Abstract
Biofuels are the most immediate, practical solution for mitigating dependence on fossil hydrocarbons, but current biofuels (alcohols and biodiesels) require significant downstream processing and are not fully compatible with modern, mass-market internal combustion engines. Rather, the ideal biofuels are structurally and chemically identical to the fossil fuels they seek to replace (i.e., aliphatic n- and iso-alkanes and -alkenes of various chain lengths). Here we report on production of such petroleum-replica hydrocarbons in Escherichia coli. The activity of the fatty acid (FA) reductase complex from Photorhabdus luminescens was coupled with aldehyde decarbonylase from Nostoc punctiforme to use free FAs as substrates for alkane biosynthesis. This combination of genes enabled rational alterations to hydrocarbon chain length (Cn) and the production of branched alkanes through upstream genetic and exogenous manipulations of the FA pool. Genetic components for targeted manipulation of the FA pool included expression of a thioesterase from Cinnamomum camphora (camphor) to alter alkane Cn and expression of the branched-chain α-keto acid dehydrogenase complex and β-keto acyl-acyl carrier protein synthase III from Bacillus subtilis to synthesize branched (iso-) alkanes. Rather than simply reconstituting existing metabolic routes to alkane production found in nature, these results demonstrate the ability to design and implement artificial molecular pathways for the production of renewable, industrially relevant fuel molecules.
Keywords: branched fatty acid biosynthesis, lux genes, metabolic engineering, synthetic biology
The demand for transport fuel currently represents 60% of global oil production (1) and is predicted to rise from 85 million barrels a day in 2007 to 104 million barrels a day by 2030 because of a combination of population growth, industrialization, and increasing prosperity (2). Moreover, the bulk of crude oil production is increasingly located in regions of global insecurity, leading to episodic disruptions in market supply and increased fuel costs. Finally, the transport sector is the second biggest source of global greenhouse gas emissions (1) and a politically acceptable target for CO2 emission mandates: within the European Union, retail fuels are mandated to contain at least 5% renewable biofuel, rising to 10% by 2020 (3), with similar blending mandates enacted in the United States, China, and Brazil. Currently, retail biofuels fall into two categories: alcohols and biodiesel. Alcohols (predominantly ethanol) are blended with gasoline (petrol), and biodiesels (fatty acid (FA) alkyl esters and hydrogenated vegetable oils) are blended with automotive gas-oil (diesel). These biofuels, however, entail substantial downstream processing and quality control costs and are not fully compatible with modern, mass-market, high-performance, low-emission engines (4–6). Simply increasing the proportion of current biofuel to petroleum distillate in retail fuels presents significant challenges for the entire transport sector. Without costly and time-consuming infrastructure and engine remodeling, the maximum blend ratio of biofuel to petroleum-distillate (the blend wall) is between 10% and 20% (4). Consequently, without a dramatic change in vehicle technology and fuel supply infrastructure, 80–90% of transport fuel demand cannot be met through replacing petroleum-derived fuels with the biofuels currently available.
To overcome the end-user blend wall, it is essential to generate precise chemical replacements to fossil fuels through sustainable means. Retail transport fuels are composed primarily of hydrocarbons (n-alkanes) of various carbon chain lengths (Cn), branched hydrocarbons (iso-alkanes), and unsaturated hydrocarbons (n-alkenes). The ideal biofuels are therefore n-alkanes, iso-alkanes, and n-alkenes that are chemically and structurally identical to the fossil fuels they are designed to replace.
Biogenic alkanes are found throughout nature (7–12), but not in the required form for direct replacement fuels; new pathways for the biological production of different alkanes must therefore be developed. The only genetically characterized alkane biosynthesis pathways are the production of very long chain alkanes (>C28) by Arabidopsis thaliana (7) and the production of pentadecane (C15:0) and heptadecane (C17:0) by cyanobacteria (8). The cyanobacterial alkane pathway has also been biochemically characterized: fatty acyl-acyl carrier protein (ACP) is reduced to fatty aldehyde (Cn) by acyl-ACP reductase (AR), which is then decarbonylated to an alkane (Cn-1) by fatty aldehyde decarbonylase (AD), primarily producing heptadecene (C17:1) in Escherichia coli and releasing formate (8, 13, 14). To advance biologically synthesized alkanes further into the fuel market, they must meet the technical requirements demanded of retail fuels, not the requirements of the source organism.
The objective of the present study was to address the problem of direct fossil fuel replacements through the de novo design and assembly of synthetic metabolic pathways, with the aim of producing linear and branched-chain alkanes and alkenes of variable, but specified, Cn. To achieve this aim, we engineered E. coli cells to use free FAs directly, rather than the FA thioesters used by the native cyanobacterial pathway (8). Exploiting free FAs as entry substrates for alkane synthesis meant that predictable alterations to the hydrocarbon output of the cells was possible by modifications to the free FA pool, either by additional pathway engineering or by exogenous supplementation of the media. Using this strategy, E. coli expressing different molecular modules produced linear tridecane (C13:0), tridecene (C13:1), pentadecane, pentadecene (C15:1), hexadecane (C16:0), hexadecene (C16:1), heptadecane, heptadecene, and branched tridecane and pentadecane, thereby replicating the chemical and structural requirements of retail diesel hydrocarbons commonly used in temperate climates. The results thus provide proof of principle for the metabolic production of industrially relevant direct petroleum-replica hydrocarbons.
Results and Discussion
Design and Assembly of an Artificial Alkane Biosynthetic Pathway in E. coli.
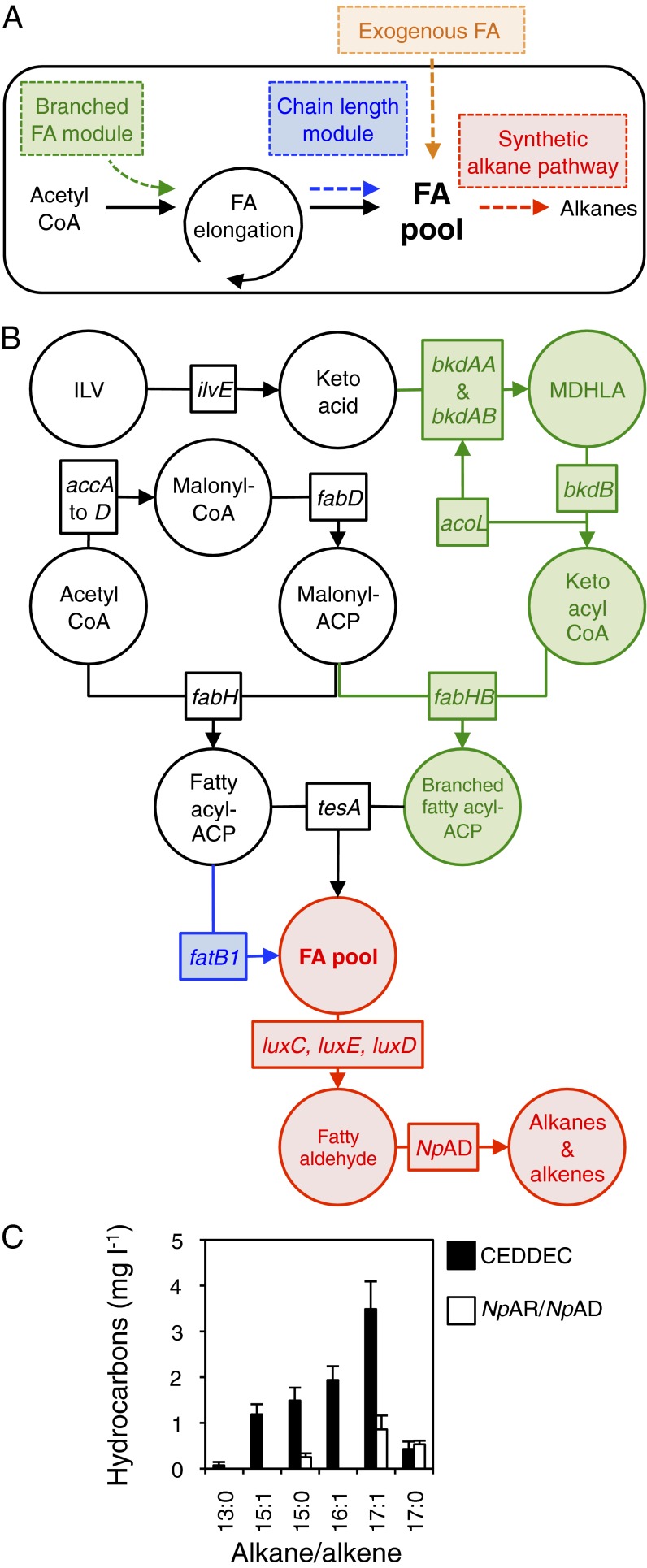
Alkane production in E. coli through the cyanobacterial biosynthetic pathway produces linear C15:0 and C17:1 aliphatic hydrocarbons from the intermediate of fatty acid biosynthesis, fatty acyl-ACP (8). We considered that conversion of free FAs, rather than fatty acyl-ACP, to alkanes was a more tractable target for bio-alkane production (Fig. 1A) for three main reasons: First, exploiting the FA pool would provide a mechanism to direct alkane biosynthesis to different Cn alkanes because heterologous expression of thioesterases in E. coli introduces control over FA Cn (15, 16); second, FAs are more abundant in cells than fatty acyl-ACP; and third, the FA pool can be substantially increased through genetic manipulation (17–19). Our first aim was therefore to exploit FAs as substrates for alkane biosynthesis. To achieve this, we generated and expressed in E. coli an artificial metabolic pathway in which the FA reductase (FAR) complex (20, 21), encoded by the genes luxC, luxE, and luxD from the bioluminescent bacterium Photorhabdus luminescens, supplied the appropriate Cn fatty aldehyde substrate to an aldehyde decarbonylase (NpAD) cloned from Nostoc punctiforme (Fig. 1B, red). Coexpression of luxC, luxE, luxD, and the aldehyde decarbonylase gene (collectively termed CEDDEC) in E. coli cells resulted in the production of alkanes and alkenes of the appropriate chain length for diesel or aviation fuel, namely tridecane, pentadecane, pentadecene, hexadecene, heptadecane, and heptadecene (Fig. 1C). By comparison, expression of AR and AD from N. punctiforme in E. coli led to the production of heptadecane, heptadecene, and pentadecane (Fig. 1C) as described previously (8). Together, these data demonstrate that the P. luminescens FAR complex provided suitable substrates for n-alkane and n-alkene biosynthesis in the engineered E. coli, as predicted.
Fig. 1.
Production of fuel-grade alkanes from FAs by engineered E. coli. (A) Overview: three independent modifications to the FA pool (two genetic and one by exogenous supplementation) resulted in predictable changes to the output of cells engineered to produce alkane fuels via the artificial pathway. (B) Details of the genetic modifications implemented: the synthetic alkane biosynthetic pathway (red) was engineered through the coexpression of the reductase (luxC), synthetase (luxE), and transferase (luxD) genes coding for the FAR complex from the P. luminescens luciferase operon and the gene encoding NpAD. For construct details, see Fig. S1. Branched-chain FAs (green) were produced by expression of genes coding for the BCKD complex (bkdAA, bkdAB, bkdB, and acoL, which code for E1α, E1β, E2, and E3 subunits, respectively) and fabHB encoding KASIII. Isoleucine, leucine, and valine (ILV) can be converted to α-keto acids through endogenous branched-chain amino acid aminotransferase activity (encoded by ilvE). MDHLA represents either S-(3-methylbutanoyl)-dihydrolipoamide E, S-(2-methylpropanoyl)-dihdrolipoamide E, or S-(2-methylbutanoyl)-dihydrolipoamide E, the products of isoleucine, leucine, or valine breakdown, respectively (Fig. S2). The E1α, E1β, and E2 subunits convert keto acids to keto acyl-CoA, whereas the E3 subunit recycles the lipoamide-E cofactor. All branched-chain components were from B. subtilis. FA chain length was altered through expression of the FatB1 thioesterase from C. camphora (blue). (C) Hydrocarbons produced by cells expressing the synthetic alkane pathway (CEDDEC) or the cyanobacterial alkane pathway (AR and AD from N. punctiforme) without modifications to the fatty acid pool. n = 6 biological replicates; error bars represent SE of mean. Cells were grown and induced as detailed in Experimental Procedures.
Modification to the FA Pool Results in Predictable Changes to the Output of the Artificial Alkane Biosynthesis Pathway.
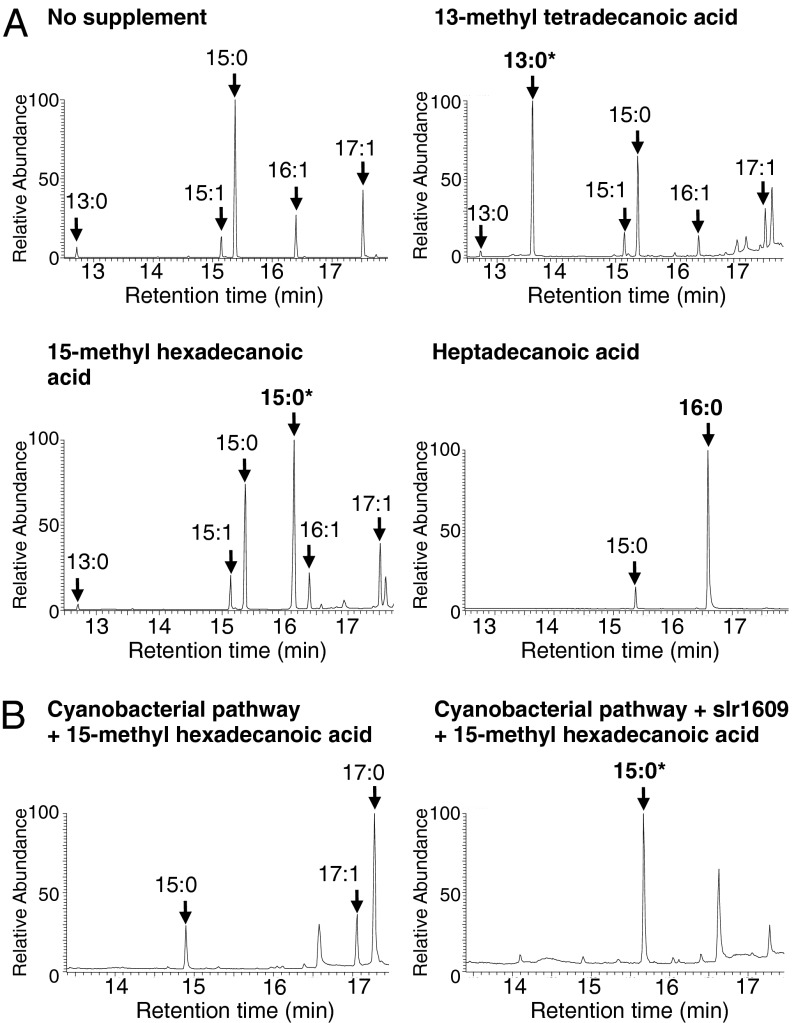
The rationale for the development of free FAs as a pool for alkane biosynthesis was that it would provide a substrate that was more tractable for fine-tuning the properties of the biofuel molecules. Such an approach has been successfully used to structurally tailor the FA ethyl ester and fatty alcohol output of engineered cells (19). To test whether the synthetic pathway CEDDEC (Fig. 1, red) was compatible with a modified FA pool and whether changes in the FA pool would translate to changes in alkane output, we predicted that the inclusion of different FAs in the growth media (Fig. 1, orange) would result in the production of different alkanes by the engineered cells; like many microbes, E. coli possesses the capacity to assimilate exogenous FAs (22). We compared the alkane products of cells engineered to express the CEDDEC synthetic pathway with those engineered to express the cyanobacterial AR/AD pathway when supplied with FAs not normally present within E. coli. When the growth medium was supplemented with 13-methyl tetradecanoic acid (a branched FA), cells expressing the CEDDEC pathway produced branched tridecane (Fig. 2A). Furthermore, cells expressing CEDDEC genes produced branched pentadecane and linear hexadecane when grown in the presence of two other FAs not present within E. coli: 15-methyl hexadecanoic acid (branched C16:0) and heptadecanoic acid (C17:0), respectively. The synthetic pathway was therefore able to use a modified free FA pool to generate specific alkane products whose nature was determined by the supplied FA. By contrast, cells expressing the N. punctiforme AR/AD pathway were unable to use exogenous 15-methyl hexadecanoic acid (Fig. 2B). To confirm that this was due to an inability of NpAR to use exogenous FA rather than an inability to use branched substrates, ORF slr1609 from Synechocystis sp. PCC 6803 was also expressed in the cells. ORF slr1609 codes for an acyl-ACP synthetase whose proposed role is the reactivation of FAs to fatty acyl-ACP (23). When ORF slr1609 and the N. punctiforme AR/AD pathway were coexpressed, cells produced branched pentadecane from exogenous branched FA. Branched substrates can therefore be used by NpAR. These results are consistent with the hypothesis that fatty acyl-ACP is the sole substrate for the cyanobacterial pathway in vivo. The results also indicate that E. coli lacks a mechanism for activating FA to fatty acyl-ACP. Importantly for this investigation, the results clearly demonstrate that the alkane biosynthetic CEDDEC pathway is capable of using branched FA substrates to generate iso-alkanes. The FA pool of E. coli cells is therefore a suitable substrate provider for the artificial CEDDEC module and is a viable target for modifying alkane output toward industrially relevant molecules.
Fig. 2.
Predictable modifications to the alkane output from engineered E. coli cells in media supplemented with exogenous fatty acids. (A) Typical GC-MS total ion chromatograms (TIC) of alkanes extracted from E. coli cells that express the CEDDEC pathway, grown and induced as described in Experimental Procedures with 100 μg⋅mL−1 FA supplementation as indicated. *Branched alkane products. Branched alkanes were identified by retention time and comparison with the mass spectral library. Branched alkanes are more likely to fragment at the position where the branch is located, giving a smaller molecular ion (e.g., m/z 197 is indicative of methyl pentadecane) with a larger molecular ion peak than straight chain alkanes. Total alkanes recovered: CEDDEC, 5.8 mg⋅L−1 ± 0.3; 13-methyl tetradecanoic acid supplement, 5.9 mg⋅L−1 ± 0.3; 15-methyl hexadecanoic acid supplement, 4.3 mg⋅L−1 ± 0.3; heptadecanoic acid supplement, 7.4 mg⋅L−1 ± 0.4. (B) GC-MS TIC of hydrocarbons extracted from E. coli cells expressing the cyanobacterial NpAR/NpAD pathway alone (Left) or in conjunction with the cyanobacterial (Synechocystis sp. PCC 6803) slr1609 ORF (Right). Cells were grown and induced as described in Experimental Procedures and supplemented with 100 μg⋅mL−1 15-methyl hexadecanoic acid. Total alkanes recovered: cyanobacterial pathway alone, 1.5 mg⋅L−1 ± 0.03; in combination with slr1609, 0.8 mg⋅L−1 ± 0.02.
LuxD Is Required for Alkane Production, but Is Likely to Be Involved in Protein–Protein Interactions Rather than as a Catalyst.
After demonstrating that E. coli cells expressing the CEDDEC pathway are capable of synthesizing alkanes from an exogenously manipulated FA pool, we aimed to express additional genetic modules to manipulate the FA pool in vivo, thereby tailoring the alkane output of the cells toward the production of compounds used in retail fuel blends. However, given that our target substrate for alkane biosynthesis was the free FA pool, it was possible that the expression of luxD was unnecessary. The luxD gene codes for the fatty acyl transferase subunit that may provide FA substrate for fatty acyl synthetase (LuxE) through cleavage of activated FAs (i.e., fatty acyl-ACP and fatty acyl CoA). The fatty acyl reductase subunit (LuxC) then uses the acyl-protein thioester product of the LuxE catalyzed reaction to produce fatty aldehyde (24). Within the FAR protein complex (a central reductase tetramer with each subunit interacting with a synthetase subunit), fatty acyl groups are directly passed between LuxE and LuxC (25). LuxD subunits interact only weakly with each synthetase subunit (24). If FAs enter the FAR complex directly through LuxE rather than being supplied by LuxD, then it may be possible to minimize the number of heterologously expressed genes (and hence decrease the cost of protein synthesis to the cells) by omitting luxD from the luxCED operon.
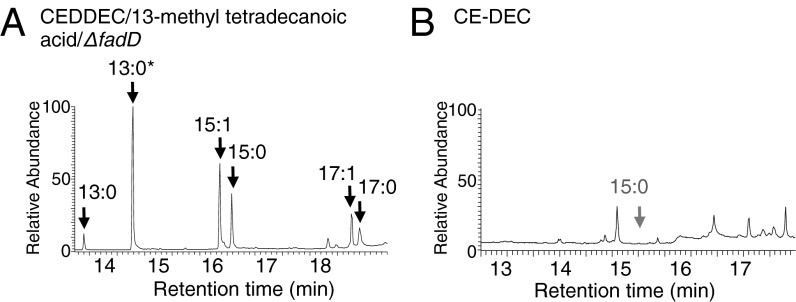
To explore this possibility, we undertook two experiments. First, we confirmed that FAs enter alkane biosynthesis through LuxE. To do this, we expressed CEDDEC in a BL21* (DE3) expression strain lacking the fadD gene: fadD codes for acyl-CoA synthetase and strains mutated in fadD cannot produce acyl-CoAs and cannot grow on exogenous FA (22). In ΔfadD cells, exogenous FAs will not be converted into their CoA thioesters. Moreover, E. coli does not appear to possess any capacity to convert exogenous FA to fatty acyl-ACP (Fig. 2B). If FAs enter the FAR complex through LuxE rather than LuxD, then exogenous FA will be converted to alkanes in a ΔfadD strain. Alternatively, if exogenous FAs need to be activated for use by LuxD, then exogenous FAs should not be converted to alkanes in a ΔfadD strain. When CEDDEC was expressed in ΔfadD E. coli supplemented with exogenous branched FA, branched alkanes were produced (Fig. 3A). We therefore concluded that FA directly enters the alkane biosynthetic pathway through LuxE.
Fig. 3.
LuxD is an essential component of the artificial alkane biosynthetic (CEDDEC) pathway. (A) GC-MS TIC of hydrocarbons extracted from E. coli cells expressing CEDDEC in ΔfadD BL21* (DE3) in the presence of 100 μg⋅mL−1 13-methyl tetradecanoic acid. (B) GC-MS TIC of hydrocarbons extracted from WT E. coli cells expressing CE-DEC (luxC, luxE, and NpAD with the luxD gene removed from the operon; Fig. S1). The absence of pentadecane is indicated by the gray arrow. All cells were grown and induced as described in Experimental Procedures.
Consequently, it may be possible to reduce the number of heterologously expressed genes in the CEDDEC module to only LuxC, LuxE, and NpAD. We therefore removed the luxD gene from the synthetic operon by digestion with HindIII and NotI (Fig. S1). Expression of the resulting module (luxC, luxE, and NpAD without luxD; termed CE-DEC) in E. coli resulted in an almost complete absence of alkane product (Fig. 3B). Although the exact nature of the interactions between LuxD and the central LuxC/LuxE complex is not fully known, LuxD is clearly a crucial component of the FAR complex and the alkane biosynthesis module. Previously, LuxD has been hypothesized to be important in FAR complex activity through protein-protein interactions between the carboxyl terminal regions of LuxD and LuxE (26) (rather than having a direct catalytic activity as a fatty acyl transferase) and our data support this proposal.
Biosynthesis of Branched Alkanes from in Vivo Biosynthesis of Branched FAs in E. coli.
Having demonstrated that supplementary FAs can be converted to their corresponding Cn-1 alkanes and alkenes (Fig. 2), we sought to manipulate the internal FA pools genetically because supplementation of exogenous FAs is not a practicable feedstock for biofuel production.
Our first aim was to produce branched alkanes biosynthetically. The presence of branched-chain molecules within fuel blends is necessary to maintain performance at low temperature and high altitude. Having previously demonstrated the capacity for incorporation of exogenous, branched FAs into the CEDDEC alkane biosynthetic pathway (Fig. 2), we designed a branched-chain FA biosynthetic module (Fig. 1, green) that would generate branched FA substrates. WT E. coli is unable to produce branched FAs endogenously because the native β-ketoacyl-ACP synthase III (KASIII) enzyme that catalyses the initial step in the FA elongation cycle accepts only linear acetyl-CoA or propionyl-CoA substrates. Moreover, the substrates required by KASIII for branched FAs are not present within E. coli (27). Many gram-positive bacteria do, however, produce branched-chain FAs (28), and branched-chain FAs can be produced in vitro by the FA elongation enzymes from E. coli if an alternative KASIII enzyme and suitable precursor molecules are present (27, 29). Both the capacity to incorporate branched starter molecules into the FA elongation cycle and the pathways to supply these substrates were therefore introduced into E. coli by expression of KASIII (FabH2) and the branched-chain α-keto acid dehydrogenase (BCKD) complex from Bacillus subtilis (Fig. 1B and Figs. S1 and S2). We reasoned that the substrates for the BCKD complex would be supplied through the endogenous activity of branched-chain amino acid aminotransferase (E.C. 2.6.1.42) in E. coli, using the branched amino acids isoleucine, leucine, and valine as substrates. Introducing B. subtilis KASIII activity into E. coli would then provide branched molecules as starter substrates for the endogenous FA elongation cycle, and thence for the CEDDEC module.
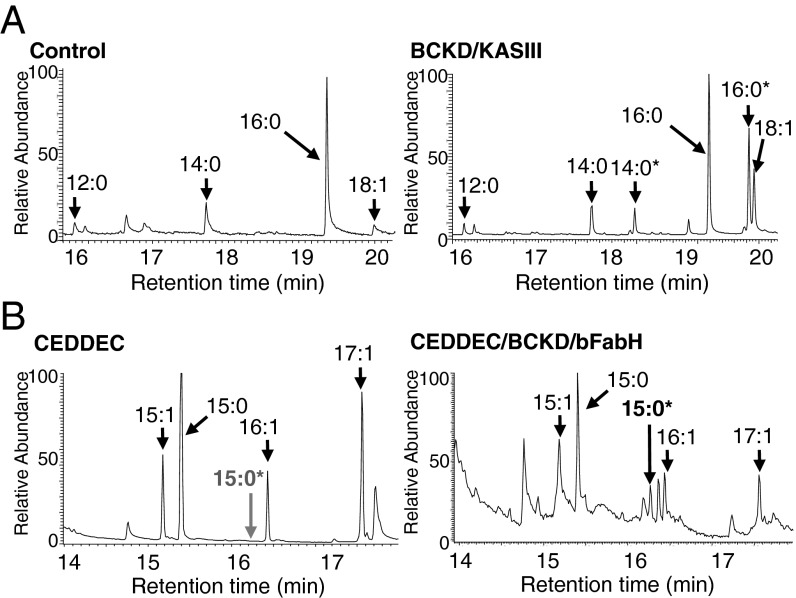
As expected, coexpression of the FabH2 KASIII and the BCKD complex from B. subtilis in E. coli resulted in the production of the branched-chain FAs 13-methyl tetradecanoic acid and 15-methyl hexadecanoic acid (Fig. 4A), thereby demonstrating branched FA biosynthesis in vivo in E. coli. To determine if it were possible to produce branched FAs and to convert them into branched alkanes in the same cells, both the branched FA biosynthesis module (Fig. 1, green) and alkane biosynthesis module CEDDEC (Fig. 1, red) were simultaneously expressed in E. coli. We observed that expressing all nine genes from both modules (bkdAA, bkdAB, bkdB, acoL, fabHB, luxC, luxE, luxD, and NpAD) simultaneously in E. coli had a negative impact on the level of recombinant protein synthesis compared with cells engineered with CEDDEC alone. Consequently, the amount of alkanes produced by these cells was also reduced compared with the baseline established by those expressing CEDDEC. Despite this limitation, a range of different alkane and alkene products was detected including the branched alkane, methyl pentadecane (Fig. 4B). The results therefore clearly demonstrate the feasibility of engineering artificial pathways for branched alkane biosynthesis in a microbial host.
Fig. 4.
De novo production of branched fatty acids and alkanes in E. coli. (A) Typical GC-MS TIC of FAs extracted from control cells without BCKD/KASIII expression (Left) or from cells expressing BCKD/KASIII (Right). *Branched FA products [methyl tetradecanoic acid (14:0*) and methyl hexadecanoic acid (16:0*)]. Total FA recovered from control cells, 0.6 mg⋅L−1 ± 0.2 in control lines; for cells expressing BCKD/KASIII, 2.5 mg⋅L−1 ± 0.4. (B) Typical GC-MS TIC of alkanes extracted from control cells expressing CEDDEC (Left) and cells coexpressing BCKD/KASIII and CEDDEC (Right). *Branched alkane product [methyl pentadecane (15:0*) absent in CEDDEC]. Mass spectral data for methyl pentadecane are given in Fig. S3.
Genetic Control over Alkane Chain Length.
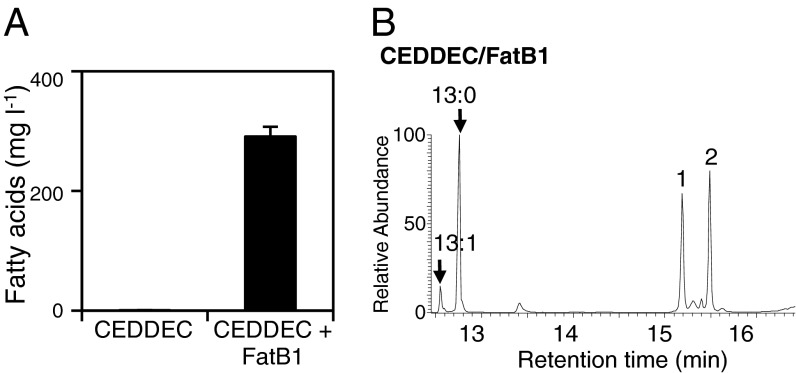
Our second aim was to tailor the Cn of the alkanes: this ability is critical for industrial applications because Cn largely defines the properties of the fuel (30). FatB1 is a thioesterase from Cinnamomum camphora (camphor) responsible for terminating FA elongation by cleaving the growing fatty acyl-ACP chain, releasing FA and an ACP molecule (15, 16). The FatB1 thioesterase from C. camphora is specific for C14 fatty acyl-ACP chains and has been used heterologously to manipulate FA pools: in E. coli, the expression of fatB1 (without the endogenous transit peptide) results in the synthesis of tetradecanoic acid (C14:0) (15). We therefore predicted that the coexpression of fatB1 and CEDDEC in E. coli would result in a modified FA pool that, because of the dominance of C14 FAs, would yield predominantly C13 alkanes. The CEDDEC/FatB1 combination resulted in a large increase in the tetradecanoic acid pool and the almost exclusive production of tridecane in the engineered E. coli (Fig. 5 and Figs. S3 and S4).
Fig. 5.
Production of tridecane in E. coli. (A) Total FAs extracted from cells expressing CEDDEC alone or CEDDEC in conjunction with the FatB1 thioesterase from C. camphora. n = 3 biological replicates; error bars represent SE of the mean. Cells were grown and induced as detailed in Experimental Procedures. Typical GC-MS TIC is given in Fig. S4. (B) Typical GC-MS TIC of hydrocarbons extracted from cells expressing CEDDEC and FatB1. Putative identification of trans-5-dodecanal or tetradecanal (peak 1) and tridecanone (peak 2) were identified in cells expressing FatB1 with and without CEDDEC. Mass spectral data for tridecane are given in Fig. S3. Total alkanes recovered from cells expressing CEDDEC/FatB1, 2.9 mg⋅L−1 ± 0.1.
Conversion of FA to Alkanes Is Limited by the in Vivo Use of Fatty Aldehyde Substrate.
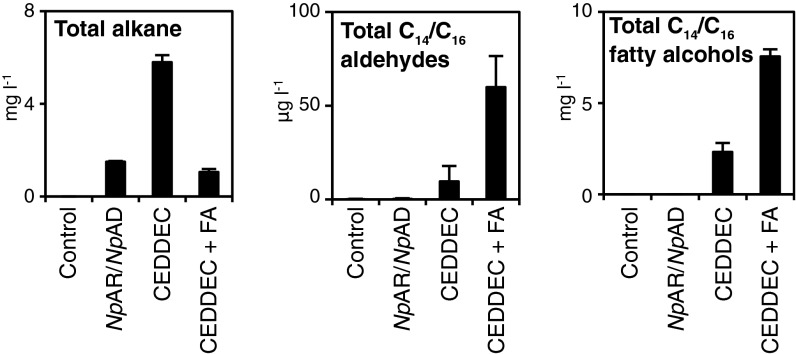
In the system presented here, alkane titers from both the cyanobacterial pathway and the artificial CEDDEC pathway were ∼2–5 mg⋅L−1 24 h after induction (Fig. 1 and Fig. S5). These values are in agreement with a recent report for expression of the cyanobacterial pathway in BL21* (DE3) cells (31). Such titers were encountered even in the presence of the high concentrations of FAs achieved through exogenous feeding (Fig. 2A) or following expression of FatB1 (Fig. 5A). Alkane titers reported previously for heterologous expression of the cyanobacterial pathway however were in the region of 10–80 mg⋅L−1 48 h postinduction (7). To determine if the FAR complex was limiting flux from FA to alkanes, we assayed the intermediate metabolite of the alkane biosynthetic pathway, fatty aldehyde. In cells expressing the CEDDEC pathway, there is a greater pool of fatty aldehydes than in control cells and in cells expressing the cyanobacterial pathway (Fig. 6). Moreover, in the presence of exogenous tetradecanoic acid, there were further significant increases in the pool of C14 fatty aldehyde (Fig. 6 and Fig. S6). The fatty aldehyde content however was small compared with total pool size for alkanes and FAs. Fatty aldehydes may be converted to fatty alcohols in E. coli cells by endogenous aldehyde reductases (32, 33). We therefore also examined the fatty alcohol content of these cells. As with fatty aldehydes, the fatty alcohol content was greater in the cells expressing the CEDDEC pathway than in those expressing the cyanobacterial pathway or in control cells (Fig. 5B). Moreover, feeding with exogenous FAs further increased the fatty alcohol content. Although we conclude that the FAR complex does not currently limit flux into this pathway (and creates a larger pool of aldehyde substrate for the decarbonylase reaction than the cyanobacterial AR), the accumulation of fatty alcohols indicates potential in vivo competition for aldehyde substrate between the decarbonylase-catalyzed reaction and aldehyde reductase reactions. Future engineering strategies will therefore need to focus on improving the flux through the decarbonylase-catalyzed reaction by increasing the ability of AD to convert aldehydes to alkanes and removal of the competing reaction(s).
Fig. 6.
Identification of alkane biosynthesis limitations in the E. coli host. Total alkane, aldehyde (C14 and C16), and fatty alcohols (C14 and C16) extracted from control cells [BL21* (DE3)], cells expressing the cyanobacterial pathway (NpAR/NpAD), the artificial CEDDEC pathway, and the artificial CEDDEC pathway supplemented with tetradecanoic acid (+ FA). n = 3 biological replicates; error bars represent SE of the mean. Cells were grown and induced as detailed in Experimental Procedures.
Conclusions
The objective of this study was to determine if it were possible to develop a unique biological route for the production of industrially-relevant alkanes and alkenes. We have engineered in E. coli pathways for the production of specified, aliphatic n- and iso-alkanes and -alkenes of various chain lengths that are exact replicas of petroleum-based molecules used in retail fuels. Rather than using fatty acyl-ACPs as substrates, the modules described here exploit the free FA pool as the primary substrate for alkane biosynthesis. This approach ensured that the hydrocarbon output of the engineered cells could be altered by rational modifications to the FA pool. This capacity for designer alkane production was demonstrated by expression of a thioesterase from C. camphora to alter alkane Cn, by expression of the branched-chain α-keto acid dehydrogenase complex and β-keto acyl-ACP synthase III from B. subtilis to synthesize branched alkanes, and by inclusion of exogenous FAs in the growth media in cells expressing a unique alkane biosynthetic pathway. One of the most important characteristics of the genetic modules described here is that there is no “blend wall” for the biologically generated fossil-fuel replicas synthesized. The identification of limitations in conversion of aldehydes to alkanes and of an alternative metabolic sink provides a focus for future engineering strategies: these may involve removing competing reactions, generating an intracellular environment in which the decarbonylase can more efficiently use the fatty aldehydes generated by the first step of the pathway or improving the efficiency of the decarbonylase itself. The size of the challenge facing advanced biofuels can be appreciated when considering the effort necessary to progress the engineered biosynthesis of semisynthetic artemisinin from the laboratory to commercial production: many changes beyond manipulating the endogenous mevalonate pathway have been required, including changes to host cell biology, fermentation conditions, and extraction procedures (34, 35). The application of engineering, life-cycle, and economic costing analysis for scale-up procedures combined with the exploitation of alternative, nonfood carbon sources to break the link between food and fuel prices (17–19, 36–39) and new approaches to engineering metabolism [exemplified by the development of dynamic sensor-regulator systems for FA-derived products (40), the remarkable reversal of the β-oxidation cycle (41), and introduction of molecular scaffolds for improving metabolic efficiency (42, 43)] will facilitate this aim. The results presented in this article, although at a very early stage in product development, contribute toward the goals of advanced biofuels by providing metabolic pathways for the production of industrially relevant, petroleum-replica fuel molecules.
Experimental Procedures
Construction of Expression Plasmids.
Genes from the P. luminescens lux operon were reverse-translated and codon-optimized for expression in E. coli using DNA2.0 GeneGPS Technology, and synthesized as a single operon by DNA2.0 into the pACYCDuet-1 expression vector (Novagen) multiple cloning site (MCS) 1. The decarbonylase gene was cloned from N. punctiforme genomic DNA as described in Fig. S1. The thioesterase amino acid sequence Q39473.1 (minus the chloroplast transit peptide) and sequences for BCKD components (NP_390285.1, NP_390284.1, NP_390283.1, and ZP_03600867.1) and for KASIII FabH2 (NP_388898.1) were reverse-translated and codon-optimized for expression in E. coli using DNA2.0 GeneGPS Technology, synthesized by DNA2.0 and cloned into pETDuet-1 MCS1 using NcoI and NotI sites. All cloning was carried out using TOP10 chemically competent cells (Invitrogen).
Metabolic Engineering.
Unless otherwise specified, E. coli BL21* (DE3) cells, transformed with the relevant expression vector, were grown in modified minimal medium (MMM) (7) with 3% (wt/vol) glucose, 0.5 g⋅L−1 yeast extract, and the relevant antibiotic. The inclusion of a small amount of yeast extract ensured the rapid and reliable growth required for laboratory procedures that we were unable to achieve using MMM. Cells were incubated at 37 °C and 180 rpm, and protein expression was induced with 20 μM isopropylthiogalactoside (IPTG) at O.D.600 of 0.6–0.8. Cells were grown for a further for 24 h.
Hydrocarbon Analysis.
Hydrocarbons (alkanes and fatty alcohols) were extracted from 8 mL of the cell suspension using an equal volume of ethyl acetate for 2.5 h as described previously (7), dried under nitrogen, and dissolved in dichloromethane. Separation and identification of hydrocarbons was performed using a ThermoQuest Finnigan Trace GC/MS 2000 equipped with a ZB1-MS column [30 m × 0.32 mm internal diameter (ID), Phenomenex]. After splitless injection, the temperature was maintained at 35 °C for 2 min and increased to 320 °C at a rate of 10 °C⋅min−1 with a subsequent incubation at 320 °C for 5 min. The injector temperature was 250 °C and the flow rate of the helium carrier gas was 1.0 mL⋅min−1. The scan range of the mass spectrometer was 30–700 m/z at a scan rate of 1.6 scans⋅s−1. Peak identification was through comparison with known standards, retention time and mass spectral comparisons with the National Institute of Standards and Technology database. Quantification of alkanes was based on heptadecane standards, quantification of alkenes was based on heptadecene standards, and quantification of fatty alcohols was based on tetradecanol standards. FAs were extracted in dichloromethane (DCM): methanol (2:1) according to the Folch method (44). Samples dissolved in DCM were derivatized using bis(trimethylsilyl)trifluoroacetamide (Supelco Analytical) 70 °C 1 h and analyzed using the same GC-MS procedure as for alkane samples. Aldehydes were extracted from 50-mL cultures by centrifugation. Pellets were sonicated for 2 h at 40 °C in 1 mL methanol containing 5 mM semicarbazide hydrochloride and 5% formic acid. Separation and identification of semicarbazide aldehyde derivatives was performed as described (45), using an Agilent 6410 Series triple quadrupole mass spectrometer interfaced with an Agilent 1200 Series liquid chromatograph. Two multiple reaction monitoring transitions were monitored for each of 11 different aldehydes (chain length from C8 to C18).
Construction of a ΔfadD BL21* (DE3) Strain.
Targeted replacement of the entire fadD gene from E. coli BL21* (DE3) with a kanamycin-selectable marker was performed using Red/ET recombination (Quick and Easy E. coli Gene Deletion Kit, Gene Bridges GmbH). Primer sequences for amplification of the deletion cassettes and confirming integration of the selectable markers were fadDF 5′ TATCATTTGGGGTTGCGATGACGACGAACACGCATTTTAGAGGTGAAGAAAATTAACCCTCACTAAAGGGCG 3′ and fadDR 5′ GCGTCAAAAAAAACGCCGGATTAACCGGCGTCTGACGACTGACTTAACGCTAATACGACTCACTATAGGGCTC 3′. The manufacturer’s protocol was adapted by the addition of an additional PCR amplification step using nested primers to increase the yield of the transforming cassettes: fadDFnest 5′ TATCATTTGGGGTTGCGATGACG 3′ and fadDRnest 5′ GCGTCAAAAAAAACGCCGG 3′. Primers for confirming integration of the selectable markers were Primer 15′ TATTATGTTAACGGCATGTATATC 3′, Primer 3 5′ TTGTTTTCTCTTTAGTGGGCGTCA 3′, and PCR primer 2 5′ CGAGACTAGTGAGACGTGCTAC 3′.
Supplementary Material
Acknowledgments
This work was supported by a grant from Shell Research Ltd. and a Biotechnology and Biological Sciences Research Council (BBSRC) Industry Interchange Partnership grant (to J.L.). D.A.P. is a visiting research fellow and R.L. is a visiting professor at the University of Exeter.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The synthetic nucleotide sequences reported in this paper have been deposited in GenBank database (accession nos. JQ901708, JQ901709, and JQ901710).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215966110/-/DCSupplemental.
References
- 1.International Energy Agency . Key World Statistics. Paris, France: 2011. [Google Scholar]
- 2.International Energy Agency . World Energy Outlook. Paris, France: 2008. [Google Scholar]
- 3. European Directive (2009) Council Directive (EC) 2009/28/EC of 23 April 2009 on the promotion of the use of energy from renewable sources and amending and subsequently repealing Directives 2001/77/EC and 2003/30/EC.
- 4.National Renewable Energy Laboratory . Biodiesel Handling and User Guide. 4th Ed. US Department of Commerce: Oak Ridge, TN; 2009. [Google Scholar]
- 5.Yuksel F, Yuksel B. The use of ethanol-gasoline blend as a fuel in an SI engine. Renew Energy. 2004;29:1181–1191. [Google Scholar]
- 6.Jeuland N, Montagne X, Gautrot X. Potentiality of ethanol as a fuel for dedicated engine. Oil Gas Sci Technol. 2004;59:559–570. [Google Scholar]
- 7.Bernard A, et al. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell. 2012;24(7):3106–3118. doi: 10.1105/tpc.112.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schirmer A, Rude MA, Li XZ, Popova E, del Cardayre SB. Microbial biosynthesis of alkanes. Science. 2010;329(5991):559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- 9.Perera MADN, et al. Biological origins of normal-chain hydrocarbons: A pathway model based on cuticular wax analyses of maize silks. Plant J. 2010;64(4):618–632. doi: 10.1111/j.1365-313X.2010.04355.x. [DOI] [PubMed] [Google Scholar]
- 10.Burger BV, Reiter B, Borzyk O, du Plessis MA. Avian exocrine secretions. I. Chemical characterization of the volatile fraction of the uropygial secretion of the green woodhoopoe, Phoeniculus purpureus. J Chem Ecol. 2004;30(8):1603–1611. doi: 10.1023/b:joec.0000042071.65335.f3. [DOI] [PubMed] [Google Scholar]
- 11.Howard RW, et al. Cuticular hydrocarbons of Reticulitermes virginicus (Banks) (Isoptera, Rhinotermitidae) and their role as potential species-recognition and caste-recognition cues. J Chem Ecol. 1982;8:1227–1239. doi: 10.1007/BF00990755. [DOI] [PubMed] [Google Scholar]
- 12.Bourre JM, Cassagne C, Larrouquère-Regnier S, Darriet D. Occurrence of alkanes in brain myelin. Comparison between normal and quaking mouse. J Neurochem. 1977;29(4):645–648. doi: 10.1111/j.1471-4159.1977.tb07781.x. [DOI] [PubMed] [Google Scholar]
- 13.Das D, Eser BE, Han J, Sciore A, Marsh ENG. Oxygen-independent decarbonylation of aldehydes by cyanobacterial aldehyde decarbonylase: A new reaction of di-iron enzymes. Angew Chem Int Ed. 2011;50:7148–7152. doi: 10.1002/anie.201101552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warui DM, et al. Detection of formate, rather than carbon monoxide, as the stoichiometric coproduct in conversion of fatty aldehydes to alkanes by a cyanobacterial aldehyde decarbonylase. J Am Chem Soc. 2011;133(10):3316–3319. doi: 10.1021/ja111607x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan L, Voelker TA, Hawkins DJ. Modification of the substrate specificity of an acyl-acyl carrier protein thioesterase by protein engineering. Proc Natl Acad Sci USA. 1995;92(23):10639–10643. doi: 10.1073/pnas.92.23.10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voelker TA, et al. Fatty acid biosynthesis redirected to medium chains in transgenic oilseed plants. Science. 1992;257(5066):72–74. doi: 10.1126/science.1621095. [DOI] [PubMed] [Google Scholar]
- 17.Youngquist JT, et al. Kinetic modeling of free fatty acid production in Escherichia coli based on continuous cultivation of a plasmid free strain. Biotechnol Bioeng. 2012;109(6):1518–1527. doi: 10.1002/bit.24420. [DOI] [PubMed] [Google Scholar]
- 18.Liu T, Vora H, Khosla C. Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metab Eng. 2010;12(4):378–386. doi: 10.1016/j.ymben.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Steen EJ, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463(7280):559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 20.Westerlund-Karlsson A, Saviranta P, Karp M. Generation of thermostable monomeric luciferases from Photorhabdus luminescens. Biochem Biophys Res Commun. 2002;296(5):1072–1076. doi: 10.1016/s0006-291x(02)02052-1. [DOI] [PubMed] [Google Scholar]
- 21.Winson MK, et al. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol Lett. 1998;163(2):193–202. doi: 10.1111/j.1574-6968.1998.tb13045.x. [DOI] [PubMed] [Google Scholar]
- 22.Rock CO. Fatty acid and phospholipid metabolism in prokaryotes. In: Vance D, Vance J, editors. Biochemistry of lipids, lipoproteins and membranes. New York: Elsevier; 2008. pp. 59–96. [Google Scholar]
- 23.Kaczmarzyk D, Fulda M. Fatty acid activation in cyanobacteria mediated by acyl-acyl carrier protein synthetase enables fatty acid recycling. Plant Physiol. 2010;152(3):1598–1610. doi: 10.1104/pp.109.148007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meighen EA. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991;55(1):123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wall L, Rodriguez A, Meighen E. Intersubunit transfer of fatty acyl groups during fatty acid reduction. J Biol Chem. 1986;261(34):15981–15988. [PubMed] [Google Scholar]
- 26.Li J, Szittner R, Meighen EA. Hyperactivity and interactions of a chimeric myristoryl-ACP thioesterase from the lux system of luminescent bacteria. Biochim Biophys Acta. 2000;1481(2):237–246. doi: 10.1016/s0167-4838(00)00131-x. [DOI] [PubMed] [Google Scholar]
- 27.Smirnova N, Reynolds KA. Branched-chain fatty acid biosynthesis in Escherichia coli. J Ind Microbiol Biotechnol. 2001;27(4):246–251. doi: 10.1038/sj.jim.7000185. [DOI] [PubMed] [Google Scholar]
- 28.Kaneda T. Iso- and anteiso-fatty acids in bacteria: Biosynthesis, function, and taxonomic significance. Microbiol Rev. 1991;55(2):288–302. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi KH, Heath RJ, Rock CO. β-ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J Bacteriol. 2000;182(2):365–370. doi: 10.1128/jb.182.2.365-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD. Microbial engineering for the production of advanced biofuels. Nature. 2012;488(7411):320–328. doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- 31.Akhtar MK, Turner NJ, Jones PR. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc Natl Acad Sci USA. 2013;110(1):87–92. doi: 10.1073/pnas.1216516110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koma D, Yamanaka H, Moriyoshi K, Ohmoto T, Sakai K. Production of aromatic compounds by metabolically engineered Escherichia coli. Appl Environ Microbiol. 2012;78:14. doi: 10.1128/AEM.01148-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng Y-N, et al. Optimization of fatty alcohol biosynthesis pathway for selectively enhanced production of C12/14 and C16/18 fatty alcohols in engineered Escherichia coli. Microb Cell Fact. 2012;11:11. doi: 10.1186/1475-2859-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westfall PJ, et al. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci USA. 2012;109(3):E111–E118. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ro DK, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 36.Wargacki AJ, et al. An engineered microbial platform for direct biofuel production from brown macroalgae. Science. 2012;335(6066):308–313. doi: 10.1126/science.1214547. [DOI] [PubMed] [Google Scholar]
- 37.Bokinsky G, et al. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc Natl Acad Sci USA. 2011;108(50):19949–19954. doi: 10.1073/pnas.1106958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Handke P, Lynch SA, Gill RT. Application and engineering of fatty acid biosynthesis in Escherichia coli for advanced fuels and chemicals. Metab Eng. 2011;13(1):28–37. doi: 10.1016/j.ymben.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 39.Dellomonaco C, Rivera C, Campbell P, Gonzalez R. Engineered respiro-fermentative metabolism for the production of biofuels and biochemicals from fatty acid-rich feedstocks. Appl Environ Microbiol. 2010;76(15):5067–5078. doi: 10.1128/AEM.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang FF, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30(4):354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 41.Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature. 2011;476(7360):355–359. doi: 10.1038/nature10333. [DOI] [PubMed] [Google Scholar]
- 42.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333(6041):470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 43.Dueber JE, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27(8):753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 44.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 45.Berdyshev EV. Mass spectrometry of fatty aldehydes. Biochim Biophys Acta. 2011;1811(11):680–693. doi: 10.1016/j.bbalip.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.