promoters controlling multiple genes, HetR could coordinate expression of these genes and facilitate initiation of the cascade that leads to differentiation.
In summary, the structure of the HetR–DNA complex obeys rules that are observed for base-specific and phosphate backbone interactions reported for other DNA-binding proteins. Glu71 interacts with three consecutive cytosines at positions 3, 4, and 5, Lys73 interacts with guanine at −5, and Arg62 interacts with adenine and guanine at −7 and −8, respectively. The interaction of Glu71 with three cytosines has not been reported previously. Other HetR interactions with bases follow the general patterns of protein-DNA specific recognition established earlier. Lys73 interacts with guanine, a preferred base, and Arg62 interacts with guanine and adenine, also preferred bases. In addition, there are many electrostatic interactions of Arg and Lys with the phosphate backbone. Particularly striking are complex networks of interactions involving the side chains, main chain, and solvent atoms, as well as α-helix dipoles. Additional interactions, mainly electrostatic, are possible in the flap region extending the DNA covered by protein by at least 6 bp.
Materials and Methods
Gene Cloning and Protein Expression.
The hetR gene from Fischerella sp. MV11 was amplified using polymerase chain reaction (PCR) with genomic DNA as a template as described previously (14). The amplified product was directly cloned in the vector pMCSG19 using a modified ligation-independent cloning protocol. All PCR-cloned genes were sequenced at the University of Chicago Cancer Research DNA Sequencing Facility. The HetR protein was produced as a fusion protein maltose binding protein (MBP)-His6-HetR in Escherichia coli BL21 (DE3) carrying plasmids pMAGIC, which encodes one rare E. coli tRNA [Arg (AGG/AGA)] and pRK1037 (Scientific Reagents). Cells were grown with ampicillin and kanamycin at 100 and 30 μg/mL, respectively, to an optical density of 0.6 at 600 nm. Isopropyl-α-d-thiogalactoside (IPTG; 0.4 mM) was added to induce expression of the fusion protein. MBP was removed in situ on production of the Tobacco Vein Mottling Virus protease encoded on plasmid pRK1037. This construct provided an N-terminal His6-tag separated from the desired gene by a Tobacco Etch Virus protease recognition sequence. HetR protein was expressed in transformed BL21 pMAGIC cells grown in Luria-Bertani broth broth at 37 °C. At A595 of 1.0, protein expression was induced with 1 mM IPTG. The cells were incubated at 18 °C overnight. The harvested cells were resuspended in lysis buffer [500 mM NaCl, 5% (vol/vol) glycerol, 50 mM Hepes, pH 8.0, 10 mM imidazole, 10 mM 2-mercaptoethanol] and stored at −80 °C.
Protein Purification.
The HetR protein from Fischerella sp. MV11 was purified using the procedure described in ref. 14. The harvested cells were resuspended in lysis buffer supplemented with 1 mg/mL lysozyme and 100 μL of protease inhibitor (P8849; Sigma) per 2 g of wet cells. This mixture was kept on ice for 20 min and then sonicated. The lysate was clarified by centrifugation at 36,000 × g for 1 h and filtered through a 0.44-μm membrane. The clarified lysate was applied to a 5-mL HiTrap nickel nitrilotriacetic acid column (GE Health Systems) on an ÄKTAxpress system (GE Health Systems). The His6-tagged protein was released with elution buffer [500 mM NaCl, 5% (vol/vol) glycerol, 50 mM Hepes, pH 8.0, 250 mM imidazole, 10 mM 2-mercaptoethanol], and the fusion tag was removed by treatment with recombinant His7-tagged TEV protease (a gift from D. Waugh, National Cancer Institute, Bethesda, MD). Ni-NTA affinity chromatography was used to remove the His6-tag, uncut protein, and His7-tagged TEV protease. The HetR protein was dialyzed against crystallization buffer containing 250 mM NaCl, 20 mM Hepes, pH 8.0, and 2 mM DTT and then concentrated to 20 mg/mL for crystallization using a Centricon with a 5,000 molecular weight cutoff (Millipore), flash-cooled, and stored in liquid nitrogen.
Preparation of DNA for Cocrystallization with Protein.
The synthetic oligonucleotides for cocrystallization were prepared according to a protocol described earlier (37). The DNA was resuspended in 100 µL of 10 mM Tris⋅HCl, pH 7.5, and 5 mM MgCl2. The concentration was determined spectrophotometrically. The DNA concentration was calculated using the extinction coefficient calculated for each DNA sequence. For crystallization, the DNA duplex in a molar ratio of 1.4 or 1.5 was added to the protein dimer.
Crystallization.
We used a specific DNA 17-bp palindromic sequence recognized by HetR to design the target for crystallization. Self-complementary and semipalindromic DNA duplexes of different lengths (from 19 to 32 bp) were prepared with various modifications on the 5′ end. The HetR protein was cocrystallized with several different DNA duplexes using sitting drop vapor diffusion at 289 K in a CrystalQuick 96-well round-bottom plate (Greiner Bio-One). A 400-nL droplet of the protein–DNA complex was mixed with a 200-nL droplet of crystallization reagent and allowed to equilibrate over 135 µL of crystallization reagent. The nanopipetting was performed using the Mosquito nanoliter liquid handling system (TTP LabTech). The finished plate was then incubated at 16 °C within a RoboIncubator automated plate storage system (RIGAKU). Automated crystal visualization (CrystalTrak; RIGAKU) was used to locate several crystals, which were cryoprotected and flash cooled in liquid nitrogen. Macroscopic crystals were obtained for at least seven oligonucleotides produced in this study. The best crystals of the complex were obtained with the 21-bp-long DNA (top strand, 5′-GTGAGGGGTTAAACCCCTCAC-3′; bottom strand, the complement except that the central T is replaced by A, making the 0 bp an AA mismatch) at a concentration of 0.47 mM and HetR at 0.31 mM from 24% (wt/vol) polyvinylpyrrolidone K15 (NH4)2HPO4, 50 mM, Hepes pH 8.0, 10 mM NaCl, and 10 mM MgCl2. The DNA to protein ratio was 1.5. The crystals belong to the P31 space group and diffracted X-rays to 2.50 Å.
The second-best crystals of the complex were obtained with the 24-base construct forming a 23-bp DNA duplex, its top strand having an extra T on its 5′ end (5′-TGGTGAGGGGTTAAACCCCTCACC-3′) at 0.60 mM and HetR at 0.43 mM from 25% (wt/vol) polyethylene glycol 4000, 50 mM Tris, pH 7.5, 160 mM Na formate, and 10 mM MgCl2. The DNA to protein ratio was 1.4. These crystals belong to the trigonal space group P31 with unit cell dimensions a = 92.93 Å, b = 92.93 Å, and c = 97.65 Å, and they diffracted X-rays to 3.00 Å in the presence of 10% glycerol as a cryoprotectant. The third crystal was obtained with the complex 29-bp DNA (5′-GCAGGCGAGGGGTCTGACCCCTCGCCTGC-3′) at 0.52 mM and of HetR at a concentration of 0.37 mM. The crystal grew at 16 °C from 0.1 M Hepes, pH 7.5, and 25% (wt/vol) polyethylene glycol monomethyl ester 2000. The DNA to protein ratio was 1.3. These crystals belong to the monoclinic space group P21 with unit cell dimensions a = 78.16 Å, b = 182.18 Å, c = 78.37 Å, and β = 91.38°, and they diffracted X-rays to 3.25 Å. All three crystals contained one HetR dimer and one DNA duplex in the asymmetric unit.
Data Collection.
Diffraction data were collected at 100 K at the 19-ID beamline of the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory (38). The single wavelength anomalous dispersion (SAD) data at 0.97931 Å (12.6605 keV) up to 2.5 Å were collected using inverse-beam geometry near the selenium absorption edge from a single protein/DNA complex crystal (0.15 × 0.05 × 0.05 mm) of HetR and 21mer bearing a hetP promoter site. The crystal was exposed for 3 s per 1.0° rotation of ω, with the crystal to detector distance of 380 mm. The data were recorded on a CCD detector Q315 from ADSC scanning a full 130° on ω. The 3.0-Å data from a HetR-24mer (23 bp with T-overhang) DNA crystal were obtained by collecting 220° (on ω) on the same detector at 0.97910 Å with the crystal to detector distance of 450 mm and 1.0° rotation per image frame of 3-s exposure. Similarly, for the HetR-29mer palindromic DNA complex crystal, the 3.25-Å data were collected at 0.91948 Å, in 200° (on ω), diffraction frames of 1° rotation over a 3-s diffraction image with a detector distance of 420 mm. The SBC-Collect program was used for data collection and visualization. Data collection strategy, integration, and scaling were performed with the HKL3000 program package (39). A summary of the crystallographic data can be found in Table S1.
Structure Determination.
The 2.50-Å structure of HetR with 21-mer DNA was determined by MR phasing using MOLREP (40) as implemented in HKL3000 and the HetR dimer structure as a starting model (PDB entry 3QOD). The first model was built by RESOLVE. Multiple cycles of manual model building and adjustment using COOT (41) together with refinement using PHENIX (42) and REFMAC 5.5 (43) on CCP4 6.3 (43) led to the final 2.50-Å model. The final model with Rcryst and Rfree of 0.182 and 0.224, respectively, contained two HetR chains and two strands of the 21-bp DNA. The 3.00-Å HetR–24mer DNA complex structure was determined by molecular replacement using MOLREP on HKL3000 using the 2.5-Å HetR–21mer DNA complex (PDB entry 4IZZ) as a search model. The structure was refined using PHENIX and refmac5.5 on CCP4 6.3. After numerous iterations of manual adjustment and restrained refinement using PHENIX, the model converged to contain two HetR molecules and two strands of the 24mer DNA (23 bp with T-overhang), with Rcryst and Rfree of 0.177 and 0.205, respectively. The 3.25-Å HetR–29mer DNA complex structure was determined by molecular replacement using the same search model (PDB entry 4IZZ, the HetR–21mer DNA complex of 2.5 Å). After multiple rounds of the refinement of alternating manual adjustment using COOT, PHENIX, and REFMAC 5.5 on CCP4 6.3, the model of the HetR–29mer DNA complex converged to contain two HetR–DNA complexes (each complex containing two HetR protein chains and a 29mer duplex DNA) with an Rcryst and an Rfree of 0.235 and 0.276, respectively. All protein residues in all three structures fell within acceptable regions of a MolProbity Ramachandran plot (44) with 97.8%, 96.6%, and 93.2% of all protein residues in favored regions for the HetR with 21mer DNA, 24mer DNA, and 29mer DNA, respectively. Refinement details are shown in Table S1.
Mutagenesis and Complementation Experiments.
To determine whether amino acid substitutions in HetR affected its ability to promote differentiation of heterocysts in vivo, strains of Anabaena were created that differed from the WT only in the nucleotides that encoded the amino acid substitution. Mutant alleles of hetR were generated by overlap-extension PCR (35), cloned into the suicide vector pDR325, and sequenced to verify their integrity as previously described (10). Allelic replacement with mutant alleles of hetR and the ΔhetR strain UHM103 was performed as previously described to yield strains with a single allele of hetR in the normal hetR chromosomal locus. The resulting strains were verified by PCR with primers that annealed outside the region of PCC 7120 chromosomal DNA used in strain construction. Presence of the desired hetR alleles was confirmed by the sizes of PCR amplification products. Growth of Anabaena strains and determination of heterocyst percentages were performed as previously described (34).
EMSA.
These assays were done with a WT Anabaena 29mer and the mutant HetR dimers shown in Fig. 4. Each assay mix contained 1 M NaCl, 100 mM Tris, pH 7.5, 10 mM DTT, 2 mM CaCl2, 50 nM oligonucleotide, and 5 μM HetR dimer. Samples were mixed at room temperature for 20 min, electrophoresed on 5% (wt/vol) TBE (Tris-Borate-EDTA) gels at 60 V for 2 h, and then stained with SYBR Green (Invitrogen). Most results were all-or-none, except for E168K shown with a downward arrow, meaning partial binding. High salt was used to keep the HetR protein in solution; we believe that binding occurs when electrophoresis begins, and the salt leaves the well before the DNA and protein.
Supplementary Material
Acknowledgments
We thank all members of the Structural Biology Center at Argonne National Laboratory for their help in conducting these experiments. Important DNA sequences were generously provided prior to publication by Tal Dagan and Robin Koch (University of Dusseldorf). This work was supported by National Institutes of Health Grant GM094585 (to A.J.), National Science Foundation Grant MCB-1121346 (to S.M.C.), the Ellison Medical Foundation (R.H.), and US Department of Energy, Office of Biological and Environmental Research, Contract DE-AC02-06CH11357 (to A.J.). The submitted manuscript has been created by UChicago Argonne, LLC, Operator of Argonne National Laboratory (“Argonne”). Argonne, a US Department of Energy Office of Science laboratory, is operated under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4IZZ, 4J00, and 4J01).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305971110/-/DCSupplemental.
References
- 1.Flores E, Herrero A. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat Rev Microbiol. 2010;8(1):39–50. doi: 10.1038/nrmicro2242. [DOI] [PubMed] [Google Scholar]
- 2.Yoon HS, Golden JW. Heterocyst pattern formation controlled by a diffusible peptide. Science. 1998;282(5390):935–938. doi: 10.1126/science.282.5390.935. [DOI] [PubMed] [Google Scholar]
- 3.Callahan SM, Buikema WJ. The role of HetN in maintenance of the heterocyst pattern in Anabaena sp. PCC 7120. Mol Microbiol. 2001;40(4):941–950. doi: 10.1046/j.1365-2958.2001.02437.x. [DOI] [PubMed] [Google Scholar]
- 4.Wolk CP. Physiological basis of the pattern of vegetative growth of a blue-green alga. Proc Natl Acad Sci USA. 1967;57(5):1246–1251. doi: 10.1073/pnas.57.5.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen YF, Motteux O, Bédu S, Li YZ, Zhang CC. Characterization of two critical residues in the effector-binding domain of NtcA in the cyanobacterium Anabaena sp. strain PCC 7120. Curr Microbiol. 2011;63(1):32–38. doi: 10.1007/s00284-011-9936-y. [DOI] [PubMed] [Google Scholar]
- 6.Llácer JL, et al. Structural basis for the regulation of NtcA-dependent transcription by proteins PipX and PII. Proc Natl Acad Sci USA. 2010;107(35):15397–15402. doi: 10.1073/pnas.1007015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao MX, et al. Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate. Proc Natl Acad Sci USA. 2010;107(28):12487–12492. doi: 10.1073/pnas.1001556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehira S, Ohmori M. NrrA, a nitrogen-regulated response regulator protein, controls glycogen catabolism in the nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. J Biol Chem. 2011;286(44):38109–38114. doi: 10.1074/jbc.M111.289124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X, Liu D, Lee MH, Golden JW. patS minigenes inhibit heterocyst development of Anabaena sp. strain PCC 7120. J Bacteriol. 2004;186(19):6422–6429. doi: 10.1128/JB.186.19.6422-6429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Risser DD, Callahan SM. Genetic and cytological evidence that heterocyst patterning is regulated by inhibitor gradients that promote activator decay. Proc Natl Acad Sci USA. 2009;106(47):19884–19888. doi: 10.1073/pnas.0909152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du Y, Cai Y, Hou S, Xu X. Identification of the HetR recognition sequence upstream of hetZ in Anabaena sp. strain PCC 7120. J Bacteriol. 2012;194(9):2297–2306. doi: 10.1128/JB.00119-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higa KC, Callahan SM. Ectopic expression of hetP can partially bypass the need for hetR in heterocyst differentiation by Anabaena sp. strain PCC 7120. Mol Microbiol. 2010;77(3):562–574. doi: 10.1111/j.1365-2958.2010.07257.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang JY, Chen WL, Zhang CC. hetR and patS, two genes necessary for heterocyst pattern formation, are widespread in filamentous nonheterocyst-forming cyanobacteria. Microbiology. 2009;155(Pt 5):1418–1426. doi: 10.1099/mic.0.027540-0. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y, et al. Structure of transcription factor HetR required for heterocyst differentiation in cyanobacteria. Proc Natl Acad Sci USA. 2011;108(25):10109–10114. doi: 10.1073/pnas.1106840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinto UM, Winans SC. Dimerization of the quorum-sensing transcription factor TraR enhances resistance to cytoplasmic proteolysis. Mol Microbiol. 2009;73(1):32–42. doi: 10.1111/j.1365-2958.2009.06730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37(17):5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundaralingam M, Pan B. Hydrogen and hydration of DNA and RNA oligonucleotides. Biophys Chem. 2002;95(3):273–282. doi: 10.1016/s0301-4622(01)00262-9. [DOI] [PubMed] [Google Scholar]
- 18.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: A three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29(13):2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spyrakis F, et al. Energetics of the protein-DNA-water interaction. BMC Struct Biol. 2007;7:4. doi: 10.1186/1472-6807-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otwinowski Z, et al. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- 21.Zhang RG, et al. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 2002;417(6892):971–974. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 22.Billeter M, Güntert P, Luginbühl P, Wüthrich K. Hydration and DNA recognition by homeodomains. Cell. 1996;85(7):1057–1065. doi: 10.1016/s0092-8674(00)81306-9. [DOI] [PubMed] [Google Scholar]
- 23.Lamoureux JS, Glover JN. Principles of protein-DNA recognition revealed in the structural analysis of Ndt80-MSE DNA complexes. Structure. 2006;14(3):555–565. doi: 10.1016/j.str.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 24.Wibowo FR, Rauch C, Trieb M, Wellenzohn B, Liedl KR. Water-mediated contacts in the trp-repressor operator complex recognition process. Biopolymers. 2004;73(6):668–681. doi: 10.1002/bip.20023. [DOI] [PubMed] [Google Scholar]
- 25.Cheng AC, Chen WW, Fuhrmann CN, Frankel AD. Recognition of nucleic acid bases and base-pairs by hydrogen bonding to amino acid side-chains. J Mol Biol. 2003;327(4):781–796. doi: 10.1016/s0022-2836(03)00091-3. [DOI] [PubMed] [Google Scholar]
- 26.Joachimiak A, Haran TE, Sigler PB. Mutagenesis supports water mediated recognition in the trp repressor-operator system. EMBO J. 1994;13(2):367–372. doi: 10.1002/j.1460-2075.1994.tb06270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain D, et al. Crystal structure of bacteriophage lambda cII and its DNA complex. Mol Cell. 2005;19(2):259–269. doi: 10.1016/j.molcel.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Brownlie P, et al. The crystal structure of an intact human Max-DNA complex: New insights into mechanisms of transcriptional control. Structure. 1997;5(4):509–520. doi: 10.1016/s0969-2126(97)00207-4. [DOI] [PubMed] [Google Scholar]
- 29.Kondo J, Westhof E. Classification of pseudo pairs between nucleotide bases and amino acids by analysis of nucleotide-protein complexes. Nucleic Acids Res. 2011;39(19):8628–8637. doi: 10.1093/nar/gkr452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Gara M, Roberts RJ, Cheng X. A structural basis for the preferential binding of hemimethylated DNA by HhaI DNA methyltransferase. J Mol Biol. 1996;263(4):597–606. doi: 10.1006/jmbi.1996.0601. [DOI] [PubMed] [Google Scholar]
- 31.Reinisch KM, Chen L, Verdine GL, Lipscomb WN. The crystal structure of HaeIII methyltransferase convalently complexed to DNA: An extrahelical cytosine and rearranged base pairing. Cell. 1995;82(1):143–153. doi: 10.1016/0092-8674(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 32.Marabotti A, et al. Energy-based prediction of amino acid-nucleotide base recognition. J Comput Chem. 2008;29(12):1955–1969. doi: 10.1002/jcc.20954. [DOI] [PubMed] [Google Scholar]
- 33.Rohs R, et al. The role of DNA shape in protein-DNA recognition. Nature. 2009;461(7268):1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borthakur PB, Orozco CC, Young-Robbins SS, Haselkorn R, Callahan SM. Inactivation of patS and hetN causes lethal levels of heterocyst differentiation in the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol. 2005;57(1):111–123. doi: 10.1111/j.1365-2958.2005.04678.x. [DOI] [PubMed] [Google Scholar]
- 35.Risser DD, Callahan SM. Mutagenesis of hetR reveals amino acids necessary for HetR function in the heterocystous cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 2007;189(6):2460–2467. doi: 10.1128/JB.01241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feldmann EA, et al. Evidence for direct binding between HetR from Anabaena sp. PCC 7120 and PatS-5. Biochemistry. 2011;50(43):9212–9224. doi: 10.1021/bi201226e. [DOI] [PubMed] [Google Scholar]
- 37.Joachimiak A, Sigler PB. Crystallization of protein-DNA complexes. Methods Enzymol. 1991;208:82–99. doi: 10.1016/0076-6879(91)08009-7. [DOI] [PubMed] [Google Scholar]
- 38.Rosenbaum G, et al. The Structural Biology Center 19ID undulator beamline: Facility specifications and protein crystallographic results. J Synchrotron Radiat. 2006;13(Pt 1):30–45. doi: 10.1107/S0909049505036721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: The integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 8):859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 40.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 12):1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2126–2132.
- 42.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.