Abstract
We described the integration of the general reversibility of glycosyltransferase-catalyzed reactions, artificial glycosyl donors, and a high throughput colorimetric screen to enable the engineering of glycosyltransferases for combinatorial sugar nucleotide synthesis. The best engineered catalyst from this study, the OleD Loki variant, contained the mutations P67T/I112P/T113M/S132F/A242I compared with the OleD wild-type sequence. Evaluated against the parental sequence OleD TDP16 variant used for screening, the OleD Loki variant displayed maximum improvements in kcat/Km of >400-fold and >15-fold for formation of NDP–glucoses and UDP–sugars, respectively. This OleD Loki variant also demonstrated efficient turnover with five variant NDP acceptors and six variant 2-chloro-4-nitrophenyl glycoside donors to produce 30 distinct NDP–sugars. This study highlights a convenient strategy to rapidly optimize glycosyltransferase catalysts for the synthesis of complex sugar nucleotides and the practical synthesis of a unique set of sugar nucleotides.
Keywords: carbohydrate, enzyme, glycobiology, protein engineering
The lack of accessibility and availability of uncommon and uniquely functionalized sugar nucleotides (NDP–sugars) continues to restrict research focused upon understanding the regulation, biosynthesis, and/or role of glycosylated macromolecules and glycosylated small molecules in biology or therapeutic development (1–7). Although there are many reported chemical, enzymatic, and chemoenzymatic strategies for NDP–sugar synthesis, those that extend beyond the reach of common biological sugars (e.g., D-glucose, D-galactose, etc.) nearly all suffer from long reaction times (>16 h), relatively low yields, and difficulties associated with product purification and/or stability (3, 4, 8, 9). Thus, the development of robust methods for sugar nucleotide synthesis directly compatible to the downstream biological processes to be studied may be advantageous.
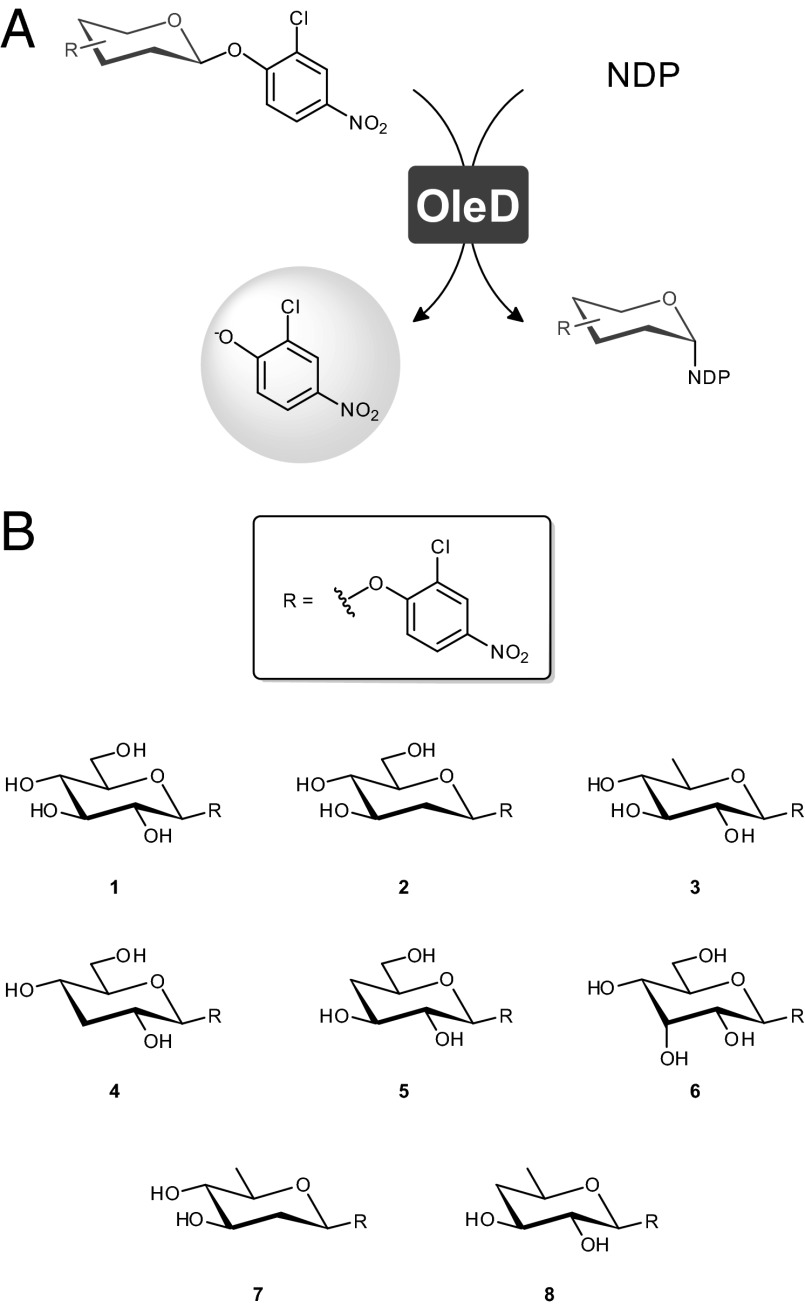
From a traditional viewpoint, NDP–sugars are used as donors by Leloir glycosyltransferases (sugar nucleotide–dependent enzymes) for formation of glycosidic bonds. However, many glycosyltransferase (GT)-catalyzed reactions are known to be readily reversible, enabling the “pirating” of unique sugars from natural products or alternative donors (resulting in generation of the respective sugar nucleotide) and one-pot sugar exchange reactions between unique natural products (4, 10–13). This general reaction feature, in conjunction with availability of highly permissive glycosyltransferases (14–18) and simple donors designed to fundamentally alter the reaction thermodynamics, recently enabled a unique platform for NDP–sugar synthesis and a high throughput colorimetric screen for NDP–sugar formation and utilization (19). While the prior platform proof-of-concept study highlighted the syntheses of 22 natural and nonnatural TDP/UDP–sugars from 11 distinct 2-chloro-4-nitrophenyl glycoside donors using a single GT catalyst (Fig. 1A) (19), the substrate specificity of the glycosyltransferase used restricted the scope of nucleoside and/or sugar functionality accessible.
Fig. 1.
(A) Basic glycosyltransferase-catalyzed reaction scheme for this engineering study. (B) Structures of 2-chloro-4-nitrophenyl glycosides donors used in this study.
Herein, we present an application of the above-mentioned high throughput colorimetric screen for enzyme evolution. Specifically, this model study focused upon the identification of catalysts capable of accommodating unique sugar functionality present among bacterial secondary metabolites [deoxysugars, dideoxysugars, and C-3 epimers of glucose (Glc)] and additional nucleoside diversity (adenosine, guanine, and cytidine). The most advantageous mutant identified (OleD Loki) was capable of recognizing six uniquely functionalized sugars (as 2-chloro-4-nitrophenyl glycoside donors) and five structurally diverse NDP acceptors to enable the combinatorial synthesis of 30 distinct NDP–sugars. The identification of the Loki variant highlights the utility of the simple 2-chloro-4-nitrophenyl glycoside-based colorimetric screen for rapid optimization of tailored catalysts to facilitate the syntheses of desired NDP–sugars. This study also highlights a robust chemoenzymatic platform for the syntheses of 30 unique sugar nucleotides that is amenable to direct coupling to a range of downstream enzymatic processes.
Results and Discussion
Selection of Assay Targets.
From the perspective of sugar modifications, the goal of this study was to focus upon a representative set of core hexose modifications reminiscent of those found appended to unique natural products. Specifically, this study focused upon deoxy- and dideoxysugars, a common sugar scaffold found attached to many natural products (1, 5–7) and the C-3 epimer of glucose, a chemical motif that is integral in the biosynthetic pathways of macrolide antibiotics (e.g., tylosin, chalcomycin) (5). Among the relevant previously tested 2-chloro-4-nitrophenyl glycoside donors in OleD-catalyzed sugar nucleotide synthesis reactions, the corresponding D-glucose and 2- and 6-deoxy-D-glucose (Fig. 1B, donors 1, 2, and 3, respectively) were found to be good substrates, 3- and 4-deoxy-D-glucose (Fig. 1B, donors 4 and 5, respectively) and D-allose (the C-3 epimer of D-glucose; Fig. 1B, donor 6) were reported as poor substrates (i.e., initial rates of formation for the respective UDP–sugars were >2 orders of magnitude slower than for the parental sugar, D-glucose) (19), and the 2,6-dideoxy and 4,6-dideoxy variants (Fig. 1B, donors 7 and 8, respectively) were not previously tested. Thus, six specific 2-chloro-4-nitrophenyl glycoside donors (Fig. 1B, donors 1 and 4–8), representing excellent (donor 1), poor (donors 4–6), or unknown (donors 7 and 8) activity, were selected for screening as part of this model study (SI Appendix).
From the perspective of nucleotide base modifications, the goal of this study was to focus upon a representative set of core bases commonly used in glycobiology including, but not limited to, those affiliated with natural product biosynthesis. Specifically, UDP/TDP–sugars (predominate in microbial natural product biosynthetic pathways) (5, 20), CDP–sugars (important to the biosynthesis of pathogenic bacteria antigens) (21, 22), GDP–sugars (used in a range of glycobiological processes) (5, 23), and ADP–sugars (important to intracellular trafficking, posttranslational modification of proteins, DNA repair, programmed cell death, and glycogen metabolism) (24-26) were targeted in this study. While UDP or TDP with 2-chloro-4-nitrophenyl glucoside were previously recognized as excellent substrates in the OleD-catalyzed sugar nucleotide syntheses reactions, ADP and GDP were identified as relatively poor substrates (i.e., seven- and 50-fold decrease in conversion for ADP and GDP, respectively, compared with equivalent reactions with UDP), while CDP was not recognized as a substrate (19). Therefore, NDPs representing good (UDP and TDP), poor (ADP and GDP), and no activity (CDP) were selected for screening.
TDP16 Saturation Library Design.
Given the large substrate set of interest, we opted to limit the number of actual OleD variants to be screened as part of this model study. Thus, a representative OleD saturation library was generated. Positions targeted for saturation mutagenesis were selected by considering both previously identified mutational “hot spots” (14–16, 18) and additional potential ligand-interacting residues based upon a model constructed from the structure of wild-type OleD (wtOleD) bound to erythromycin and UDP (27), wherein UDP–2-deoxy-2-fluoroglucose from the UGT72B1 (a GT possessing a similar GT-B fold to that of OleD; Protein Data Bank ID code 2VCE) ligand-bound structure (28) was substituted for UDP. Based upon this assessment, seven putative sugar donor-interacting positions (amino acids 67, 74, 85, 111–113, and 134) and eight potential NDP acceptor-interacting positions (amino acids 132, 242, 243, 268, 290, 309, 330, and 331) were selected for saturation mutagenesis to present the potential of 285 possible mutants to be generated for screening with each desired substrate pairing. Of these positions, five (amino acids 67, 112, 132, 242, and 268) were previously identified as impacting upon OleD permissivity and/or proficiency (9–14). Special care was taken to ensure that the substrate binding pockets for both the putative 2-chloro-4-nitrophenyl glycoside donors (donors 1 and 4–8) and NDP acceptors were equally represented (Fig. 1; SI Appendix, Fig. S1). The parental sequence selected as the basis for this saturation library was OleD TDP16, a mutant previously characterized as being the most permissive and proficient toward substrates in both forward and reverse reactions compared with other OleD variants (18, 19). Compared with wtOleD, OleD TDP16 contains four beneficial mutations (P67T, S132F, A242L, and Q268V; discussed in detail below).
Creation of Saturation Libraries.
Using standard molecular biology techniques (SI Appendix), a total of 15 saturation libraries were generated based upon the parental sequence of the gene that encodes for the OleD variant TDP16 [oleD(P67T/S132F/A242L/Q268V)] (18). A total of 269 out of a possible 285 variants (94% coverage) were generated and confirmed by DNA sequencing (SI Appendix). Single isolates of Escherichia coli BL21(DE3)pLysS transformed with pET28a vectors containing these individual sequence variants and a number of previously reported OleD variants (specifically OleD ASP, 1C9, and 3–1H12, all variants identified for catalytic enhancements in either promiscuity and/or proficiency under various reaction conditions) (14–18) were used to generate three separate 96-well master plates (SI Appendix, Fig. S2) for high throughput screening.
Saturation Library Expression Test.
To assess the overall expression and solubility of the TDP16 saturation library members, strains containing empty pET28a vector, pET28a/oleD[TDP16], or plasmid variants possessing mutations (either alanine or glycine) at each of the targeted saturation positions within the parental gene sequence were selected for expression testing. An expression analysis of this representative variant subset by SDS/PAGE demonstrated that mutations could be introduced into each of the 15 selected saturation positions without detriment to the overall expression or solubility of the enzyme (SI Appendix, Fig. S3).
High Throughput Screening.
Master plates containing the saturation libraries were grown, induced for protein expression, processed, and the resulting cell lysate screened against a total of 13 distinct substrate combinations in reverse GT reactions for the production of NDP–sugars following our previously described protocol (19). Two main sets of screens were conducted: (i) 2-chloro-4-nitrophenyl β-D-glucopyranoside (donor 1) as donor with variant NDP acceptor (UDP, TDP, CDP, ADP, or GDP) and (ii) UDP as acceptor with variant 2-chloro-4-nitrophenyl glycosides (donors 4–8) as donor (Fig. 1). Following the addition of cell lysate to the prepared reaction mixtures, absorbance at 410 nm was monitored over a period of several hours. Representative data from a single assay of an individual master plate is provided in SI Appendix, Figs. S4 and S5. Following statistical analysis of the primary data, a subset of hits identified as top performers across multiple assays or substrates types was selected for additional screening.
Secondary Screening.
A total of 19 “hits” identified from the primary screen (7% of variants screened) were expressed and purified for secondary screening using purified catalysts (SI Appendix). Analysis by SDS/PAGE demonstrated the isolated enzymes to exhibit purity of >95% homogeneity (SI Appendix, Fig. S6). All purified catalysts were screened against a total of 10 assays with either 2-chloro-4-nitrophenyl-β-D-glucopyranoside (donor 1) as donor and various NDPs as acceptors or various 2-chloro-4-nitrophenyl glycosides (donors 4–8) as donors and UDP as acceptor. Following the addition of equal amounts of catalyst, the absorbance at 410 nm for each reaction was followed for a minimum of 3 h. Analysis of the initial slopes for each assay set revealed four specific mutations (I112P, T113M, L242I, and V268Q) (in the context of the TDP16 parental sequence) to be most advantageous overall to the formation of the targeted NDP–glucose and/or UDP–sugar combinations.
Recombination and Tertiary Screening.
The four mutations identified through secondary screening were recombined in the context of the TDP16 parental sequence to yield eight additional variants (five out of six potential double mutants, two out of four potential triple mutants, and one quadruple mutant) (SI Appendix). The recombinants were expressed and purified from 0.5 L cultures (SI Appendix) and analyzed by SDS/PAGE for purity (SI Appendix, Fig. S7). Following, the hits from the primary screen (i.e., I112P, T113M, L242I, and V268Q) and all of the obtained recombinant catalysts were screened in a total of 10 assays with either 2-chloro-4-nitrophenyl-β-D-glucopyranoside (donor 1) as donor and various NDPs as acceptors or various 2-chloro-4-nitrophenyl glycosides (donors 4–8) as donors and UDP as acceptor. Following the addition of equal amounts of catalyst, the absorbance at 410 nm for each reaction was monitored for a minimum of 3 h. Product formation, as initially assessed by the colorimetric assay, was also confirmed by liquid chromatography-MS at this stage (SI Appendix, Table S1).
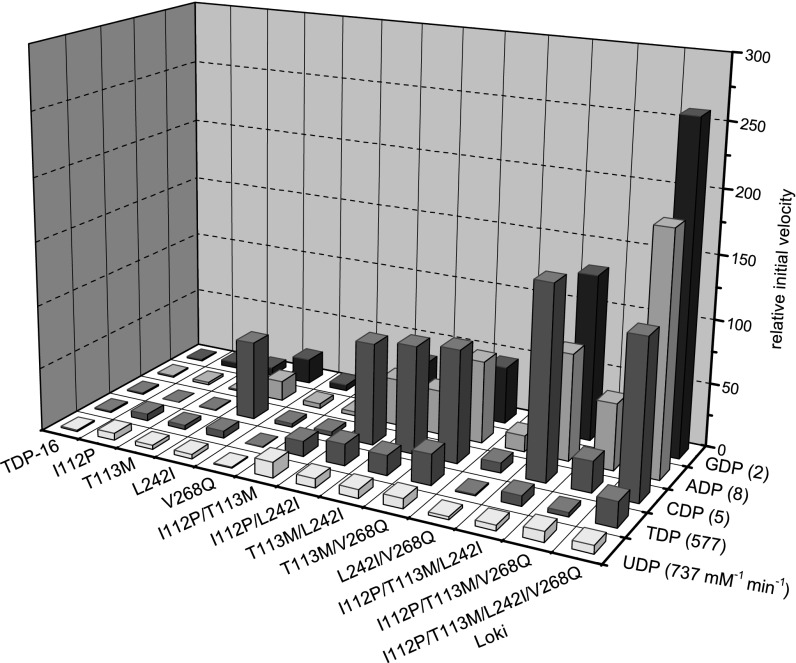
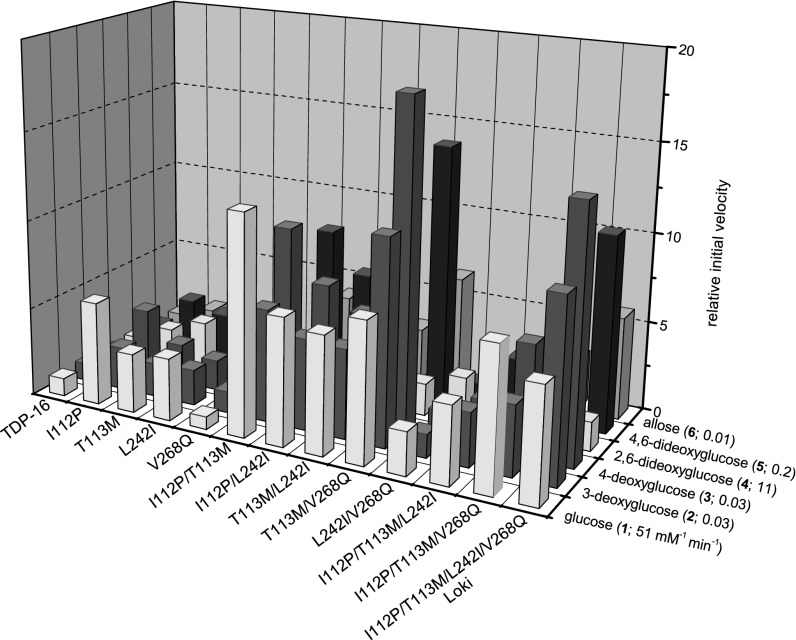
Comparison of the initial slopes for each reaction revealed a few key combinations to be particularly beneficial for NDP–glucose turnover [e.g., I112P/T113M with a 12-fold improvement for UDP–glucose (donor 9)], or UDP–sugar conversion [e.g., T113M/V268Q with an 18-fold improvement for UDP–4-deoxyglucose (donor 15)]. However, the recombination of I112P, T113M, L242I, and V268Q (in the context of TDP16) (i.e., the Loki variant, named after the Norse god of trickery and deception) served as the best overall “generalist” with an average improvement across the five NDP–glucose and six UDP–sugar combinations targeted in the screen of 119-fold and eightfold, respectively (Figs. 2 and 3). The most dramatic improvements were observed in the context of purine-based NDPs with Loki-based ADP–glucose (donor 12) and GDP–glucose (donor 13) turnover improvements of 188-fold and 259-fold, respectively (Fig. 2). While the goal of this engineering effort was to identify the best overall general catalyst for NDP–sugar formation, these results support the contention that the best generalist may not be the best “specialist” for a particular targeted reaction (29, 30). Overall, compared with wtOleD, the Loki variant contains a total of five mutations (P67T/I112P/T113M/S132F/A242I; Fig. 4), and the possible contributions of each mutation are briefly discussed below.
Fig. 2.
Tertiary screening results with NDPs. Relative initial velocities of OleD variants with 2-chloro-4-nitrophenyl glucoside (donor 1) as donor and NDPs as acceptors. The relative initial velocity for TDP-16 was set equal to 1 for all NDPs. The label for each variant are the amino acid mutation(s) introduced into the parental gene sequence of oleD[P67T/S132F/A242L/Q268V] (the template for OleD variant TDP-16). The determined kcat(Km)–1 for TDP-16 with donor 1 saturating and the respective NDP varied are listed after each NDP label. SDs of absorbance readings for TDP-16 (n = 2) typically varied by <5% for each data point over the course of each assay.
Fig. 3.
Tertiary screening results with varied sugar donors. Relative initial velocities of OleD variants with various 2-chloro-4-nitrophenyl glycoside donors (donors 1 and 4–8) and UDP as acceptor. The relative initial velocity for TDP-16 was set equal to 1 for all glycoside donors. The mutations listed are the amino acid mutations introduced into the parental gene sequence of oleD[P67T/S132F/A242L/Q268V] (generates OleD variant TDP-16). The determined kcat(Km)–1 for TDP-16 with UDP saturating and the respective 2-chloro-4-nitrophenyl glycoside (donors 1 and 4–8) varied are listed after the label of each donor sugar. SDs of absorbance readings for TDP-16 (n = 2) typically varied by <5% for each data point over the course of each individual assay.
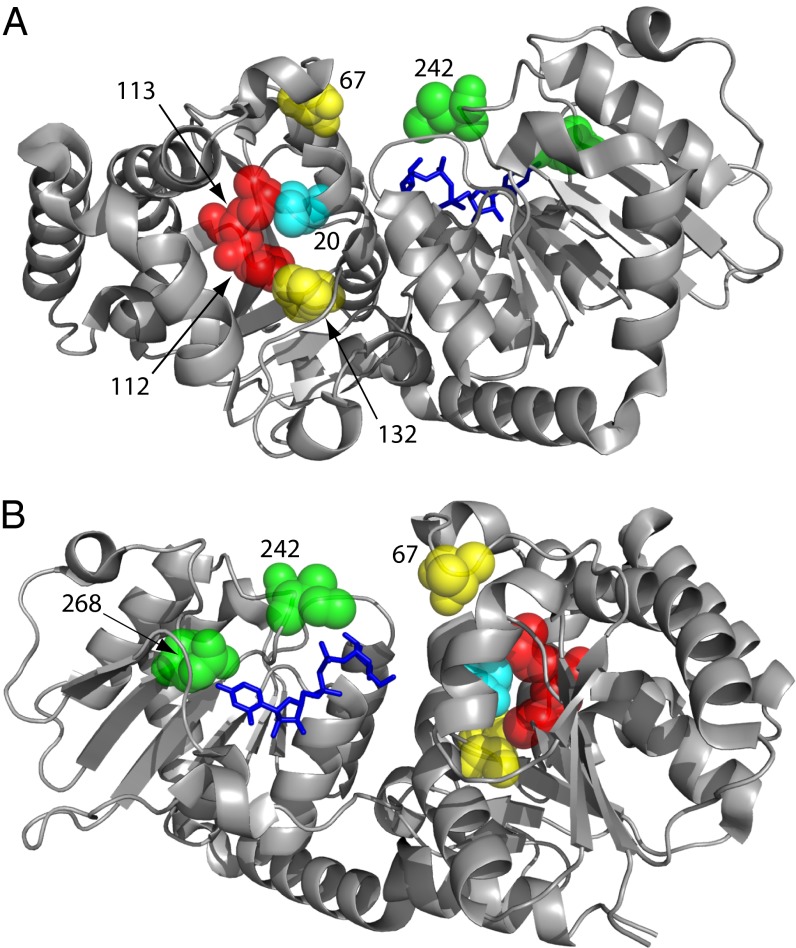
Fig. 4.
Crystal structure representation of divergence from wtOleD within the OleD variant Loki (i.e., P67T/I112P/T113M/S132F/A242I). In the context of a GT-catalyzed reverse reaction, the different views highlight the (A) donor pocket or (B) “acceptor” pocket of the OleD variant Loki. UDP–2-fluoroglucose (modeled) and the catalytic residue H20 are highlighted in blue and cyan, respectively.
Discussion of Identified Mutations.
While it is important to note that how the five mutations within Loki uniquely synergize cannot be gleaned from existing data, the potential roles of each position identified via this study are briefly highlighted below.
The P67T mutation was initially identified from a random mutagenesis library screened in the context of a GT-catalyzed forward reaction (14) and has been further studied in conjunction with additional OleD variants (15–19). Within the reported OleD crystal structure (27), residue 67 is situated amid a loop region (amino acids 60–76, loop N3), which is hypervariable in other GTs possessing the GT-B fold and contributes to forming the “donor” site in the context of GT-catalyzed reverse reactions (14, 31). Studies with other GT-B fold GTs have highlighted additional positions within the N3 loop as impacting upon enzyme specificity (32).
While mutations at position 112 have been previously characterized (14, 15, 18), advantageous mutations of position 113 within OleD have not been reported. The single mutation I112T was initially reported as having no effect upon the OleD-catalyzed glucosylation of 4-methylumbelliferone (14), but a subsequent study noted substantial improvement of kcat/Km upon the OleD-catalyzed glucosylation of novobiocic acid by both I112T and I112K (15). In the context of a GT-catalyzed reverse reaction, residues 112 and 113 are located deep within the donor pocket (Fig. 4), where they provide potential interactions with each other, the catalytic residue His20, and the targeted 2-chloro-4-nitrophenyl glycoside donors.
The alteration of position 132 from serine to phenylalanine was initially reported by Williams et al. (14), and the same alteration was present in a number of other characterized OleD variants (15–19). The OleD Ser132 equivalent in the plant flavonoid GT VvGT1 (Thr141) forms a hydrogen bond with the C-6 hydroxyl group of UDP–2-deoxy-2-fluoroglucose (33, 34). Lack of a hydrogen bond donor/acceptor at this position may explain the large increases in activity observed in previous studies (17) and in this investigation, particularly with C-6 modified sugars.
Substitution of residue Ala242 with hydrophobic replacements (Val, Leu, and Ile) has been reported in several cases to be advantageous in the context of both forward and reverse GT-catalyzed reactions (14–19). In the OleD crystal structure, Ala242 follows Ser241, which forms a hydrogen bond to the α-phosphate of the UDP acceptor in the context of a GT-catalyzed reverse reaction (this interaction was also observed in the VvGT1 structure) (14, 27, 33). Thus, hydrophobic mutations at the adjacent residue may influence NDP acceptor and/or sugar moiety interactions after transfer from a 2-chloro-4-nitrophenyl glycoside donor.
The residue Gln268 interacts directly with the UDP uridine C-4′ carbonyl (27). The Q268V mutation within TDP16 was originally identified in a screen for utilization of TDP–glucose (donor 10) in forward OleD-catalyzed reactions (18). However, reversion of the Val back to Gln is beneficial to GT-catalyzed reverse reactions, particularly in the context of purine NDPs and the glycoside donors screened in this study (Figs. 2–4).
Kinetic Evaluation.
The kinetic parameters of the parental TDP16 and the Loki variant were compared using all substrates identified in this study (SI Appendix, Figs. S8–S11 and Tables S24 and S25). Overall, the greatest increases in efficiency were observed for purine-based NDPs. For example, under saturating 2-chloro-4-nitrophenyl glucoside (donor 1), the catalytic efficiency (i.e., kcat/Km) of Loki with ADP or GDP (for formation of donors 12 and 13, respectively) was improved >400-fold over TDP-16 (SI Appendix, Table S24). Interestingly, there also was a notable change to the rank order of the NDPs when comparing kcat/Km across these variants (UDP > TDP > ADP > CDP > GDP for TDP-16; ADP > TDP > CDP > UDP > GDP for Loki) (SI Appendix, Table S24). Modest increases in catalytic efficiency ranging from ∼two- to 16-fold were observed for all sugar substrates (SI Appendix, Table S25). There was no substantial change to the rank order of the substrates screened when comparing kcat/Km for TDP16 and Loki as the parental sugar D-glucose remains the preferred substrate for the Loki variant by at least an order of magnitude. A comparison of the kinetic parameters for wtOleD- versus Loki-catalyzed “reverse” (sugar nucleotide forming) reactions reveals Loki to impart major improvements in Km for both UDP and donor 1 (SI Appendix, Table S26), while a similar comparison in the context of a model forward reaction (using UDP–Glc as donor and 4-methyl umbelliferone as the acceptor) revealed Loki improvements in both kcat and Km (SI Appendix, Table S27). While it is possible that the screening approach described here would identify variants with hydrolytic activity, HPLC analysis hydrolysis of 2-chloro-4-nitrophenyl glycoside donors was only observed at high enzyme concentrations (typically, >10-fold excess of that used for in the screening method).
Combinatorial NDP–Sugar Synthesis.
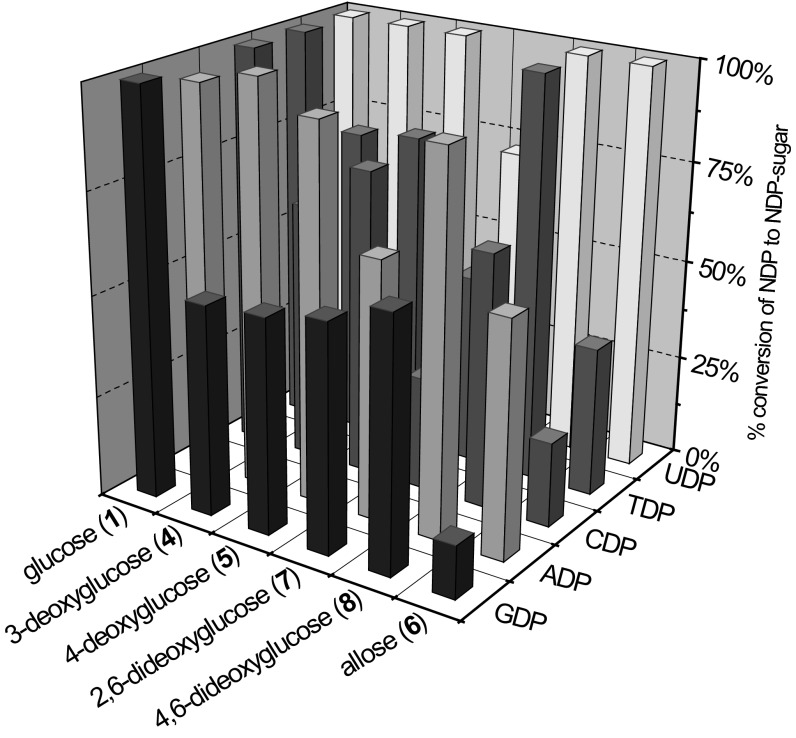
To assess the combinatorial potential of the Loki variant as a general catalyst for NDP–sugar formation, individual OleD–Loki–catalyzed reactions, covering all possible NDP–donor pairings from six glycoside donors and five NDP acceptors, were analyzed for product formation. Surprisingly, meaningful formation of all 30 expected NDP–sugar products was observed in this cumulative analysis (Fig. 5; SI Appendix, Fig. S12 and Table S2). The average conversion across all 30 products was 74 ± 27%, with 17 examples demonstrating yields >70%. The best overall conversions were for formation of NDP–glucoses (donors 9–13) and UDP–sugars (donors 9 and 14–18), with average observed yields of 99 ± 1.7% and 96 ± 11%, respectively. NDP–alloses displayed the poorest conversions with TDP–, CDP–, and GDP–allose (donors 23, 28, and 38, respectively), averaging an observed yield of 24 ± 12%.
Fig. 5.
Percent conversions of NDP to NDP–sugar with the OleD variant Loki. Reactions contained 0.5 mM of 2-chloro-4-nitrophenyl glycoside donor (donors 1 and 4–8), 1.0 mM NDP, and 5.25 μM (50 μg) of purified Loki variant in Tris⋅HCl buffer (50 mM, pH 8.0) and a final volume of 200 μL. Absorbance at 410 nm was followed and reactions were halted when they reached completion, increase in absorbance halted, or total reaction time reached 24 h (SI Appendix). Percent conversions were determined by HPLC. The SD for production of UDP–α-D-glucose (donor 9) across multiple runs (n = 3) was <10%.
Taken together with previous results (19), NDP–sugars based upon D-glucose, D-allose, and D-xylose, along with sugars containing 2-, 3-, 4-, or 6-deoxy, 2,6- or 4,6-dideoxy, and 6-modified motifs (e.g., 6-bromo, 6-thio-, 6-azido-, etc.) are now readily accessible through OleD-catalyzed reactions. This strategy compares favorably to the state of the art for sugar nucleotide synthesis. For example, the synthesis of TDP–D-olivose (a precursor to many 2,6-dideoxysugar–containing natural products) (2, 5) derived from donor 7 (five simple steps from peracetylated 2-deoxy-D-glucose) to give a 47% observed yield in the final OleD-catalyzed reverse reaction (Fig. 5). For comparison, Minami et al. reported a nine-step chemical synthesis of TDP–D-olivose from 2-deoxy-D-glucose with a 2.5% overall isolated yield (35); Amann et al. reported a chemoenzymatic approach from D-glucose-6-phosphate requiring five enzymes with a 15% overall yield and a putative mixture of C-4 epimers (36); while Wang et al. reported an eight-enzyme chemoenzymatic method from α-D-glucose-1-phosphate with an overall yield of 61% (37). Importantly, the OleD-catalyzed method described uses stable donors, can be directly coupled to downstream sugar nucleotide-using reactions (including GT-catalyzed forward reactions) (19), is a robust single enzyme process (thus eliminating feedback and feed-forward inhibition commonly observed in multienzyme systems), and uniquely regenerates sugar nucleotide in the presence of NDP (a product of GT-catalyzed forward reactions) that, when directly coupled to GT-catalyzed glycosylation reaction, serves as a driving force.
Preliminary Assessment of Loki in a Model Coupled Forward Reaction.
While this study clearly reveals the Loki mutant to provide dramatic improvements for sugar nucleotide production, the ability of Loki to catalyze a coupled forward reaction was also assessed using a previously developed small molecule model. Specifically, the ability of Loki to catalyze the overall transglycosylation reaction from 2-chloro-4-nitrophenyl glucoside to 4-methyl-umbelliferone (mediated via UDP–Glc formation and consumption) was assessed via a simple 4-methyl-umbelliferone fluorescence assay (14, 16). This study revealed the specific activity of Loki, based upon the coupled 4-methyl-umbelliferone glucosylation assay, to be 32-fold greater than that of TDP16 assessed in parallel (SI Appendix, Fig. S13). This preliminary study indicates the Loki catalyst, in comparison with progenitors, to be not only a superior catalyst for sugar nucleotide synthesis but also superior as a potential catalyst for subsequent small molecule glycosylation (17, 39–41).
Conclusions.
Inspired by ability of simple “activated” donors to modulate the thermodynamics of GT-catalyzed reactions, this study highlights an application of a corresponding 2-chloro-4-nitrophenyl glycoside-based high throughput screen in glycosyltransferase--directed engineering toward enhanced catalysts for sugar nucleotide synthesis. While many specialists (catalysts enhanced for specific nonnative substrate pairings) were observed, the most productive generalist identified (OleD Loki) was capable of using all possible NDP–donor combinations of five NDP acceptors and six sugar donors to give 30 distinct NDP–sugars (9–38). The impressive aglycon malleability of OleD (17–19), the demonstrated ability to expand the sugar scope of this catalyst, and the demonstrated superior ability of Loki to also catalyze “forward” coupled transglycosylation reactions suggest a range of exciting opportunities. Examples include extending applications toward (i) modified nucleoside-based NDPs (3) (reminiscent of the kinase “bump and hole” strategies) (38); (ii) specific sugar–drug or sugar–natural product pairings (39–41); (iii) a broadened scope of deoxy, dideoxy, and/or uniquely functionalized sugars (e.g., sugars bearing amino-, N-akyl, O-alkyl, C-alkyl-, nitro, nitroso-, thio-modifications) (2, 4, 7, 10, 15, 18, 19); and (iv) other important biomolecules (e.g., proteins, polysaccharides) (42–45).
Materials and Methods
Primary High Throughput Screen.
Glycerol master plates (SI Appendix) were thawed at 4 °C, and 10 μL was transferred to 96-deep-well microtiter plates containing 1 mL of Luria–Bertani (LB) media in each well supplemented with 50 μg⋅mL−1 kanamycin. The glycerol master plates were refrozen at –80 °C while the culture plates were sealed with Breathe-Easy Sealing Membrane (Research Products International) and incubated at 37 °C and 350 rpm overnight. Following, 50 μL of each culture was transferred to a fresh deep-well plate containing 950 μL of LB medium supplemented with 50 μg⋅mL−1 kanamycin.
The freshly inoculated plates were incubated for 3 h at 37 °C and 350 RPM. Expression of the N-terminal His6-tagged OleD variants was induced via the addition of isopropyl β-D-1-thiogalactopyranoside to a final concentration of 0.4 mM, and the plates were incubated for 12–15 h at 28 °C and 350 RPM. Cells were harvested by centrifugation at 3,000 × g for 20 min at 4 °C, the cell pellets were thoroughly resuspended in 250 μL of 50 mM Tris⋅HCl (pH 8.0) containing 5 mg⋅mL–1 lysozyme (Sigma Aldrich) at 4 °C, and the plates were subjected to a single freeze-thaw cycle. Cell debris was then collected by centrifugation at 3,000 × g for 20 min at 4 °C, and either 25 or 100 μL of the cleared supernatant was used for the various enzyme assays (SI Appendix, Table S3).
All assays were conducted in 50 mM Tris⋅HCl buffer (pH 8.0) and a final volume of 200 μL. For the assays, cleared lysate was added to various concentrations and combinations of 2-chloro-4-nitrophenyl glycoside donor and nucleotide diphosphate acceptor (SI Appendix, Table S3). Upon mixing, the production of 2-chloro-4-nitrophenolate was followed by absorbance at 410 nm over a period of up to 48 h at 25 °C.
Analysis of High Throughput Data.
The initial slope and area under the curve for the absorbance data were calculated individually for each OleD variant. Area under the curve for each reaction was determined with Eq. 1:
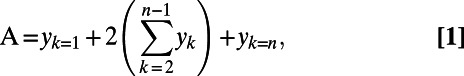 |
where A equals the total area of the curve and yk equals the absorbance value at the kth time point. Hits that performed above the TDP16 parental sequence and ranked high under both initial rate and area under the curve for individual substrate sets were selected for secondary screening and validation. Intraplate SDs for the TDP16 parental sequence absorbance data across all substrate combinations were typically <15% (n = 2) over the course of the respective assay.
Secondary and Tertiary Screening.
In a final volume of 200 μL, reactions containing 2-chloro-4-nitrophenyl glycoside donor, NDP acceptor, and purified catalyst (SI Appendix) in Tris⋅HCl buffer (50 mM, pH 8.0) were prepared in 96-well microtiter plates (SI Appendix, Tables S4 and S5). Following the addition of equivalent amounts of each catalyst, absorbance readings at 410 nm were recorded for a minimum of 3 h. Assay conditions were optimized for each substrate pair to maximize the difference in absorbance signal between the parental catalyst TDP16 (n = 2) and the best performing variants (n = 1). Analysis of the raw absorbance data (see above) was used to identify and quantify the top performers. SDs of absorbance readings for TDP16 (n = 2) typically varied by <5% for each data point over the course of the assay.
Combinatorial NDP–Sugar Synthesis.
Reactions containing 5.25 μM (50 μg) of Loki variant and each individual combination of 0.5 mM 2-chloro-4-nitrophenyl donor (donors 1 and 4–8) and 1 mM NDP acceptor (UDP, TDP, CDP, ADP, or GDP) were prepared in Tris⋅HCl buffer (50 mM, pH 8.0) with a final volume of 200 μL in a flat bottom 96-well microtiter plate. Following the addition of catalyst, the change in absorbance was followed at 410 nm. Identical reactions containing triethylammonium acetate buffer (50 mM, pH 7.0) in place of Tris⋅HCl buffer were run simultaneously for LC–MS analysis.
When reactions with Tris⋅HCl buffer reached completion (determined by comparing the absorbance reading to a standard curve), absorbance readings remained constant over two or more readings, or when the total reaction time reached 24 h, reactions with triethylammonium acetate buffer were treated with 20 U of alkaline phosphatase (Roche) for 30 min at RT. Following, reactions from both buffer systems were frozen at –80 °C. After thawing at 4 °C, all samples were filtered through a MultiScreen Filter plate with Ultracel-10 Membrane (Millipore) according to the manufacturer’s instructions.
The cleared supernatant from each sample with Tris⋅HCl was evaluated for formation of the expected NDP–sugars with analytical reverse phase HPLC. Formation of UDP– (donors 9 and 14–18), TDP– (donors 10 and 19–23), ADP– (donors 12 and 29–33), and GDP–sugars (donors 13 and 34–38) was analyzed with a 250 mm × 4.6 mm Gemini-NX 5 μ C18 column with a gradient of 0–50% CH3CN (solvent B) over 25 min [solvent A = 50 mM PO4–2, 5 mM tetrabutylammonium bisulfate, 2% acetonitrile (pH adjusted to 6.0 with KOH); flow rate = 1 mL⋅min−1; A254 nm]. CDP–sugars (donors 11, 24–26, and 28) were analyzed with a 250 mm × 4.6 mm Inertsil octadecylsilane-4 3 μ C18 column using a gradient of 0% B for 30 min, 0–100% B over 3 min, and 100% B for 22 min [solvent A = 100 mM PO4–2, 8 mM tetrabutylammonium bisulfate (pH adjusted to 6.4 with KOH); solvent B = 70% buffer A, 30% CH3CN (after mixing components, pH was adjusted to 6.4); flow rate = 0.5 mL min−1; A254 nm]. Formation of donor 27 was confirmed on a 250 mm × 4.6 mm Gemini-NX 5 μ C18 column with a gradient of 0% CH3CN (solvent B) for 15 min, 0–25% B over 10 min, and 25–90% B over 5 min (A = 50 mM triethylammonium acetate buffer; flow rate = 1 mL⋅min−1; A254 nm). The SD for production of UDP–α-D-glucose (donor 9) across multiple runs (n = 3) was <10%.
The cleared supernatant from each sample with triethylammonium acetate buffer was lyophilized and resuspended in water (3 × 500 μL). Following the final lyophilization, the crude reactions were resuspended in 30 μL of dH2O, diluted 1:10 with 1 mM ammonium acetate in 85% CH3CN/15% dH2O (pH 5.5), and then 5 μL was injected for LC–MS analysis. The LC–MS method consisted of an Acquity bridged ethane hybrid 1.7 μm Amide column (2.1 × 100 mm; Waters Corp.) with a gradient of 75–25% B over 5 min, 25% B for 3 min, 25–75% B over 0.2 min, and 75% B for 10 min [solvent A = 1.0 mM ammonium acetate in 65% CH3CN/35% dH2O (pH 5.5); solvent B = 1.0 mM ammonium acetate in 85% CH3CN/15% dH2O (pH 5.5); flow rate = 1.0 mL⋅min−1; A254 nm) while HRMS spectra were collected over the course of the separation (SI Appendix). Retention times for NDP–sugars were typically 4–7 min.
Supplementary Material
Acknowledgments
We thank the School of Pharmacy Analytical Instrumentation Center (University of Wisconsin, Madison, WI) for analytical support. This work was supported by National Institutes of Health MERIT Award AI52218 (to J.S.T.) and National Center for Advancing Translational Sciences UL1TR000117. R.W.G. is an American Foundation for Pharmaceutical Education Fellow.
Footnotes
Conflict of interest statement: J.S.T. is a cofounder of Centrose, Madison, WI.
This article is a PNAS Direct Submission. S.G.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220220110/-/DCSupplemental.
References
- 1.Varki A, Sharon N. In: Essentials of Glycobiology. Varki A, et al., editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2009. pp. 1–100. [Google Scholar]
- 2.Thibodeaux CJ, Melançon CE, III, Liu HW. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446(7139):1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- 3.Wagner GK, Pesnot T, Field RA. A survey of chemical methods for sugar-nucleotide synthesis. Nat Prod Rep. 2009;26(9):1172–1194. doi: 10.1039/b909621n. [DOI] [PubMed] [Google Scholar]
- 4.Gantt RW, Peltier-Pain P, Thorson JS. Enzymatic methods for glyco(diversification/randomization) of drugs and small molecules. Nat Prod Rep. 2011;28(11):1811–1853. doi: 10.1039/c1np00045d. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka H, Yoshimura Y, Jørgensen MR, Cuesta-Seijo JA, Hindsgaul O. A simple synthesis of sugar nucleoside diphosphates by chemical coupling in water. Angew Chem Int Ed Engl. 2012;51(46):11531–11534. doi: 10.1002/anie.201205433. [DOI] [PubMed] [Google Scholar]
- 6.Mohamady S, Desoky A, Taylor SD. Sulfonyl imidazolium salts as reagents for the rapid and efficient synthesis of nucleoside polyphosphates and their conjugates. Org Lett. 2012;14(1):402–405. doi: 10.1021/ol203178k. [DOI] [PubMed] [Google Scholar]
- 7.Thibodeaux CJ, Melançon CE, 3rd, Liu HW. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew Chem Int Ed Engl. 2008;47(51):9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rupprath C, Schumacher T, Elling L. Nucleotide deoxysugars: Essential tools for the glycosylation engineering of novel bioactive compounds. Curr Med Chem. 2005;12(14):1637–1675. doi: 10.2174/0929867054367167. [DOI] [PubMed] [Google Scholar]
- 9.Pérez M, et al. Combinatorial biosynthesis of antitumor deoxysugar pathways in Streptomyces griseus: Reconstitution of “unnatural natural gene clusters” for the biosynthesis of four 2,6-D-dideoxyhexoses. Appl Environ Microbiol. 2006;72(10):6644–6652. doi: 10.1128/AEM.01266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang C, et al. Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions. Science. 2006;313(5791):1291–1294. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]
- 11.Lairson LL, Wakarchuk WW, Withers SG. Alternative donor substrates for inverting and retaining glycosyltransferases. Chem Commun (Camb) 2007;(4):365–367. doi: 10.1039/b614636h. [DOI] [PubMed] [Google Scholar]
- 12.Lougheed B, Ly HD, Wakarchuk WW, Withers SG. Glycosyl fluorides can function as substrates for nucleotide phosphosugar-dependent glycosyltransferases. J Biol Chem. 1999;274(53):37717–37722. doi: 10.1074/jbc.274.53.37717. [DOI] [PubMed] [Google Scholar]
- 13.Minami A, Kakinuma K, Eguchi T. Algycon switch approach toward unnatural glycosides from natural glycoside with glycosyltransferase VinC. Tetrahedron Lett. 2005;46(37):6187–6190. [Google Scholar]
- 14.Williams GJ, Zhang C, Thorson JS. Expanding the promiscuity of a natural-product glycosyltransferase by directed evolution. Nat Chem Biol. 2007;3(10):657–662. doi: 10.1038/nchembio.2007.28. [DOI] [PubMed] [Google Scholar]
- 15.Williams GJ, Goff RD, Zhang C, Thorson JS. Optimizing glycosyltransferase specificity via “hot spot” saturation mutagenesis presents a catalyst for novobiocin glycorandomization. Chem Biol. 2008;15(4):393–401. doi: 10.1016/j.chembiol.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams GJ, Thorson JS. A high-throughput fluorescence-based glycosyltransferase screen and its application in directed evolution. Nat Protoc. 2008;3(3):357–362. doi: 10.1038/nprot.2007.538. [DOI] [PubMed] [Google Scholar]
- 17.Gantt RW, Goff RD, Williams GJ, Thorson JS. Probing the aglycon promiscuity of an engineered glycosyltransferase. Angew Chem Int Ed Engl. 2008;47(46):8889–8892. doi: 10.1002/anie.200803508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams GJ, Yang J, Zhang C, Thorson JS. Recombinant E. coli prototype strains for in vivo glycorandomization. ACS Chem Biol. 2011;6(1):95–100. doi: 10.1021/cb100267k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gantt RW, Peltier-Pain PP, Cournoyer WJ, Thorson JS. Using simple donors to drive the equilibria of glycosyltransferase-catalyzed reactions. Nat Chem Biol. 2011;7(10):685–691. doi: 10.1038/nchembio.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh S, Phillips GN, Jr, Thorson JS. The structural biology of enzymes involved in natural product glycosylation. Nat Prod Rep. 2012;29(10):1201–1237. doi: 10.1039/c2np20039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu HW, Thorson JS. Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu Rev Microbiol. 1994;48:223–256. doi: 10.1146/annurev.mi.48.100194.001255. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, et al. Biochemical characterization of the CDP-D-arabinitol biosynthetic pathway in Streptococcus pneumoniae 17F. J Bacteriol. 2012;194(8):1868–1874. doi: 10.1128/JB.06487-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16(12):158R–184R. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 24.Ballicora MA, Iglesias AA, Preiss J. ADP-glucose pyrophosphorylase, a regulatory enzyme for bacterial glycogen synthesis. Microbiol Mol Biol Rev. 2003;67(2):213–225. doi: 10.1128/MMBR.67.2.213-225.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schreiber V, Dantzer F, Amé J-C, de Murcia G. Poly(ADP-ribose): Novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7(7):517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 26.Abd Elmageed ZY, Naura AS, Errami Y, Zerfaoui M. The poly(ADP-ribose) polymerases (PARPs): New roles in intracellular transport. Cell Signal. 2012;24(1):1–8. doi: 10.1016/j.cellsig.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 27.Bolam DN, et al. The crystal structure of two macrolide glycosyltransferases provides a blueprint for host cell antibiotic immunity. Proc Natl Acad Sci USA. 2007;104(13):5336–5341. doi: 10.1073/pnas.0607897104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brazier-Hicks M, et al. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci USA. 2007;104(51):20238–20243. doi: 10.1073/pnas.0706421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reetz MT, Bocola M, Carballeira JD, Zha D, Vogel A. Expanding the range of substrate acceptance of enzymes: Combinatorial active-site saturation test. Angew Chem Int Ed Engl. 2005;44(27):4192–4196. doi: 10.1002/anie.200500767. [DOI] [PubMed] [Google Scholar]
- 30.Nair NU, Denard CA, Zhao H. Engineering of enzymes for selective catalysis. Curr Org Chem. 2010;14(17):1870–1882. [Google Scholar]
- 31.Mulichak AM, Lu W, Losey HC, Walsh CT, Garavito RM. Crystal structure of vancosaminyltransferase GtfD from the vancomycin biosynthetic pathway: Interactions with acceptor and nucleotide ligands. Biochemistry. 2004;43(18):5170–5180. doi: 10.1021/bi036130c. [DOI] [PubMed] [Google Scholar]
- 32.Hoffmeister D, et al. Engineered urdamycin glycosyltransferases are broadened and altered in substrate specificity. Chem Biol. 2002;9(3):287–295. doi: 10.1016/s1074-5521(02)00114-x. [DOI] [PubMed] [Google Scholar]
- 33.Shao H, et al. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell. 2005;17(11):3141–3154. doi: 10.1105/tpc.105.035055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Offen W, et al. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006;25(6):1396–1405. doi: 10.1038/sj.emboj.7600970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minami A, Eguchi T. Substrate flexibility of vicenisaminyltransferase VinC involved in the biosynthesis of vicenistatin. J Am Chem Soc. 2007;129(16):5102–5107. doi: 10.1021/ja0685250. [DOI] [PubMed] [Google Scholar]
- 36.Amann S, Dräger G, Rupprath C, Kirschning A, Elling L. (Chemo)enzymatic synthesis of dTDP-activated 2,6-dideoxysugars as building blocks of polyketide antibiotics. Carbohydr Res. 2001;335(1):23–32. doi: 10.1016/s0008-6215(01)00195-1. [DOI] [PubMed] [Google Scholar]
- 37.Wang G, Kharel MK, Pahari P, Rohr J. Investigating Mithramycin deoxysugar biosynthesis: Enzymatic total synthesis of TDP-D-olivose. ChemBioChem. 2011;12(17):2568–2571. doi: 10.1002/cbic.201100540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407(6802):395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 39.Peltier-Pain P, Marchillo K, Zhou M, Andes DR, Thorson JS. Natural product disaccharide engineering through tandem glycosyltransferase catalysis reversibility and neoglycosylation. Org Lett. 2012;14(19):5086–5089. doi: 10.1021/ol3023374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou M, et al. Probing the regiospecificity of enzyme-catalyzed steroid glycosylation. Org Lett. 2012;14(21):5424–5427. doi: 10.1021/ol3024924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou M, Hamza A, Zhan C-G, Thorson JS. Assessing regioselectivity of OleD-catalyzed glycosylation with a diverse set of acceptors. J Nat Prod. 2013;76(2):279–286. doi: 10.1021/np300890h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peter-Katalinić J. Methods in enzymology: O-glycosylation of proteins. Methods Enzymol. 2005;405:139–171. doi: 10.1016/S0076-6879(05)05007-X. [DOI] [PubMed] [Google Scholar]
- 43.Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol. 2005;3(3):225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 44.Schiller B, Hykollari A, Yan S, Paschinger K, Wilson IB. Complicated N-linked glycans in simple organisms. Biol Chem. 2012;393(8):661–673. doi: 10.1515/hsz-2012-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baskin JM, Dehnert KW, Laughlin ST, Amacher SL, Bertozzi CR. Visualizing enveloping layer glycans during zebrafish early embryogenesis. Proc Natl Acad Sci USA. 2010;107(23):10360–10365. doi: 10.1073/pnas.0912081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.