Abstract
X-linked congenital generalized hypertrichosis (Online Mendelian Inheritance in Man 307150) is an extremely rare condition of hair overgrowth on different body sites. We previously reported linkage in a large Mexican family with X-linked congenital generalized hypertrichosis cosegregating with deafness and with dental and palate anomalies to Xq24-27. Using SNP oligonucleotide microarray analysis and whole-genome sequencing, we identified a 389-kb interchromosomal insertion at an extragenic palindrome site at Xq27.1 that completely cosegregates with the disease. Among the genes surrounding the insertion, we found that Fibroblast Growth Factor 13 (FGF13) mRNA levels were significantly reduced in affected individuals, and immunofluorescence staining revealed a striking decrease in FGF13 localization throughout the outer root sheath of affected hair follicles. Taken together, our findings suggest a role for FGF13 in hair follicle growth and in the hair cycle.
Keywords: congenital hypertrichosis, excessive hair growth
Inherited hypertrichoses are rare human disorders characterized by excessive hair growth that does not depend on androgen stimulation and is independent of age, sex, and ethnicity (1). Hypertrichosis syndromes fall under the larger umbrella of ectodermal dysplasias, or abnormal development of the hair, skin, nails, teeth, and/or eccrine glands, and are often associated with additional anomalies including gingival hyperplasia, deafness, cardiomegaly, and bone abnormalities (2). It has been suggested that inherited hypertrichoses represent examples of atavisms, or the recurrence of an ancestral phenotype, where the genes that promote a full coat of hair in other mammals and were silenced throughout evolution have become “reactivated” in human hypertrichosis, invoking unusual genetic mechanisms to explain their occurrence (3, 4).
We and others have identified genetic defects in two forms of autosomal dominant congenital generalized hypertrichosis (CGH) associated with copy number variants on chromosome 17q24 and with rearrangements on chromosomes 3, 7, and 8 (5–7). We previously reported a position effect on the zinc-finger transcription factor Trichorhinophalangeal syndrome 1 (TRPS1) associated with Ambras syndrome congenital hypertrichosis, which was recapitulated in the koala (Koa) mouse hypertrichosis model (8). Likewise, we recently demonstrated a position effect on SRY-related HMG box gene 9 (SOX9) associated with CGH terminalis (9). Importantly, our findings provided evidence for position effects—instances in which a change in gene expression results from altering the location of a gene relative to its native chromosomal position (10)—as mechanisms contributing to inherited hypertrichoses (8).
In this work, we sought to identify the genetic mechanism associated with X-linked CGH in a family in which we previously reported linkage to chromosome Xq24-27 (11). We identified a large interchromosomal insertion that leads to decreased expression of a distant gene, Fibroblast Growth Factor 13 (FGF13), which is expressed in the hair follicle, providing evidence to support a position effect as the underlying genetic basis of X-linked hypertrichosis.
Results and Discussion
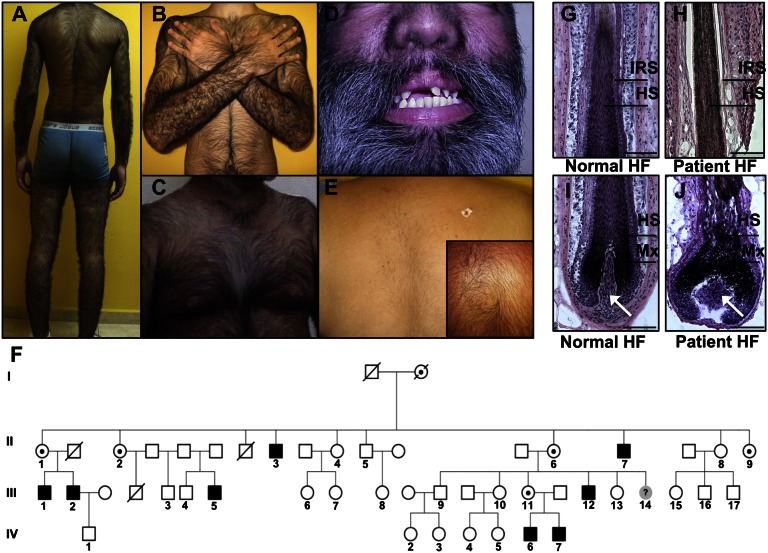
We ascertained a large kindred from Mexico with X-linked CGH (Online Mendelian Inheritance in Man 307150) cosegregating with deafness and with dental and palate anomalies (Fig. 1 A–E) (11). Affected males have approximately three times the number of normal hairs on the scalp and exhibit excessive growth of highly pigmented terminal hairs (medullated) on the scalp, back, shoulders, chest, arms, legs as well as on the face (Fig. 1 A–D), whereas hemizygous carrier females have mild hypertrichosis uniformly distributed across the body (Fig. 1E).
Fig. 1.
Clinical features, pedigree, and histology of hair follicles in a Mexican family with X-linked CGH, deafness, and palate and dental anomalies. (A–D) Clinical photos of affected males with excessive hair growth on the back, shoulders, arms (A and B), chest (C), and (D) face. (D) Dental anomalies and a bulbous nose are also evident on patients. (E) Moderate hair growth is observed on the back of a female carrier. (Inset) A close-up image of a cowlick on the back of another carrier. (F) Pedigree of a four-generation family, three of whom are obligate carriers and eight of whom are affected. (G–J) Histology of a normal hair follicle and affected hair follicle, both from males, revealed that the hairs are of the terminal type, as they are medullated and highly pigmented. Affected hair follicles have a widened dermal papilla (white arrows), matrix (Mx), and hair shaft (HS) compared to control hair follicles. (Scale bars, 100 μm.)
Histological analysis of affected hair follicles using hematoxylin and eosin staining confirmed that the hairs are of the terminal type because they are medullated, pigmented, and penetrate deep within the dermis (Fig. 1 H and J). Affected individuals have an increased density in the number of hair follicles and a transformation from vellus (fine, nonmedullated, unpigmented) to terminal hair follicles on multiple body sites, which cause an excessive hair overgrowth phenotype. Morphometric analysis of affected hair follicles revealed a widened dermal papilla (threefold increase; P = 0.0000343), matrix (1.9-fold increase; P = 0.0000642), and hair shaft (1.25-fold increase; P = 0.036) in hair follicles from three affected individuals compared with controls (Fig. 1 G–J and Fig. S1).
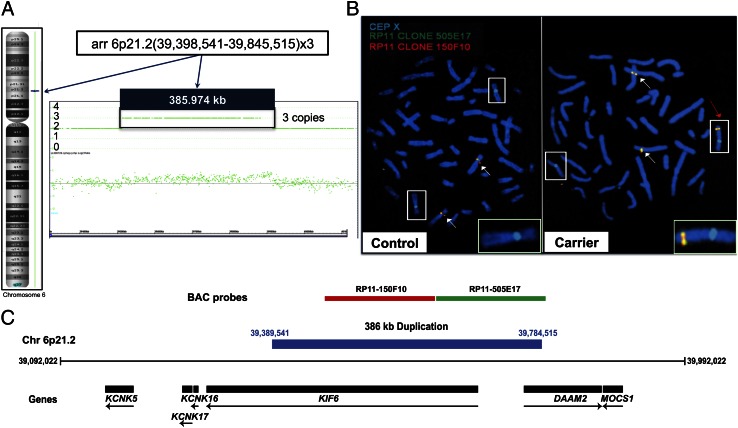
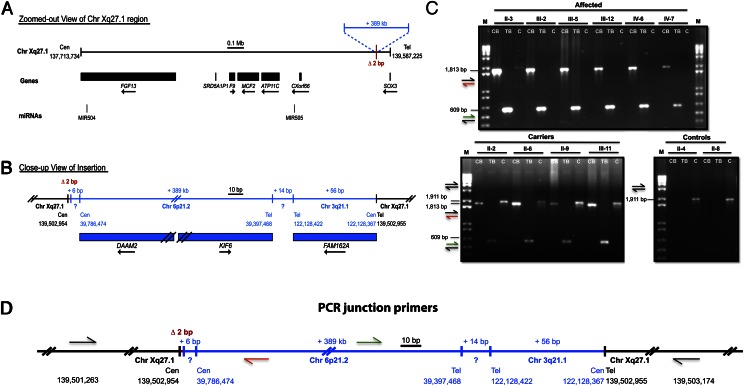
In our previous work on this family, we performed linkage analysis, which defined a 19-Mb region on Xq24-27 that spans ∼82 genes and completely cosegregates with the disease (11). Sequencing the coding exons of every gene in the interval proved unsuccessful in identifying the mutation. To identify the genetic defect, we next performed SNP oligonucleotide microarray analysis (SOMA) using the Affymetrix Cytogenetics Whole-Genome 2.7M array, which revealed a 386-kb duplication of chromosome 6p21.2 (Fig. 2A). We then visualized the duplication at the cytogenetic level using fluorescence in situ hybridization (FISH) with two nonoverlapping BAC probes spanning the chromosome 6p21.2 duplication, which revealed a third signal for chromosome 6 present on the X chromosome (Fig. 2 B and C). To identify the insertion breakpoints as well as their content, we performed whole-genome sequencing (WGS), which revealed a large interchromosomal insertion at an extragenic palindromic sequence on chromosome Xq27.1, consisting of a 386-kb duplication of chromosome 6p21.2 and 56 bp of chromosome 3q21.1 in the reverse orientation, separated by 14 bp of unknown origin (Fig. 3 A and B). This complex insertion leads to a 2-bp deletion within the Xq27.1 palindromic site and contains sequences from two genes on chromosome 6p21.2 [Dishevelled associated activator of morphogenesis 2 (DAAM2) and Kinesin family member 6 (KIF6)] and one gene from chromosome 3q21.2 [Family with sequence similarity 162, member A (FAM162A)], none of which have reported functions in the skin or hair follicle. To confirm that the insertion cosegregated with the disease phenotype, we used PCR amplification of the centromeric and telomeric breakpoint junctions on genomic DNA from control, carrier, and affected individuals, which revealed junction bands present only in affected and carrier individuals, whereas DNA from controls produced an amplicon representative of an unaffected X chromosome (Fig. 3 C and D).
Fig. 2.
SOMA identified a 386-kb duplication of chromosome 6, and FISH revealed its insertion on the X chromosome. (A) SOMA performed on an affected individual using the Affymetrix Cytogenetics Whole-Genome 2.7M array revealed a 386-kb duplication of chromosome 6p21.2 encompassing the KIF6 and DAAM2 genes (as shown in C). (B) FISH on control and carrier metaphase chromosomes revealed the insertion of the chromosome 6 duplication onto the X chromosome at the cytogenetic level. Boxes indicate X chromosomes, white arrows indicate chromosome 6, and the red arrow indicates the X chromosome containing the insertion. Insets are magnified images of the unaffected and affected X chromosome from control and carrier individuals, respectively. (C) Nonoverlapping BAC clones used to span the chromosome 6 duplication include the KIF6 and DAAM2 genes (drawn to scale). Genomic coordinates reference the UCSC Genome Browser human reference genome build hg19.
Fig. 3.
Whole-genome sequencing revealed a 389-kb interchromosomal insertion at Xq27.1 that cosegregates with the X-linked hypertrichosis phenotype. (A) Chromosome Xq27.1 in X-linked hypertrichosis. The genes and microRNAs encoded in the surrounding region are shown as black boxes with arrows indicating the direction of transcription. (B) WGS was used to determine the breakpoints and content of the interchromosomal insertion (shown in blue), including the 386-kb duplication from chromosome 6p21.2, 14 bp of unknown origin, and 56 bp of chromosome 3q21.2. (C) PCR amplification of the centromeric and telomeric junctions of the insertion on DNA from control, carrier, and affected individuals demonstrated segregation of the X-linked phenotype in the family at the genomic level. CB, TB, and C represent centromeric breakpoint, telomeric breakpoint, and controls, respectively. M, marker (1 kb + ladder). (D) Primer design of the centromeric and telomeric junctions; colored arrows correspond to the amplicons produced as shown in C. All images are drawn to scale. Genomic coordinates reference the UCSC Genome Browser human reference genome build hg19.
To gain insight into whether the insertion at Xq27.1 affects X-chromosome inactivation in female carriers, because skewed X inactivation is a phenomenon reported in several X-linked disorders (12, 13), we performed the X-chromosome inactivation (XCI) assay and did not observe significant skewing (≥80%) in four of five carriers (Table S1 and SI Materials and Methods).
Importantly, two other recently reported families with X-linked CGH contain linkage to chromosome Xq24-27, consistent with our findings in which one family contains a 300-kb interchromosomal insertion from chromosome 4q31.2 and the other family contains a 125-kb interchromosomal insertion from chromosome 5q35.3 (14) (Fig. S2). Interestingly, the insertion events in all three families occur at the same human-specific extragenic palindrome sequence at Xq27.1, suggesting that the presence of the insertion (rather than its content) may be responsible for the excessive hair overgrowth phenotype by disruption of the chromosomal architecture in the region.
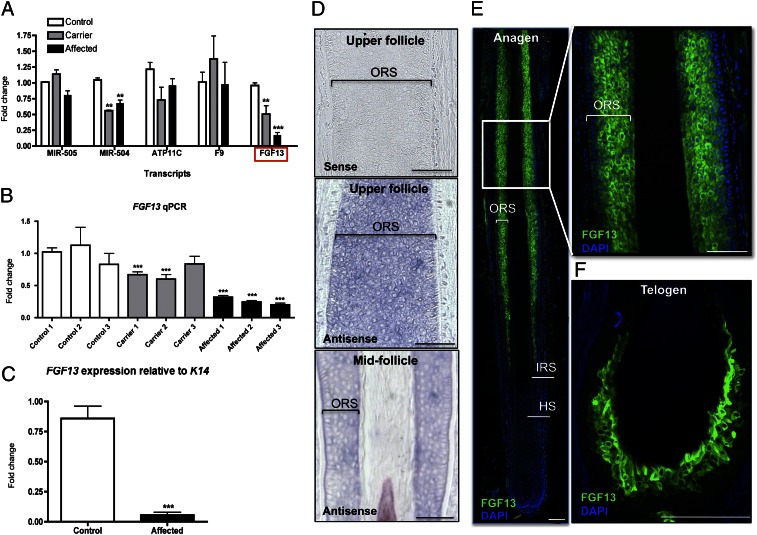
Despite the identification of different insertions in these three cases, their impact on the expression of the surrounding genes has not been thoroughly investigated. Therefore, we analyzed the expression of several neighboring genes using quantitative RT-PCR (qRT-PCR) on RNA isolated from control, carrier, and affected skin biopsies. Unexpectedly, we found that FGF13 levels were significantly reduced in affected individuals by approximately fourfold (P = 0.0007) relative to controls and observed a clear dosage effect when comparing levels between carrier and affected individuals (Fig. 4 A and B). To verify that the change in FGF13 expression observed in affected skin biopsies was not due to differences in the number of hair follicle cells present, we normalized FGF13 expression to that of Keratin 14 (KRT14), which marks the outer root sheath and basal layer of the epidermis, and found that FGF13 levels were dramatically reduced (18.1-fold; P = 0.00006) in affected hair follicle cells (Fig. 4C).
Fig. 4.
FGF13 levels are reduced in X-linked hypertrichosis, and FGF13 is expressed in the human hair follicle. (A) qRT-PCR of candidate genes surrounding the insertion on control, carrier, and affected skin biopsies reveals that FGF13 levels are reduced by approximately fourfold in affected individuals relative to controls. (B) FGF13 qRT-PCR in control, carrier, and affected individuals reveals a dosage effect. (C) FGF13 expression normalized to K14 reveals that FGF13 levels are dramatically reduced by eighteen fold in affected cells relative to that of controls. qRT-PCR results are representative of the averaged values of three independent experiments on three controls, three carriers, and three affected individuals. Values are relative to the unaffected samples and standardized to the housekeeping gene GAPDH (in A and B). A Student t test was performed comparing each value to control 1 with a cutoff P value of 0.05 for statistical significance; **P < 0.01; ***P < 0.001. Error bars represent the SEM. (D) In situ hybridization of FGF13 in anagen hair follicles reveals expression in the outer root sheath (ORS) within the middle and upper portions of the hair follicle, where the sense probe produced no signal. (E) Immunofluorescence staining reveals that FGF13 localizes to the outer root sheath (magnified image) within the middle and upper portions of the human anagen hair follicle (n = 5). ORS, outer root sheath; IRS, inner root sheath; HS, hair shaft. (F) FGF13 expression is detected in the trichilemma (ORS) of telogen club-hair follicles by immunofluorescence staining. (Scale bar, 100 μm.)
Importantly, the mRNA levels of additional neighboring genes were not significantly changed, and SOX3 and Chromosome X open reading frame 66 (Cxorf66) expression levels were undetectable in control and affected individuals (Fig. 4A). To further examine the expression levels of the genes surrounding the 389-kb insertion, we performed RNA sequencing (RNA-seq) on RNA from the skin of one control and one affected individual, which verified the decrease in FGF13 expression (eightfold; P = 0.012) and revealed that the expression levels of most of the genes in the surrounding region over a distance of ∼3 Mb on either side of the insertion were not significantly changed (Table S2 and SI Materials and Methods). The expression of the microRNA, miR-504, intronic to FGF13, was reduced by approximately 1.5-fold in the carrier and affected individuals by qRT-PCR (P = 0.00831) (Fig. 4A). FGF13 is not a predicted target gene of miR-504, yet there are several predicted targets with known roles in hair follicle development and cycling whose expression levels were altered in X-linked hypertrichosis, detected by RNA-seq (Table S3 and SI Materials and Methods). Although these changes in gene expression may simply reflect a difference in the number of hair follicle cells present, it remains possible that the reduction of FGF13–miR-504 transcripts either directly or indirectly leads to increased expression of some of these downstream genes.
FGF13 is a plausible candidate gene to be the target of the position effect because its expression was previously detected in the hair follicle bulge, which is the stem cell compartment of the hair follicle (15, 16). However, it was unclear whether these were the only FGF13-expressing cells in the human hair follicle or whether expression was more widespread. Using in situ hybridization and immunofluorescence staining on hair follicles in the growth stage of the hair cycle, anagen, we detected expression of FGF13 in the outer root sheath within the middle and upper portions of the human hair follicle (Fig. 4 D and E). We also observed FGF13 expression in the trichilemma, or outer root sheath compartment of the club hair, during the resting stage of the hair cycle, telogen, by immunofluorescence staining (Fig. 4F). In anagen hairs, FGF13 localizes to the outer root sheath (ORS) where KRT14 is expressed and also localizes to the companion layer that separates the outer from the inner root sheath, as evidenced by overlapping expression with KRT75 (companion layer marker) (Fig. S3 A and B). However, we did not observe FGF13 localization to the bulge region of the anagen human hair follicle, through costaining with CD200, a well-characterized bulge marker (Fig. S3C).
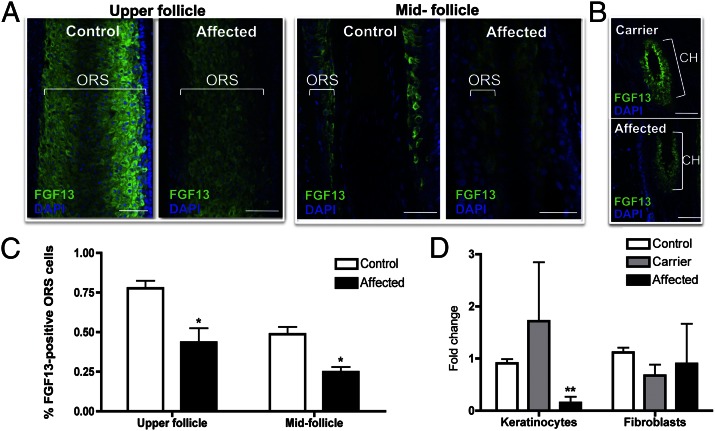
To gain insight into which cells exhibited decreased FGF13 levels, we performed FGF13 immunofluorescence staining on control, carrier, and affected hair follicles and observed a decrease in the intensity of expression and the number of FGF13-positive cells throughout the outer root sheath of affected anagen follicles compared with controls and carriers (Fig. 5 A and C). Moreover, a comparison of affected and carrier telogen follicles revealed a decrease in expression throughout the affected hair follicle, recapitulating the dosage effect observed at the mRNA level (Fig. 5B).
Fig. 5.
Immunofluorescence staining reveals that FGF13 expression is dramatically reduced in affected hair follicles compared with control. (A) Immunofluorescence staining in control and affected anagen hair follicles reveals a decrease in FGF13 localization throughout the outer root sheath (ORS) in the mid and upper portions of the hair follicle. (B) Immunofluorescence staining in carrier and affected telogen hair follicles reveals decreased FGF13 expression in the affected hair follicle, recapitulating the dosage effect seen at the mRNA level. Z-stack images were taken using identical settings and a consistent Z-stack interval between control, carrier, and affected samples. CH, club hair of a telogen follicle. (C) Quantification of the percentage of FGF13-expressing ORS cells in control and affected hair follicles reveals a decrease in the number of FGF13-expressing cells within the upper and midfollicle regions of the ORS (P < 0.05). Data represent the averaged value of three independent experiments, where images taken at a 40× magnification were used to quantify the number of FGF13-positive cells relative to the total number of ORS cells. For immunofluorescence studies, hair follicles were stained from three control and two affected skin biopsies. (D) qRT-PCR revealed that FGF13 expression is reduced in keratinocytes but not in fibroblasts grown from skin biopsies. A Student t test was performed with a cutoff P value of 0.05 for statistical significance; *P < 0.05, **P < 0.01. Error bars represent the SEM. (Scale bar, 100 μm.)
To further investigate the selective decrease of FGF13 expression in affected keratinocytes, we performed qRT-PCR on keratinocytes and fibroblasts cultured from control, carrier, and affected skin biopsies and observed a significant decrease in FGF13 expression in affected keratinocytes of 6.7-fold (P = 0.00193) (Fig. 5D), but not in the fibroblasts. Consistent with our previous observations, our findings localize the defect to the keratinocyte compartment.
To determine Fgf13 expression during murine hair follicle morphogenesis, we performed whole-mount and section in situ hybridization on embryonic day 12.5 (E12.5)–E16.5 embryos and observed strong expression in placodes (the sites of newly forming follicles), as well as in the dermal condensate beneath the placodes that becomes the dermal papilla of the hair follicle at E14.5 within the developing whisker pad and guard hair pelage follicles (Fig. S4A). Immunofluorescence staining of vibrissae follicles during morphogenesis at E16.5 revealed that Fgf13 localizes to the outer root sheath, similar to the postnatal localization pattern of the human FGF13 protein in anagen follicles (Fig. S4B). Moreover, immunofluorescence staining on postnatal skin revealed that Fgf13 localizes to the bulge, isthmus region, and outer root sheath of the hair follicle (Fig. S4 C and D). Taken together, our results suggest a potential role for Fgf13 in regulating hair follicle growth and cycling.
In mice and in humans, five-prime alternative splicing of FGF13 and use of different transcription start sites generates transcripts with distinct 5′ exons referred to as 1S, 1U, 1V, 1Y, and 1V+1Y, where exons 2–5, encoding the conserved core region of the protein, are common to all transcripts. Isoform-specific PCR revealed that isoforms 1S, 1V, 1Y, and 1V+1Y are expressed in human scalp skin (Fig. S5). To gain insight into the mechanism by which the interchromosomal insertion alters FGF13 transcript levels in X-linked hypertrichosis, we used the RNA-seq data to test for differentially expressed isoforms using Cuffdiff (SI Materials and Methods), but did not observe differential expression between the FGF13 isoforms, suggesting that the interchromosomal insertion disrupts the transcription of all isoforms rather than altering the use of a particular transcription start site.
Position effects on single genes have been reported in several other human genetic diseases associated with large chromosomal rearrangements where the distances of the farthest breakpoints from the target genes were as large as 1.0 Mb [sonic hedgehog (SHH) in preaxial polydactylyl II] (17, 18), 1.3 Mb (SOX9 in camptomelic dysplasia) (19, 20), and 7.3 Mb (TRPS1 in Ambras syndrome) (8). Because FGF13 lies 1.2 Mb away from the insertion and its expression was selectively reduced, we postulate that the interchromosomal insertion at Xq27.1 separates the gene from a tissue- or temporal-specific modifier element (such as an enhancer) required for proper FGF13 expression during hair follicle morphogenesis and cycling. Consistent with this notion, we found that FGF13 expression was selectively reduced in a tissue-specific manner, as transcript levels were decreased in affected keratinocytes, but not in fibroblasts.
Our data indicate that FGF13 is primarily expressed throughout the outer root sheath of human hair follicles, and findings from the clinical, histological, and morphometric analyses revealed increased width of hair follicles in X-linked hypertrichosis (Fig. 1 H and J and Fig. S1). Several genes expressed in the outer root sheath of hair follicles have been reported to have noncell autonomous effects on other compartments, indirectly or directly leading to changes in hair follicle width and/or length. In the case of Dishevelled 2 (Dvl2), an effector of wingless-type MMTV integration site family, member 3 (Wnt3) signaling normally expressed in the outer root sheath and the precortical and precuticle cells of the hair shaft, its overexpression in the outer root sheath induces a short-hair phenotype by altering the differentiation of hair shaft precursor cells (21). Similarly, overexpression of Vegf in the outer root sheath, where it is normally expressed, induces perifollicular vascularization of the hair follicle, resulting in accelerated hair regrowth and in increased size of hair shafts (22).
In our mouse expression studies, we found that Fgf13 is expressed during hair follicle induction and morphogenesis, suggesting that it may play an important role in these processes. Affected individuals in the X-linked hypertrichosis family possess an increased density of hairs; thus, it is possible that dysregulation of FGF13 levels in X-linked hypertrichosis leads to the formation of extra hairs, but further studies using Fgf13-deficient mice would be needed to directly implicate a role for Fgf13 in hair follicle morphogenesis and cycling. Interestingly, Fgf13 expression has been demonstrated in the dental mesenchyme and developing tooth bud (23), an additional site of pathology for X-linked hypertrichosis patients who have dental and palate anomalies, suggesting a potential role for this gene in odontogenesis.
Several FGFs and their receptors are known to play important roles in hair regrowth. Although canonical FGFs are known to signal through their respective receptors to control hair growth via stem cell activation and quiescence, FGF13 is the first noncanonical FGF to be implicated in hair follicle morphogenesis and cycling. As FGF13 has been reported to bind the MAPK scaffolding protein islet brain 2 (IB2) (24), leading to activation of a stress-induced MAPK that lies downstream of the canonical FGF-signaling pathway (24), one possibility is that FGF13 internally modulates the transcriptional output of canonical FGF signaling to control hair growth. Consistent with this notion, we found the expression levels of several FGFs to be dysregulated in X-linked hypertrichosis (Table S4). Among these was FGF5, a known regulator of the anagen-to-catagen transition and responsible for the excessive hair overgrowth phenotype in angora mice, dogs, goats, and rabbits (25–28). A second possibility is that FGF13 acts as a microtubule-stabilizing protein in the hair follicle, similar to its role in neurons (29), to regulate additional signaling molecules active in the developing follicle. Further functional studies on FGF13 will reveal the mechanism by which it regulates hair follicle growth and distribution.
In this study, we identified a 389-kb interchromosomal insertion at Xq27.1 that completely cosegregates with the X-linked CGH phenotype and found that, among the genes surrounding the insertion, FGF13 expression was selectively and profoundly reduced. FGF13 lies 1.2 Mb away from the insertion, revealing a position effect on a distant gene as a result of the chromosomal insertion at Xq27.1. Although it has been suggested that these large interchromosomal insertions may mediate pathogenic effects by introducing novel regulatory elements, it is more likely that the presence of the insertions (rather than their content) is responsible for the hair overgrowth phenotype because the sequences contained within each insertion are different among the families (14). Moreover, these insertions occur at an extragenic palindrome sequence and do not disrupt the coding region of a gene in the surrounding region (Fig. S2).
The density of hair follicles covering the human body is markedly reduced compared with other primates, and as such, the excessive hair phenotype observed in hereditary hypertrichosis has been suggested to be reminiscent of an atavism (3). Various examples of atavisms involving several body parts have been reported in mammals, including the occurrence of complete ulnas and fibulas in miniature horses, reptile-like coronary circulation and myocardial architecture in humans, and the development of the ancestral tooth primordia in retinoic acid receptor-deficient mice (30–32). Importantly, the occurrence and prevalence of these ancestral features is highly suggestive of a genetic basis, one involving unusual mechanisms. Here, we report a position effect on FGF13 in X-linked hypertrichosis that alters the spatiotemporal expression of the gene in the hair follicle. In light of our findings, we suggest that the altered FGF13 expression in affected hair follicles influences important downstream signaling pathways, ultimately leading to the terminal hair overgrowth phenotype of X-linked hypertrichosis.
Materials and Methods
Ethics Statement.
Informed consent was obtained from all subjects and approval for this study was provided by the Institutional Review Board of Columbia University in accordance with the Declaration of Helsinki Principles.
Patient Materials, Histological Analysis, and RNA Extraction.
DNA was previously collected from 26 members of a family, 3 of whom are obligate carriers and 8 of whom are affected (11). Whole-skin biopsies taken from the back were then obtained from three female carriers and three affected male individuals. Biopsies were divided into three separate pieces for RNA extraction, cell culture [see SI Materials and Methods for details), and optimal cutting temperature compound (OCT) embedding for histological and morphometric analysis (see SI Materials and Methods for details]. We obtained control hair follicles from occipital scalp biopsies from discarded tissue following hair transplant surgeries. Samples were designated as non-human subject research under 45 Code of Federal Regulations (CFR) Part 46, and we therefore received an institutional review board exemption to use these materials. RNA extraction was performed using the Qiagen RNeasy Mini Kit following the manufacturer’s instructions. Total RNA was used for first-strand cDNA synthesis, as previously described (8).
SOMA and WGS.
DNA from an affected individual was prepared and hybridized as per the manufacturer’s instructions on the Affymetrix Cytogenetics Whole-Genome 2.7M array, and data were analyzed with the Affymetrix Chromosome Analysis Suite. WGS was performed on one affected male of this Mexican family using the methods described in SI Materials and Methods. All coordinates reference the UCSC Genome Browser human reference genome build hg19.
Cytogenetic Analysis, Amplification of Genomic DNA, and qRT-PCR.
FISH analysis was performed on metaphase chromosome spreads prepared from phytohemagglutinin (PHA)-stimulated cultured peripheral blood cells using standard techniques. The RPCI-11 clone 505E17 [labeled with Orange 5-TAMRA (carboxytetramethylrhodamine) dUTP] and RPCI-11 clone 150F10 (labeled with green 5-fluorescein dUTP) from Empire Genomics were used as FISH probes. Hybridization and posthybridization washing were performed as per the manufacturer’s instructions. To test cosegregation of the insertion with the disease phenotype, 100 ng of DNA was used for PCR amplification of the centromeric and telomeric breakpoints as well as of the control region of the unaffected X chromosome (details listed in the SI Materials and Methods). qRT-PCR was performed as previously described (8) using the Delta-Delta-Ct (ddCt) method, and primers are listed in the SI Materials and Methods.
In Situ Hybridization and Immunofluorescence Staining.
Whole-mount in situ hybridization was performed as previously described (8) (see SI Materials and Methods for details). Immunofluorescence staining was performed on human control, carrier, and affected 12-μm hair follicle sections as well as on Swiss Webster dorsal skin sections (10 μm) from postnatal day 30 (anagen) and 50 (telogen) mice using the conditions described in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Antonio Sobrino, Jim Russo, Ming Chen, Barbara Ross, and Conrad Gilliam for their collaboration and stimulating discussions; Dr. Geoffrey Pitt for kindly providing the FGF13 antibody; Mr. Ming Zhang for technical assistance; and members of the A.M.C. laboratory for helpful discussions. We appreciate the support and expert assistance of the Skin Disease Research Center in the Department of Dermatology at Columbia University [National Institutes of Health (NIH)/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) Grant P30AR44535]. G.M.D. was supported by Columbia University Department of Genetics and Development Grant T32GM007088, and K.A.F. was supported by Columbia University Genetic Mechanisms of Skin Disease Grant T32AR007605. This work was supported in part by NIH/NIAMS Grant R01AR44924 (to A.M.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216412110/-/DCSupplemental.
References
- 1.Beighton P. Congenital hypertrichosis lanuginosa. Arch Dermatol. 1970;101(6):669–672. [PubMed] [Google Scholar]
- 2.Garcia-Cruz D, Figuera LE, Cantu JM. Inherited hypertrichoses. Clin Genet. 2002;61(5):321–329. doi: 10.1034/j.1399-0004.2002.610501.x. [DOI] [PubMed] [Google Scholar]
- 3.Cantú JM, Ruiz C. On atavisms and atavistic genes. Ann Genet. 1985;28(3):141–142. [PubMed] [Google Scholar]
- 4.Hall BK. Development mechanisms underlying the formation of atavisms. Biol Rev Camb Philos Soc. 1984;59(1):89–124. doi: 10.1111/j.1469-185x.1984.tb00402.x. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, et al. Ambras syndrome in a Korean patient with balanced pericentric inversion (8)(p11.2q24.2) J Dermatol Sci. 2010;59(3):204–206. doi: 10.1016/j.jdermsci.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Tadin M, et al. Complex cytogenetic rearrangement of chromosome 8q in a case of Ambras syndrome. Am J Med Genet. 2001;102(1):100–104. doi: 10.1002/1096-8628(20010722)102:1<100::aid-ajmg1396>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 7.Sun M, et al. Copy-number mutations on chromosome 17q24.2-q24.3 in congenital generalized hypertrichosis terminalis with or without gingival hyperplasia. Am J Hum Genet. 2009;84(6):807–813. doi: 10.1016/j.ajhg.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fantauzzo KA, et al. A position effect on TRPS1 is associated with Ambras syndrome in humans and the Koala phenotype in mice. Hum Mol Genet. 2008;17(22):3539–3551. doi: 10.1093/hmg/ddn247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fantauzzo KA, Kurban M, Levy B, Christiano AM. Trps1 and its target gene Sox9 regulate epithelial proliferation in the developing hair follicle and are associated with hypertrichosis. PLoS Genet. 2012;8(11):e1003002. doi: 10.1371/journal.pgen.1003002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinjan DJ, van Heyningen V. Position effect in human genetic disease. Hum Mol Genet. 1998;7(10):1611–1618. doi: 10.1093/hmg/7.10.1611. [DOI] [PubMed] [Google Scholar]
- 11.Tadin-Strapps M, et al. Congenital universal hypertrichosis with deafness and dental anomalies inherited as an X-linked trait. Clin Genet. 2003;63(5):418–422. doi: 10.1034/j.1399-0004.2003.00069.x. [DOI] [PubMed] [Google Scholar]
- 12.Plenge RM, Stevenson RA, Lubs HA, Schwartz CE, Willard HF. Skewed X-chromosome inactivation is a common feature of X-linked mental retardation disorders. Am J Hum Genet. 2002;71(1):168–173. doi: 10.1086/341123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knudsen GP, et al. Increased skewing of X chromosome inactivation in Rett syndrome patients and their mothers. Eur J Hum Genet. 2006;14(11):1189–1194. doi: 10.1038/sj.ejhg.5201682. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H, et al. X-linked congenital hypertrichosis syndrome is associated with interchromosomal insertions mediated by a human-specific palindrome near SOX3. Am J Hum Genet. 2011;88(6):819–826. doi: 10.1016/j.ajhg.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohyama M, et al. Characterization and isolation of stem cell-enriched human hair follicle bulge cells. J Clin Invest. 2006;116(1):249–260. doi: 10.1172/JCI26043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawano M, Suzuki S, Suzuki M, Oki J, Imamura T. Bulge- and basal layer-specific expression of fibroblast growth factor-13 (FHF-2) in mouse skin. J Invest Dermatol. 2004;122(5):1084–1090. doi: 10.1111/j.0022-202X.2004.22514.x. [DOI] [PubMed] [Google Scholar]
- 17.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12(14):1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 18.Lettice LA, et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci USA. 2002;99(11):7548–7553. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leipoldt M, et al. Two novel translocation breakpoints upstream of SOX9 define borders of the proximal and distal breakpoint cluster region in campomelic dysplasia. Clin Genet. 2007;71(1):67–75. doi: 10.1111/j.1399-0004.2007.00736.x. [DOI] [PubMed] [Google Scholar]
- 20.Velagaleti GV, et al. Position effects due to chromosome breakpoints that map approximately 900 Kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am J Hum Genet. 2005;76(4):652–662. doi: 10.1086/429252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millar SE, et al. WNT signaling in the control of hair growth and structure. Dev Biol. 1999;207(1):133–149. doi: 10.1006/dbio.1998.9140. [DOI] [PubMed] [Google Scholar]
- 22.Yano K, Brown LF, Detmar M. Control of hair growth and follicle size by VEGF-mediated angiogenesis. J Clin Invest. 2001;107(4):409–417. doi: 10.1172/JCI11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kettunen P, Furmanek T, Chaulagain R, Kvinnsland IH, Luukko K. Developmentally regulated expression of intracellular Fgf11-13, hormone-like Fgf15 and canonical Fgf16, -17 and -20 mRNAs in the developing mouse molar tooth. Acta Odontol Scand. 2011;69(6):360–366. doi: 10.3109/00016357.2011.568968. [DOI] [PubMed] [Google Scholar]
- 24.Schoorlemmer J, Goldfarb M. Fibroblast growth factor homologous factors and the islet brain-2 scaffold protein regulate activation of a stress-activated protein kinase. J Biol Chem. 2002;277(51):49111–49119. doi: 10.1074/jbc.M205520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hébert JM, Rosenquist T, Götz J, Martin GR. FGF5 as a regulator of the hair growth cycle: Evidence from targeted and spontaneous mutations. Cell. 1994;78(6):1017–1025. doi: 10.1016/0092-8674(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 26.Housley DJ, Venta PJ. The long and the short of it: Evidence that FGF5 is a major determinant of canine ‘hair’-itability. Anim Genet. 2006;37(4):309–315. doi: 10.1111/j.1365-2052.2006.01448.x. [DOI] [PubMed] [Google Scholar]
- 27.Liu HY, Yang GQ, Zhang W, Zhu XP, Jia ZH. [Effects of FGF5 gene on fibre traits on Inner Mongolian cashmere goats] Yi Chuan. 2009;31(2):175–179. doi: 10.3724/sp.j.1005.2009.00175. [DOI] [PubMed] [Google Scholar]
- 28.Li CX, Jiang MS, Chen SY, Lai SJ. [Correlation analysis between single nucleotide polymorphism of FGF5 gene and wool yield in rabbits] Yi Chuan. 2008;30(7):893–899. doi: 10.3724/sp.j.1005.2008.00893. [DOI] [PubMed] [Google Scholar]
- 29.Wu QF, et al. Fibroblast growth factor 13 is a microtubule-stabilizing protein regulating neuronal polarization and migration. Cell. 2012;149(7):1549–1564. doi: 10.1016/j.cell.2012.04.046. [DOI] [PubMed] [Google Scholar]
- 30.Tyson R, Graham JP, Colahan PT, Berry CR. Skeletal atavism in a miniature horse. Vet Radiol Ultrasound. 2004;45(4):315–317. doi: 10.1111/j.1740-8261.2004.04060.x. [DOI] [PubMed] [Google Scholar]
- 31.Walia I, Arora HS, Barker EA, Delgado RM, III, Frazier OH. Snake heart: A case of atavism in a human being. Tex Heart Inst J. 2010;37(6):687–690. [PMC free article] [PubMed] [Google Scholar]
- 32.Peterkova R, Lesot H, Peterka M. Phylogenetic memory of developing mammalian dentition. J Exp Zoolog B Mol Dev Evol. 2006;306(3):234–250. doi: 10.1002/jez.b.21093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.