Abstract
Somatic hypermutation (SHM) requires not only the expression of activation-induced cytidine deaminase, but also transcription in the target regions. However, how transcription guides activation-induced cytidine deaminase in targeting SHM to the Ig genes is not fully understood. Here, we found that the “facilitates chromatin transcription” (FACT) complex promotes SHM by RNAi screening of transcription elongation factors. Furthermore, FACT and histone H3.3, a hallmark of transcription-coupled histone turnover, are enriched at the V(D)J region, 5′ flanking sequence of the Sμ switch region and the light chain Jκ 5 segment region in the Ig loci. The regions with the most abundant deposition of FACT and H3.3 were also the most efficient targets of SHM. These results demonstrate the importance of histone-exchanging dynamics at the chromatin of SHM targets, especially in Ig genes.
Keywords: AID, SSRP1
Somatic hypermutation (SHM) is the process by which antigen-stimulated germinal-center B cells accumulate point mutations in the Ig genes, which leads to the generation of higher affinity antibodies. Both the expression of activation-induced cytidine deaminase (AID) and transcription of the target genomic regions are required for SHM (1). As genomic mutations by AID can lead to genomic instability and possible tumorigenesis (2), the targets of SHM are restricted almost exclusively to the rearranged V(D)J regions and the switch region in the Ig genes. This highly specific SHM targeting can be partially explained by special DNA structures, such as the R-loop, G-quartet, and non-B structure, that can be produced during transcription at the target regions (1, 3, 4). In fact, mutation targets are flanked by non–B-prone DNA sequences, such as tandem repeats and inverted repeats (3, 5). It has been proposed that non-B DNA structure can cause irreversible DNA cleavage by DNA Topoisomerase I, leading to generation of the DNA lesions responsible for class switch recombination (CSR) and SHM (3, 6, 7).
In addition to DNA structure, recent studies revealed the importance of transcription elongation in SHM targeting (8, 9). Transcription elongation by RNA polymerase II (RNAPII) is a dynamic process that is controlled by a group of molecules called transcription elongation factors (10). In addition, elongation factors are involved in various transcription-coupled processes including chromatin modifications (10). Studies by many groups including ourselves have revealed that several transcription elongation factors play important roles in the antibody diversification processes. Pavri et al. (9) proposed that suppressor of Ty 5 homolog (Spt5), the large subunit of elongation factor DSIF (5,6-dichloro-1-beta-d-ribofuranosylbenzimidazole sensitivity-inducing factor), promotes both CSR and SHM by recruiting AID through physical interaction and by inducing RNAPII stalling. We previously showed that Spt5 is involved in the generation of H3K4me3 marking on chromatin, which is required for DNA cleavage by AID (11). In addition, we found that Spt5 promotes an end-joining repair process during CSR (11). Spt6 is another elongation factor that we found promotes CSR (12). We originally identified Spt6 based on its physical interaction with AID but later found that their interaction is not important for SHM or CSR (8). A recent work indicates that another elongation factor, the polymerase associated factor 1 (Paf1) complex, can serve as a binding platform for AID on target chromatin (13).
The facilitates chromatin transcription (FACT) complex is a histone chaperone-type elongation factor that was originally discovered by its biochemical activity to promote RNAPII transcription elongation on the nucleosomal DNA template (14). FACT is proposed to evict nucleosomal histones and deposit them at the site of transcription by RNAPII, so that the polymerase can proceed beyond the nucleosomes (15). We previously discovered that FACT is required to induce CSR (16). FACT knockdown blocks CSR at the DNA-cleavage step without affecting the level of the Ig heavy chain gene (Igh) transcription, and FACT is important for inducing trimethylation on histone H3K4 (H3K4me3), which seems to be recognized by a protein complex with DNA-cleaving activity. Therefore, we proposed that the H3K4me3 chromatin modification is a marker for CSR. We later showed that H3K4me3 also accumulates at SHM-targeted genomic regions, indicating that this histone modification is important for the induction of both CSR and SHM (5).
Nonetheless, the elongation factors and chromatin modifications that are enriched specifically at the mutable regions on Ig genes have yet to be identified. Spt5 binding is almost proportional to the level of RNAPII at genetic loci (9, 17). Similarly, H3K4me3 serves as a general marker of active chromatin (18), and the Paf1 complex acts as a scaffold for the H3K4 trimethylase complex and is required for the elongation-promoting activity of the DSIF complex (19). Recent data for the chromatin occupancy of AID also failed to fully explain the specificity of SHM, as no specificity of AID binding to the Ig locus was found (20).
In the present study, we identified FACT as an SHM-promoting transcription elongation factor, by siRNA screening using an SHM reporter in human Burkitt's lymphoma (BL)2 cells. Consistent with this finding, we found gene-specific enrichment of FACT and the histone H3 variant H3.3, a marker for replication-independent histone turnover, in the V(D)J and 5′ flanking sequence of the Sμ switch (5′Sμ) regions. Furthermore, we showed that the genomic regions that were occupied by abundant FACT and H3.3 are efficient targets for SHM. We discuss the possible functional relevance of the FACT enrichment on SHM targets and of histone exchanging dynamics in SHM.
Results
Identification of FACT as an SHM-Promoting Elongation Factor.
To search for SHM-coupled transcription elongation factor(s), we performed small interference RNA (siRNA) screening in human BL2 cells expressing the Hygromycin-GFP fusion reporter gene (HygGFP) and a C-terminally truncated AID mutant (JP8Bdel) fused with the estrogen-binding domain (JP8Bdel-ER). JP8Bdel has four times stronger SHM activity than wild-type AID, although it has little CSR activity (21). The estrogen analog 4-hydroxyl Tamoxifen (OHT) was added to the culture medium to activate JP8Bdel-ER, and SHM was quantified by the number of cells that lost the GFP fluorescence of HygGFP.
To identify transcription elongation factors involved in SHM, we depleted representative subunits of 12 known transcription elongation factors by introducing siRNA oligonucleotides into the BL2 cells before the OHT addition. The knockdown of each elongation factor was carried out by three different siRNAs. The GFP-negative cell population and efficiencies of depletion were then evaluated by FACS and quantitative (q)RT-PCR, respectively.
Each factor was knocked down reproducibly with more than one siRNA oligonucleotide. The knockdown of most of the elongation factors, including DSIF, Spt6, and the Paf1 complex, which were previously reported to be involved in CSR, did not reduce the GFP-negative cell population. This inconsistency can be explained by either a mechanistic difference between CSR and SHM or a target difference between Ig genes and the HygGFP reporter construct.
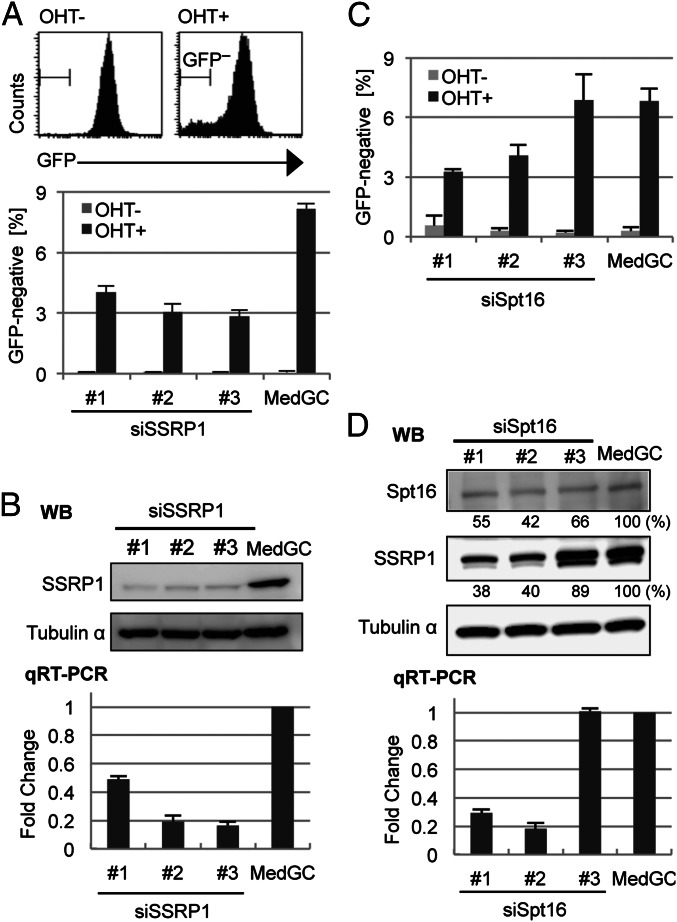
Among the twelve elongation factors tested, we found that the depletion of FACT subunit structure specific recognition protein 1 (SSRP1) reduced the GFP-negative population much more than depletion of any of the others (Fig. S1). All three siRNAs against SSRP1 reduced the GFP-negative population by more than half (Fig. 1A, Lower). Western blot analysis showed that all three siRNAs had depleted the SSRP1 protein efficiently (Fig. 1B, Upper). We determined the actual knockdown efficiency of the siRNA oligonucleotides against SSRP1 by qRT-PCR and found that the SSRP1 knockdown efficiency was correlated with the level of GFP loss (Fig. 1B, Lower).
Fig. 1.
Depletion of the FACT complex inhibits the induction of a GFP-negative cell population by the AID transgene. (A) Flow cytometric analysis for the GFP-negative cell population in SSRP1-depleted cells. (Upper) Example histograms of GFP fluorescence in OHT-treated (OHT+) and untreated (OHT−) cells. The population within the “GFP−“ area was defined as GFP-negative. (Lower) Percentages of GFP-negative population in anti-SSRP1 Stealth siRNA (#1–#3) and MedGC control-treated cells. (B) Confirmation of SSRP1 knockdown with Western blot (WB, Upper) and qRT-PCR (Lower). Samples were prepared from cells 48 h after siRNA transfection. In WB, Tubulin-α protein was used for the loading control. In qRT-PCR, the expression level of Ssrp1 relative to the MedGC was calculated and normalized to the Gapdh expression level. The primers for qRT-PCR are shown in Table S4. (C and D) The same experiments as in A and B were performed for Spt16 and are shown in C and D, respectively. (D) Densitometric evaluation of the band intensities is shown below the respective blots. The lower band in the SSRP1 blot is the degraded form. All the graph data are the mean and SD of three independent experiments.
We next knocked down the other FACT subunit Spt16 with three siRNAs, among which two of them reduced the GFP-negative population, consistent with the mRNA depletion levels (Fig. 1 C and D, Lower). Although the reduction of Spt16 protein was not clearly correlated with the mRNA levels, the effective oligos (oligos 1 and 2) clearly depleted the SSRP1 protein (Fig. 1D, Upper). It appears that Spt16 knockdown cannot be separated from SSRP1 knockdown as the Spt16 knockdown causes destabilization of the SSRP1 protein.
To further confirm our findings, we also used a different SHM assay system, which estimates SHM by the recovery of GFP fluorescence in a GFP construct that contains a stop codon (22). As expected, we observed a significant reduction of the AID-induced GFP recovery in the SSRP1-depleted cells (Fig. S2).
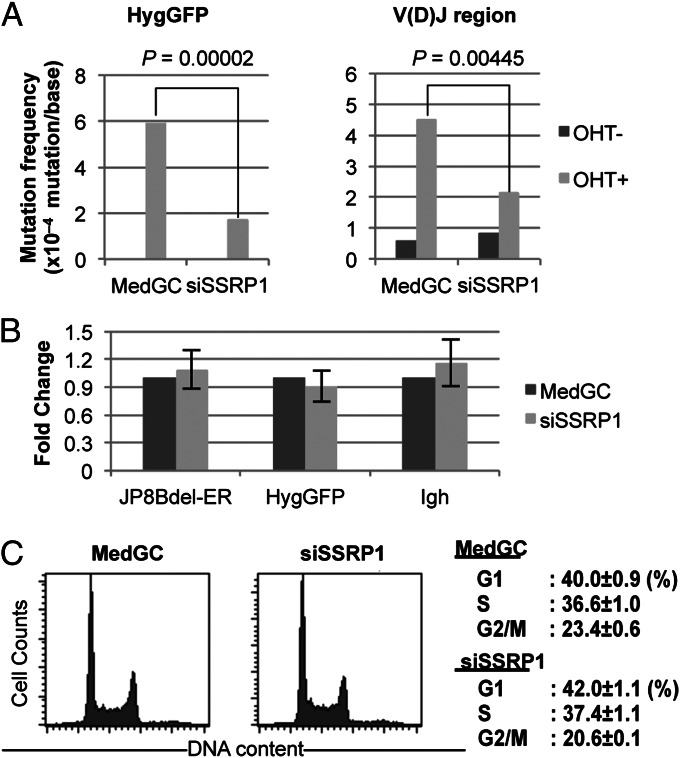
To verify the SHM reduction by SSRP1 knockdown, we analyzed mutations by DNA sequencing. The genomic DNA of the control medium GC (MedGC) and SSRP1#3 samples was purified after measuring the GFP signal-loss levels by FACS. The HygGFP transgene and V(D)J region were amplified by PCR. Sequencing of these regions showed that SSRP1 knockdown reduced the mutation frequency on the HygGFP gene to nearly one-third the level of the control (MedGC) (Fig. 2A, Left). The V(D)J region of the Igh locus was also significantly less mutated under the SSRP1-depleted condition (Fig. 2A, Right). Note that, in the V(D)J region, OHT-independent mutations were observed in both the depleted and control samples. These mutations may have been caused by leaky JP8Bdel-ER activity or by the endogenous AID activity of the BL2 cells. Mutation spectra did not reveal any particular bias for specific bases by SSRP1 depletion (Fig. S3 C and D).
Fig. 2.
SSRP1 depletion inhibits SHM. Cells treated with either anti-SSRP1#1 siRNA or MedGC were subjected to sequencing analysis. (A) Results of the mutation analysis for HygGFP and the V(D)J region. P values for the significance of reduction by SSRP1 knockdown are shown in each graph (Fisher’s exact test). The detailed results are shown in Fig. S3. (B) Transcript levels of JP8Bdel-ER, HygGFP, and Igh were unaffected by SSRP1 knockdown. Total RNAs purified 48 h after transfection were analyzed by qRT-PCR, and the transcript levels relative to the MedGC were calculated. Data shown are the mean and SD from three independent experiments. (C) Cell-cycle distribution was unaltered by SSRP1 knockdown during AID activation. (Left) Cell-cycle histograms of representative samples. (Right) Fractions of cells in the G1, S, and G2/M of triplicated samples. Values are the mean and SD.
To exclude the possibility that SSRP1 knockdown reduced SHM by inhibiting transcription, we assessed the levels of the HygGFP, Igh, and JP8Bdel-ER transcripts by qRT-PCR and found that the expression levels of these genes were not affected by SSRP1 knockdown (Fig. 2B). In addition, we analyzed the cell-cycle distribution of control and SSRP1-depleted BL2 cells by propidium iodide staining. The cell-cycle distribution of the BL2 cells 48 h after siRNA transfection was essentially identical between the two samples (Fig. 2C), excluding the possibility that an altered cell cycle was responsible for the SHM reduction. Taken together, these data show that FACT is a transcription elongation factor required for SHM.
Accumulation of FACT on the SHM Target Regions of Igh.
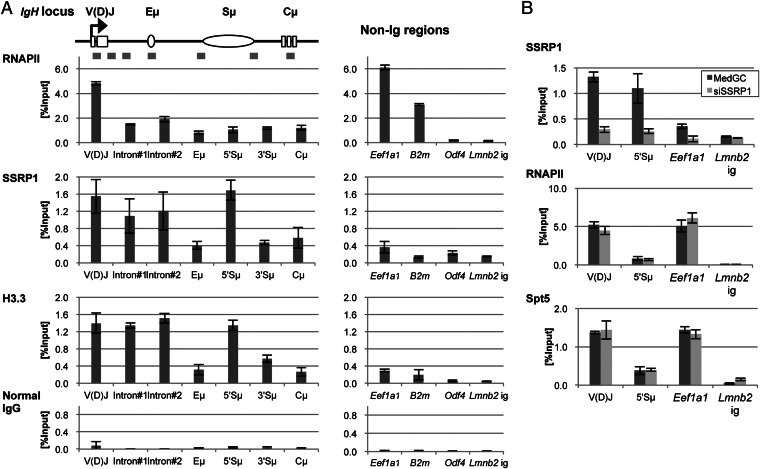
It is well known that the V(D)J and 5′Sµ regions are the primary targets of SHM. Even though SHM occurs in several non-Ig loci, their mutation rates are much lower than that of the V(D)J region (23), suggesting that the SHM targets in the Ig genes are specifically enriched with SHM-promoting factor(s). To test whether FACT is concentrated at the V(D)J and 5′Sµ regions, we performed chromatin immunoprecipitation (ChIP) assays using antibodies against SSRP1 and several related factors. The distribution of each factor was examined for seven regions across Igh. In addition, two highly transcriptionally active [promoters for eukaryotic translation elongation factor 1 alpha 1 (Eef1a1) and beta-2 microglobulin (B2M)] and two inactive [the outer dense fiber of sperm tails 4 (ODF4) promoter and the intergenic region close to the lamin B2 (Lmnb2) locus] nonimmunoglobulin (non-Ig) regions were examined for comparison.
We found that SSRP1 was specifically concentrated on the SHM target regions of Igh. Strong SSRP1 binding was observed from the 5′ side of the V(D)J region to the 5′ side of Sµ region, with disappearance at the Eμ intronic enhancer region (Fig. 3A, Left). SSRP1 binding decreased from the 3′ side of the Sµ region and was further reduced at the Cμ constant region. This pattern of SSRP1 distribution was strikingly similar to the known profile of hypermutation distribution on Igh (24). Moreover, the SSRP1 occupancy on transcriptionally active non-Ig loci was much lower than that on Igh, even though all three loci had strong RNAPII occupancy (Fig. 3A, Left and Right), suggesting a specific association of FACT enrichment with SHM induction on Igh. Note that the ChIP results with normal IgG showed the background signal levels of each tested region (Fig. 3A, bottom graphs).
Fig. 3.
Enriched FACT occupancy in the SHM target regions of Igh with histone H3.3 in JP8Bdel-ER BL2 cells. (A) ChIP analysis for Igh and non-Ig regions by qPCR. Schematic of the positions of PCR amplicons on Igh used for the ChIP assay is shown at Top. The occupancies of RNAPII, SSRP1, H3.3, and normal IgG were calculated as % Input and are shown in graphs for each factor. Data are the mean and SD of three independent experiments. (B) ChIP analysis with SSRP1-knocked down cells. Forty-eight hours after the transfection of MedGC control or anti-SSRP1#1 stealth siRNA, the cells were subjected to a ChIP assay. The occupancies of SSRP1, RNAPII, and Spt5 on the indicated regions were then measured by qPCR. Data are the mean and SD of three independent experiments. The primers and antibodies used for the ChIP assay are shown in Tables S6 and S7, respectively.
It has been proposed that Spt5, the component of another elongation factor DSIF, promotes SHM (9). To test whether SSRP1 knockdown affects Spt5 loading, we tested the Spt5 occupancy with SSRP1-depleted cells. We confirmed that 48 h of siRNA treatment dramatically decreased SSRP1’s binding to the V(D)J, Sµ, and Eef1a1 regions (Fig. 3B, Top). The ChIP signal at the Lmnb2 intergenic region was unchanged by SSRP1 knockdown, showing that this signal was background. Under this condition, we found that the Spt5 occupancy remained intact at all loci tested (Fig. 3B, Bottom). Similarly, the RNAPII occupancy was unaffected by SSRP1 depletion (Fig. 3B, Middle). As the Spt5 occupancy was proportional to the RNAPII occupancy, Spt5 appears to be more generally distributed than FACT.
In summary, these data revealed that FACT is specifically enriched on SHM target regions in Igh and promotes SHM independently from Spt5.
Similar Distribution of Histone H3.3 and FACT.
FACT is proposed to be involved in exchanging nucleosomal histones during transcription elongation (15). To determine whether the genomic regions with high FACT occupancy also had a high histone-exchange rate, we examined the occupancy of the histone H3 variant H3.3, the marker for replication-independent histone turnover. As shown in Fig. 3A, we found that the distribution of histone H3.3 was similar to that of FACT, suggesting that the nucleosomes in FACT-enriched regions are rapidly exchanged.
To confirm the association of FACT and H3.3 enrichment in the Ig genes in primary B cells, we performed a ChIP assay with mouse spleen cells (Fig. S4). Consistent with the data in human BL2 cells, we detected SSRP1 and H3.3 enrichment in the JH4 segment and 5′Sµ region in the Igh locus as well as in the Jκ5 segment in the kappa light chain locus. In this experiment, similar to H3.3 and FACT, the H3K4me3 modification was enriched on the SHM target regions in Ig genes, but it was also detected strongly in the non-Ig loci, showing that H3.3 and FACT are more specific than H3K4me3 for the Ig genes.
Relationship Between H3.3 Deposition and FACT Enrichment.
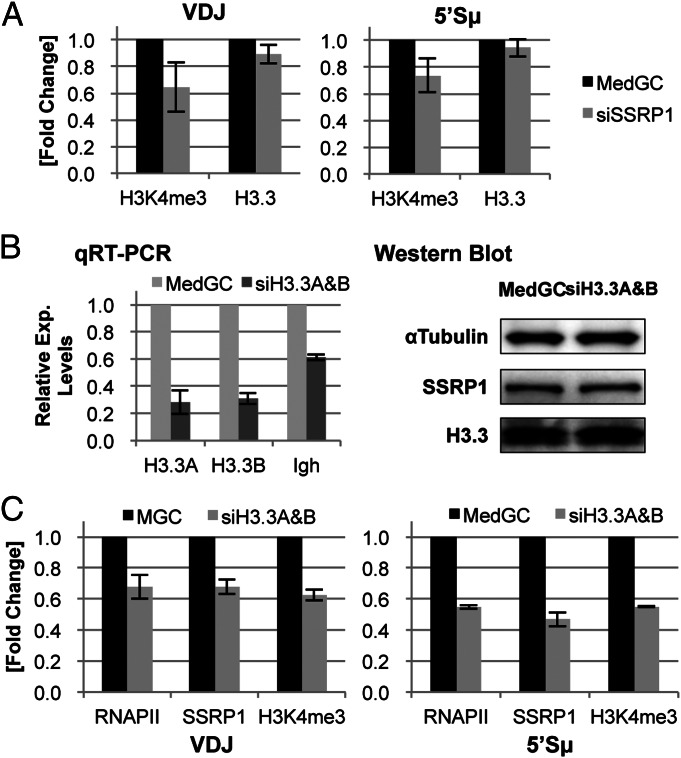
The concordant enrichment patterns of FACT and histone H3.3 suggest a possible relationship between the high histone-exchange rate and SHM targeting in the Ig genes. Because FACT has been reported to promote H3.3 deposition in Drosophila (25), we examined whether FACT regulates H3.3 deposition in the FACT-enriched regions. As shown in Fig. 4A, the SSRP1 knockdown did not alter the H3.3 deposition levels in the V(D)J and Sµ regions, but it reduced the H3K4me3 occupancy, in agreement with our previous findings in SSRP1-depleted murine B lymphoma cells (16), indicating that FACT is not required for H3.3 deposition into Igh chromatin.
Fig. 4.
Relationship between FACT enrichment and histone H3.3 deposition. (A) Effect of SSRP1 depletion on the H3K4me3 and H3.3 occupancies. The siSSRP1#1 oligo was used for SSRP1 knockdown, and the relative occupancy of H3K4me3 and H3.3 were analyzed by q-PCR. (B) H3.3 double knockdown. (Left) The relative expression level of the indicated transcripts was analyzed by qRT-PCR. (Right) Western blot of the control and double-knockdown cells. Whole-cell extracts were prepared from the same cell culture used for the qRT-PCR at Left and subjected to Western blotting with the indicated antibodies. (C) Relative occupancy levels of RNAPII, SSRP1, and H3K4me3 by H3.3 double knockdown at the V(D)J and 5′Sµ regions. In the experiments in A to C, siRNA was transfected 48 h before the analysis, and the values shown are the mean and SD of three independent experiments.
We then examined the alternative possibility that the FACT enrichment is affected by H3.3 deposition. Because histone H3.3 is encoded by two independent genes in distinct sequences in the human genome (H3.3A and -B), we introduced two separate siRNA oligonucleotides into cells to deplete both transcripts and found that the H3.3A and H3.3B transcripts were successfully depleted (Fig. 4B, Left). Notably, the H3.3 double knockdown reduced the Igh transcript by nearly half, indicating that, unlike FACT depletion, H3.3 depletion inhibited Igh transcription. Unexpectedly, this siRNA treatment did not reduce the H3.3 protein, indicating that the half-life of H3.3 protein is too long to cause a visible reduction in Western blot (Fig. 4B, Right). The SSRP1 protein expression was not affected either. Interestingly, the ChIP assay showed that the H3.3 double knockdown nonetheless reduced the occupancies of SSRP1 and H3K4me3 in the V(D)J and Sµ regions (Fig. 4C). The RNAPII occupancy in these regions was also similarly reduced, consistent with the reduced Igh transcript level. These results suggest that newly synthesized H3.3 protein has an important role in Igh transcription. It is therefore not clear whether the H3.3 deposition at the SHM target regions directly affects FACT enrichment.
Correlation Between Chromatin Marks and SHM Efficiency.
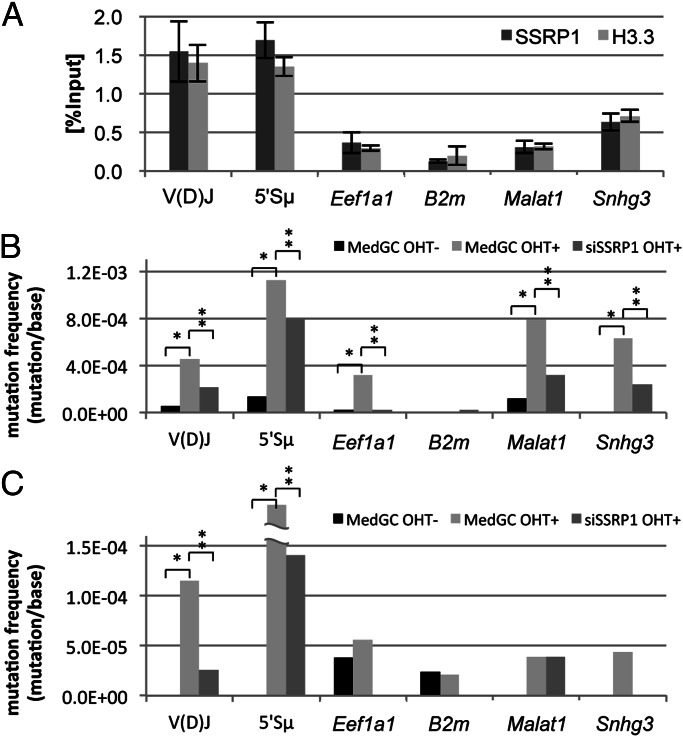
In BL2 cells, the non-Ig genes metastasis associated lung adenocarcinoma transcript 1 (Malat1) and small nucleolar RNA host gene 3 (Snhg3) are mutated at a similar level as the V(D)J region by the JP8Bdel-ER AID transgene (5). To evaluate the quantitative correlation between chromatin marking by FACT and H3.3 enrichment and SHM, we examined the FACT and H3.3 occupancies in these non-Ig SHM target regions. As shown in Fig. 5A, FACT occupancy at the mutable regions of Malat1 and Snhg3 was detected, but it was much lower than that at the V(D)J and 5′Sµ regions, and similar to the level at Eef1a1. The H3.3 occupancy in Malat1 and Snhg3 was completely proportional to the FACT occupancy.
Fig. 5.
Comparison between the FACT occupancy level and mutation rate. (A) FACT and H3.3 occupancies in the indicated regions. Data except for the Malat1 and Snhg3 are the same as the data shown in Fig. 3A. Data are the mean and SD of three independent experiments. (B and C) Mutation frequencies of the indicated regions in JP8Bdel-ER (B) and AID-ER BL2 cells (C) with or without OHT treatment. Note that the data of the V(D)J mutations in B are the same as shown in Fig. 2A. The detailed information for the sequencing analysis is shown in Table S1. *P < 0.05 compared with MedGC OHT−; **P < 0.05 compared with MedGC OHT+ (Fisher’s exact test).
SHM was significantly induced by JP8Bdel-ER in Malat1 and Snhg3, to a level comparable to or even stronger than in the V(D)J region (Fig. 5B). The highest mutation frequency was observed in the 5′Sµ region. SHM induction was also found in Eef1a1, but not in the B2m promoter-proximal region, where the FACT and H3.3 depositions were at background levels, confirming that SHM requires these chromatin marks. The strong transcriptional activity may make the Eef1a1 gene a preferred target of SHM despite the weak FACT loading. Importantly, all of the detected SHM induction was significantly reduced by SSRP1 depletion. The RNAPII occupancy at the SHM target regions of Malat1 and Snhg3 was not changed by SSRP1 knockdown (Fig. S5B). These data suggest that there is a good correlation between the FACT occupancy and SHM by JP8Bdel-ER.
Several studies have suggested that the enhanced SHM activity of JP8Bdel may broaden the target specificity of SHM (26, 27). We therefore performed the same experiment with the wild-type AID transgene fused with the estrogen-binding domain (AID-ER) (Fig. 5C). Although much weaker than by JP8Bdel-ER, we found clear SHM induction in the V(D)J and 5′Sµ regions by AID-ER. On the other hand, the non-Ig genes SHM-targeted by JP8Bdel-ER were not detectably mutated by AID-ER. In addition, we again observed significant SHM reduction by SSRP1 knockdown in the V(D)J and 5′Sµ regions. The distribution of SSRP1 and H3.3 was found unchanged in AID-ER BL2 cells (Fig. S5A). Thus, the AID-ER-induced SHM was limited to the chromatin regions with high deposition of both FACT and H3.3. Taken together, our findings indicated that there is a quantitative correlation between FACT and H3.3 chromatin marking with SHM targeting.
Discussion
Role of FACT-Mediated Histone Exchange in SHM.
The present study uncovered the importance of site-specific enrichment with the FACT complex and histone H3.3 in SHM, suggesting that transcription-coupled histone exchanging activity is required in this process. By comparison with non-Ig genes bearing similar RNAPII occupancy, the magnitude of the FACT recruitment on the V(D)J and 5′Sµ regions appears to be much greater than that required to maintain basic transcription. The FACT enrichment thus seems to be a locus-dependent phenomenon. The FACT enrichment was found to be associated with histone H3.3, the replication-independent histone turnover marker. Transcription-coupled histone exchange consists of histone turnover and recycling (28). The transcription-coupled turnover uses chaperones like histone cell cycle regulation defective homolog A (HIRA), which is recruited to a transcriptionally active region and deposits a histone dimer composed of H3.3 and H4 (29). In addition to turnover, nucleosomes in transcribed regions are subjected to the histone recycling activity of transcription elongation factors such as FACT and Spt6. These factors disassemble nucleosomes to help the elongation of RNAPII and reassemble them after passage of the polymerase (10). Thus, the enrichment of both FACT and H3.3 in the V(D)J and 5′Sµ regions indicates that these regions are subjected to frequent histone exchange in terms of both the recycling and turnover of histones.
How can rapid histone exchange promote SHM? One plausible explanation is that nucleosomes act as barriers against SHM by limiting the accessibility of DNA to the cleaving enzyme. Indeed, Kodgire et al. reported that insertion of the nucleosome positioning sequence, which is tightly folded by the nucleosome, into the variable region of the Ig light chain gene reduces SHM in and around the inserted region (30). Another explanation involves the fact that unwinding DNA from nucleosome produces one negative supercoil (31). It is known that excessive negative supercoil behind the transcription elongation complex tends to form irregular DNA structures called non-B DNA (3, 32), which is proposed to be the substrate for DNA-cleaving enzymes such as DNA topoisomerase 1 (6). Such cleavage can induce SHM during repair by error-prone polymerases (33). Therefore, we speculate that the more frequently DNA is unwound from the nucleosome, the higher the chance of the DNA being hypermutated. We previously reported that H3K4me3 serves as a marker for SHM and CSR (5, 16). In CSR, FACT contributes to the DNA break by promoting the trimethylation of H3K4 (16). Regarding SHM, we reported that H3K4me3 is enriched in SHM target regions (5). However, as H3K4me3 is a common modification on active chromatin, the targeting mechanism for CSR and SHM cannot be fully explained by the level of H3K4me3. We found that normal AID can induce SHM in the Igh locus, which is marked by H3K4me3 as well as FACT and H3.3. On the other hand, certain non-Ig regions without strong FACT and H3.3 deposition were mutated only by the hyperactive AID mutant. We propose here that, in addition to H4K4me3, rapid histone exchange promotes SHM by increasing the accessibility of the genomic region and the competency of the DNA structure for DNA cleavage induction during SHM. The augmented AID activity may not require the rapid histone exchange at the target and may broaden the range of SHM targets to other H3K4me3-rich regions.
Mechanism of the FACT and H3.3 Deposition in SHM Targets.
Although the functional relationship between FACT and H3.3 is not yet clearly understood, these two factors appear to accumulate at the same loci. A genome-wide study showed that FACT tends to accumulate at intragenic enhancer-like regions together with H3.3 in Drosophila cells (34). Furthermore, our ChIP analysis revealed that the distribution of SSRP1 and H3.3 on various genomic loci was highly overlapping. Another Drosophila study showed that FACT promotes H3.3 deposition at the white gene enhancer after being recruited by the GAGA transcription factor (25). We did not observe a change in H3.3 accumulation by FACT knockdown. Although H3.3 knockdown reduced the FACT deposition, it also inhibited Igh transcription. We therefore could not conclude that the recruitment of FACT was directly regulated by H3.3. It is likely that the loading mechanism of FACT and H3.3 differs between Drosophila and mammals and possibly between different loci. It has been proposed that H3.3 is generally deposited to transcribed regions by the H3.3-specific histone chaperone HIRA (29, 35). However, we previously showed that HIRA knockdown does not affect CSR (8). It seems likely that site-specific H3.3 accumulation, such as that we observed at the V(D)J and 5′Sµ regions, is regulated by another mechanism(s) (36). Further analysis is required to elucidate the mechanism by which FACT and H3.3 accumulate together at the SHM target regions in Ig genes.
Are Other Elongation Factors Required for CSR and SHM?
In the RNAi screening, we did not find SHM-promoting activity of elongation factors that were previously reported to be involved in SHM and/or CSR. A factor with CSR-promoting activity would not necessarily have SHM-promoting activity because CSR requires additional recombination activity. In addition, hypermutation of the reporter transgene may not fully mimic the SHM at the V(D)J region, as we reported previously that Spt6 reduced SHM of the V(D)J region but not of the HygGFP transgene (8, 12). Moreover, reduction of the GFP reporter expression by elongation factor knockdown could perturb the SHM assay results. Thus, we do not exclude the possibility that we have overlooked elongation factors that promote SHM but also promote HygGFP expression. Indeed, in Spt5-depleted cells, GFP-negative population was observed even under AID-inactive condition, indicating the transcription defect in HygGFP (Fig. S1). Moreover, sequencing analysis revealed that Spt5 knockdown reduces the mutation frequency in HygGFP (Table S2). Our previous finding indicate that Spt5 is required for DNA break induction in addition to DNA repair during CSR (11). Therefore, we assume that Spt5 is also required for SHM. However, the roles in SHM may be different between Spt5 and FACT, as their distributions are different from each other. In the Drosophila heatshock protein 70 gene, FACT accumulates more in the distal transcribed region whereas Spt5 is enriched at the promoter-proximal region in association with RNAPII (37). In genome-wide studies, Spt5 occupancy appears to be proportional to the RNAPII occupancy (9, 17) whereas FACT enrichment is not necessarily associated with RNAPII (34). In the present study, we show that the site-specific accumulation of FACT and H3.3 at the SHM target regions in Ig genes may at least partly explain the mechanism of SHM targeting.
In summary, we found that the FACT complex promotes SHM and is enriched at the V(D)J and 5′Sµ regions. We propose that the rapid histone-exchange status, characterized as FACT-high and H3.3-high, makes a region a sensitive substrate for SHM. Future study will elucidate the detailed molecular mechanism for this enrichment and its roles in antibody diversification processes.
Materials and Methods
siRNA Oligonucleotide Transfection.
Stealth RNAi oligo (Invitrogen), a chemically modified short double-stranded RNA, was introduced into BL2 cells by the Nucleofector 96-well electroporation system (Lonza) with the DS-150 program. As a negative control, Stealth siRNA Negative Control Med GC (Invitrogen) was used. The Stealth siRNAs used in this study are listed in Table S3.
SHM Reporter Assay.
The SHM assay was initiated by adding 1 mM 4-hydroxyl-Tamoxifen (OHT) to cultured BL2 JP8Bdel-ER HygGFP cells. Twenty-four hours later, the OHT was removed by washing, and the cells were incubated for 48 h. The cells were then subjected to flow cytometric analysis with a FACSCalibur (Becton Dickinson) to measure the GFP-negative cell population.
Mutation Analysis by Sequencing.
Three independent genomic DNA samples were pooled, and the target genomic regions were then amplified with PrimeSTAR polymerase (TaKaRa). The primer sequences are shown in Table S5. The PCR products were cloned into a pGEM-T easy vector (Promega). The plasmid DNA was isolated and sequenced with an ABI3130xl Genetic Analyzer (Applied Biosystems). The sequence data were analyzed with Sequencher software (Gene Codes).
Other materials and methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. H. Nagaoka for developing the GFP-based SHM reporter assay system in JP8Bdel-ER BL2 cells; Ms. Y. Shiraki for preparing the manuscript; and members of the T.H. laboratory for critical reading of the manuscript. The anti-Spt5 antisera and anti-SSRP1 antibody were gifts from Dr. H. Handa and Biolegend Inc., respectively. This work was supported by Grant-in-Aid for Specially Promoted Research 17002015 (to T.H.), Grant-in-Aid for Scientific Research (C) 24590352 (to N.A.B.), and the Global Centers of Excellence program (M.A.) of the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305859110/-/DCSupplemental.
References
- 1.Kinoshita K, Honjo T. Linking class-switch recombination with somatic hypermutation. Nat Rev Mol Cell Biol. 2001;2(7):493–503. doi: 10.1038/35080033. [DOI] [PubMed] [Google Scholar]
- 2.Okazaki IM, Kotani A, Honjo T. Role of AID in tumorigenesis. Adv Immunol. 2007;94:245–273. doi: 10.1016/S0065-2776(06)94008-5. [DOI] [PubMed] [Google Scholar]
- 3.Begum NA, Honjo T. Evolutionary comparison of the mechanism of DNA cleavage with respect to immune diversity and genomic instability. Biochemistry. 2012;51(26):5243–5256. doi: 10.1021/bi3005895. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4(7):541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 5.Kato L, et al. Nonimmunoglobulin target loci of activation-induced cytidine deaminase (AID) share unique features with immunoglobulin genes. Proc Natl Acad Sci USA. 2012;109(7):2479–2484. doi: 10.1073/pnas.1120791109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi M, et al. AID-induced decrease in topoisomerase 1 induces DNA structural alteration and DNA cleavage for class switch recombination. Proc Natl Acad Sci USA. 2009;106(52):22375–22380. doi: 10.1073/pnas.0911879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi M, et al. Decrease in topoisomerase I is responsible for activation-induced cytidine deaminase (AID)-dependent somatic hypermutation. Proc Natl Acad Sci USA. 2011;108(48):19305–19310. doi: 10.1073/pnas.1114522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begum NA, Stanlie A, Nakata M, Akiyama H, Honjo T. The histone chaperone Spt6 is required for activation-induced cytidine deaminase target determination through H3K4me3 regulation. J Biol Chem. 2012;287(39):32415–32429. doi: 10.1074/jbc.M112.351569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavri R, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143(1):122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: The short and long of it. Genes Dev. 2004;18(20):2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 11.Stanlie A, Begum NA, Akiyama H, Honjo T. The DSIF subunits Spt4 and Spt5 have distinct roles at various phases of immunoglobulin class switch recombination. PLoS Genet. 2012;8(4):e1002675. doi: 10.1371/journal.pgen.1002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okazaki IM, et al. Histone chaperone Spt6 is required for class switch recombination but not somatic hypermutation. Proc Natl Acad Sci USA. 2011;108(19):7920–7925. doi: 10.1073/pnas.1104423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willmann KL, et al. A role for the RNA pol II-associated PAF complex in AID-induced immune diversification. J Exp Med. 2012;209(11):2099–2111. doi: 10.1084/jem.20112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92(1):105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 15.Belotserkovskaya R, et al. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301(5636):1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 16.Stanlie A, Aida M, Muramatsu M, Honjo T, Begum NA. Histone3 lysine4 trimethylation regulated by the facilitates chromatin transcription complex is critical for DNA cleavage in class switch recombination. Proc Natl Acad Sci USA. 2010;107(51):22190–22195. doi: 10.1073/pnas.1016923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130(1):77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, et al. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 2009;23(23):2765–2777. doi: 10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamane A, et al. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12(1):62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito S, et al. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci USA. 2004;101(7):1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshikawa K, et al. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 2002;296(5575):2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- 23.Liu M, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451(7180):841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 24.Martin A, Scharff MD. AID and mismatch repair in antibody diversification. Nat Rev Immunol. 2002;2(8):605–614. doi: 10.1038/nri858. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama T, Nishioka K, Dong YX, Shimojima T, Hirose S. Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev. 2007;21(5):552–561. doi: 10.1101/gad.1503407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doi T, et al. The C-terminal region of activation-induced cytidine deaminase is responsible for a recombination function other than DNA cleavage in class switch recombination. Proc Natl Acad Sci USA. 2009;106(8):2758–2763. doi: 10.1073/pnas.0813253106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robbiani DF, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36(4):631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elsaesser SJ, Goldberg AD, Allis CD. New functions for an old variant: No substitute for histone H3.3. Curr Opin Genet Dev. 2010;20(2):110–117. doi: 10.1016/j.gde.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116(1):51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 30.Kodgire P, Mukkawar P, North JA, Poirier MG, Storb U. Nucleosome stability dramatically impacts the targeting of somatic hypermutation. Mol Cell Biol. 2012;32(10):2030–2040. doi: 10.1128/MCB.06722-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Germond JE, Hirt B, Oudet P, Gross-Bellark M, Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci USA. 1975;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. J Biol Chem. 2004;279(46):47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 33.Seki M, Gearhart PJ, Wood RD. DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 2005;6(12):1143–1148. doi: 10.1038/sj.embor.7400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471(7339):480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140(5):678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res. 2011;21(3):421–434. doi: 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saunders A, et al. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301(5636):1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.