Abstract
The c-rings of ATP synthases consist of individual c-subunits, all of which harbor a conserved motif of repetitive glycine residues (GxGxGxG) important for tight transmembrane α-helix packing. The c-ring stoichiometry determines the number of ions transferred during enzyme operation and has a direct impact on the ion-to-ATP ratio, a cornerstone parameter of cell bioenergetics. In the extreme alkaliphile Bacillus pseudofirmus OF4, the glycine motif is replaced by AxAxAxA. We performed a structural study on two mutants with alanine-to-glycine changes using atomic force microscopy and X-ray crystallography, and found that mutants form smaller c12 rings compared with the WT c13. The molar growth yields of B. pseudofirmus OF4 cells on malate further revealed that the c12 mutants have a considerably reduced capacity to grow on limiting malate at high pH. Our results demonstrate that the mutant ATP synthases with either c12 or c13 can support ATP synthesis, and also underscore the critical importance of an alanine motif with c13 ring stoichiometry for optimal growth at pH >10. The data indicate a direct connection between the precisely adapted ATP synthase c-ring stoichiometry and its ion-to-ATP ratio on cell physiology, and also demonstrate the bioenergetic challenges and evolutionary adaptation strategies of extremophiles.
Keywords: F1Fo-ATP synthase rotor, membrane protein complex
ATP synthases are conserved, membrane-anchored enzyme complexes that use a rotary-mechanical mechanism to produce ATP, the cell’s universal energy currency. The enzymes consist of two major subcomplexes, F1 (α3β3γδε) and Fo (ab2c8–15), which are intimately coupled and driven by the transmembrane electrochemical ion gradient to synthesize three ATP molecules per complete rotation of the rotor, one each in the three (β3) catalytic F1 sites (1–4). One part of the enzyme’s rotor cnγε, the c-ring (Fo rotor ring), is involved in shuttling ions from the outside to the inside of the cell across the membrane by reversible binding and release of ions (H+ or Na+). This converts the energy of the proton motive force (pmf) into kinetic energy that is ultimately required for ATP synthesis in F1.
The c-ring is an oligomeric assembly of c-subunit copies. Its stoichiometry was initially proposed to be adapted to cell environmental conditions (5), but later structural work pointed instead to a constant stoichiometry within one species but variable stoichiometry in different species, ranging from 8 to 15 (6, 7). In addition, several high-resolution structures of isolated rotor rings have demonstrated an overall conserved structural appearance and functionality (8). The c-ring has a cylindrical shape, a constriction in the middle, and a central pore (9, 10). Each c-subunit is able to coordinate one ion via a conserved carboxylate group from a glutamate or aspartate side chain. The number of ions translocated per 360-degree rotation is determined by the c-ring stoichiometry (cn); thus, the ion-to-ATP ratio, i, of the enzyme is i = cn/β3. This bioenergetic cornerstone parameter is known to vary from 2.7 to 5 (11).
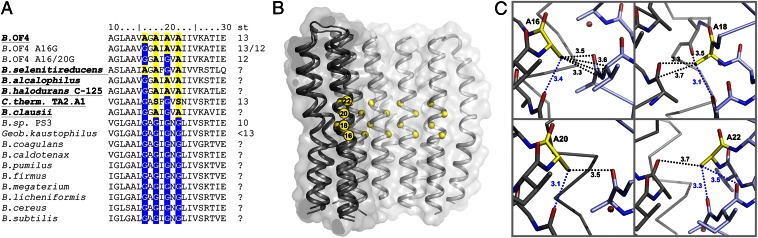
Alkaliphilic bacteria grow in environments that are largely devoid of protons and thus are confronted with a particular challenge with respect to cell energy metabolism (12, 13). The synthesis of ATP by the alkaliphile ATP synthase at high pH is challenged by the presence of a low pmf, because when the medium is highly alkaline, these bacteria must maintain a cytoplasmic pH significantly lower than the outside. This reversed chemical gradient of protons, in the opposite orientation of the gradient that powers bioenergetic work, reduces the total bulk pmf significantly. The effective pmf at the membrane surface may be larger than the bulk pmf (14–16), thus partially addressing the energetic challenge. However, alkaliphilic bacteria have also developed several adaptation strategies, which in combination support robust growth at high pH (12, 13, 17, 18). These adaptations include alkaliphile-specific features of the ATP synthase, including changes at the c-ring’s ion-binding site (19) as well as its size and stoichiometry (19–21). The N-terminal α-helix of the c-subunits contains a typical membrane protein-packing motif (22) of repetitive glycine residues (GxGxGxG) (Fig. 1A). This motif causes a tight packing of the c-ring’s inner ring of α-helices (23), essentially established by hydrophobic contacts at the c/c-subunit interfaces. The motif is altered in the family of alkaliphilic Bacillus species that grow at pH >10 (13). For example, in the alkaliphilic soil bacterium Bacillus pseudofirmus OF4, the complete motif consists of only alanines (AxAxAxA) (Fig. 1A). Mutants within this motif exhibit altered c-ring migration profiles, suggesting reduced c-subunit stoichiometry (24). Concurrent with this observation, it was recently structurally demonstrated that replacement of glycine with alanine can augment the c-ring stoichiometry (25).
Fig. 1.
The alanine motif (AxAxAxA) in the c13 ring of B. pseudofirmus OF4 ATP synthase. (A) Amino acid alignment of the alanine motif (highlighted in color) in selected c-subunits from neutralophilic and alkaliphilic (in bold) Bacillus species. c-ring stoichiometries (st) are indicated. (B) A c6-section of the c13 ring showing the location of the alanine motif on the inner, N-terminal helix. (C) Interactions (<3.7 Å, dashed lines) of A16, A18, A20, and A22 (yellow) with nearby amino acid residues (gray). The c-subunits are in different colors. Intra– and inter–c-subunit interactions are indicated by blue and black dashed lines, respectively. A16 shows the most complex interaction network with its neighborhood.
We hypothesized that the presence of the AxAxAxA in the B. pseudofirmus OF4 c-subunit is a crucial determinant of c-ring stoichiometry, and that it critically influences the cell’s capability to grow at alkaline pH >10. Thus, we performed a structural analysis using atomic force microscopy (AFM) and X-ray crystallography with purified ATP synthase c-rings to image individual mutant protein complexes with potentially altered stoichiometries (24). We also performed concomitant growth studies, the results of which underscore the importance of c-ring stoichiometry for the cell’s capacity for growth at high pH under ATP synthase-dependent conditions. Our data connect c-ring stoichiometries directly with cell bioenergetics and cellular adaptation to environmental challenges (i.e., growth at alkaline pH) using a paradigmatic example of an extreme alkaliphile, B. pseudofirmus OF4.
Results
Analysis of the AxAxAxA Motif in the B. pseudofirmus OF4 c-Ring.
A section through a c6 segment of the B. pseudofirmus OF4 c13 ring with marked Cα positions of the alanines within the alkaliphile-specific 16AxAxAxA22 stretch shows this motif as a notable feature and the tight c-subunit packing (Fig. 1B). Structurally (19), one of the motif's central elements is the hydrophobic interaction network of the alanine methyl side chains with several atoms from neighboring residues (Fig. 1C). Alanines 18, 20, and 22 (A18, A20, and A22) share between two and four interactions, whereas alanine 16 (A16) shares a total of five interaction partners. Within these, A16 has four partners on the adjacent c-subunit, compared with only one or two for the others. We hypothesized that a change of these alanine residues to glycines alters important c2-dimer contacts within the protein complex interaction network and causes steric rearrangements within the c/c-subunit packing. In particular, a disturbance of the interaction network seen at residue A16 would, by prediction (Fig. 1C), cause the greatest effect on the structure compared with other alanines of this packing motif.
In a previous site-directed mutagenesis study, we showed that selected alanine-to-glycine mutations, all of which included an A16G change, shifted the c-ring migration profiles on SDS gels (24, 26). To uncover structural evidence that these mutant B. pseudofirmus OF4 ATP synthases have altered c-ring stoichiometries, we analyzed their c-rings on the single protein complex level by AFM and at the c/c-subunit contacting interface by X-ray crystallography.
Purification and AFM of B. pseudofirmus OF4 Mutant c-Rings.
We examined four different B. pseudofirmus OF4 c-rings: WT (control), extended WT (extWT; see the next paragraph), and two mutants, extA16G and extA16/20G (24). For this, the WT B. pseudofirmus OF4 c13 ring was purified and densely reconstituted in monogalactosyldiacylglycerol (MGDG) vesicles. However, few small, quasi-2D crystalline lattices were obtained showing c-rings packed in different orientations (Fig. S1). Some areas showed only one orientation of the c-rings, whereas in very rare cases both periplasmic and cytoplasmic sides were visible (23, 27), but those vesicles were too small to permit closer AFM imaging. One of the sides contained a central mass and represents phospholipids bound in the middle c-ring pore at the periplasmic c-ring side (28). In all cases, the quality of the AFM topographs was not sufficient to unambiguously determine the stoichiometry of the rings from the unprocessed images, which is a prerequisite for determining a potentially heterogeneous distribution (25). The quality was hampered by the rather small size of the vesicles (∼100–200 nm) and rather weak packing of the c-rings.
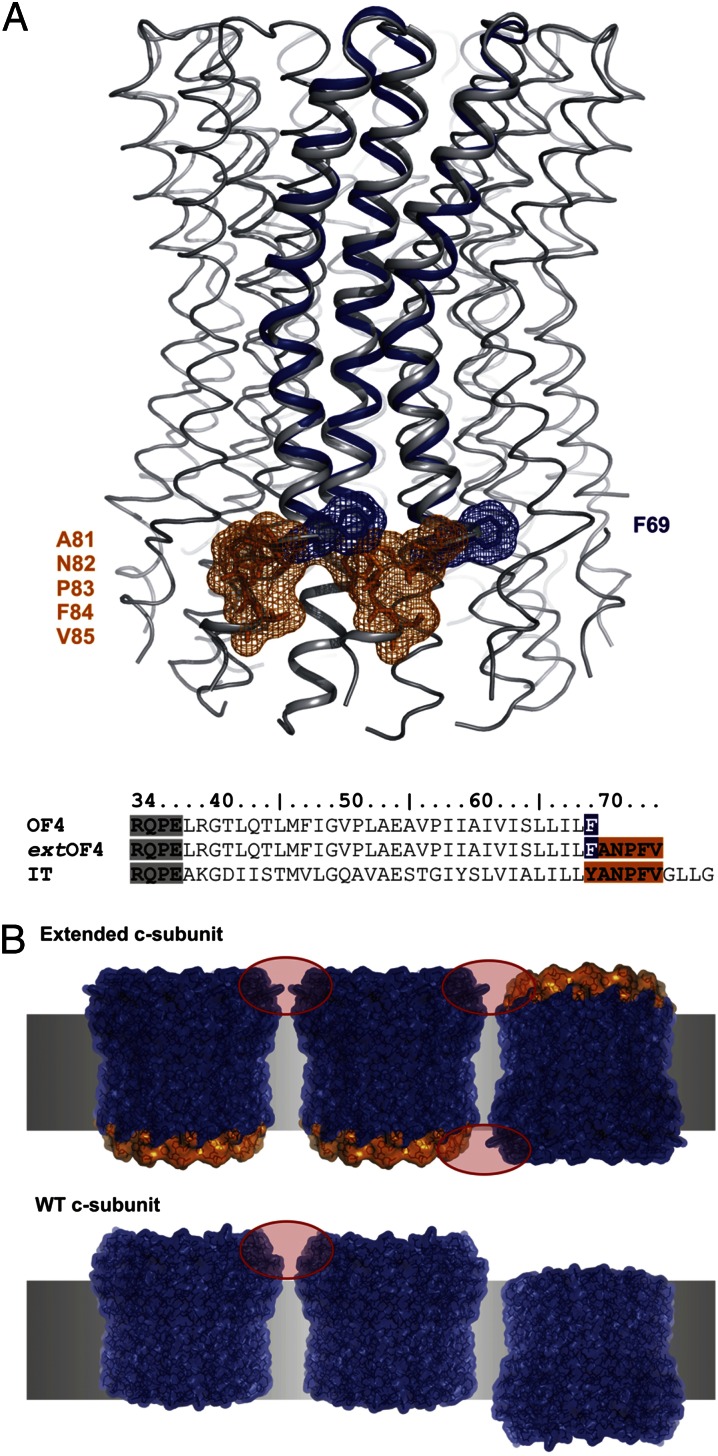
To improve the image quality, we aimed to optimize the c-ring/c-ring crystal contacts through a rational approach (Fig. 2, orange region). The Ilyobacter tartaricus c-subunit has an extension of nine amino acids relative to B. pseudofirmus OF4. Bearing the 2D crystal contacts of the I. tartaricus c11 ring in mind (23), we genetically engineered the c-subunit encoding atpE gene in the genome of B. pseudofirmus OF4 and extended its sequence by adding the coding sequence for five amino acids from the I. tartaricus atpE-terminal sequence (29), to finally obtain a c-subunit with an I. tartaricus C-terminal extension (ext) that provides more and better crystal contacts than the WT.
Fig. 2.
Two-dimensional crystal contact optimization by C-terminal extension of the B. pseudofirmus OF4 c-subunit. (A) Overlay of the I. tartaricus (IT) c-ring (gray) with the B. pseudofirmus OF4 (OF4) c-subunit (blue). The protein’s C-terminus (F69) is highlighted in blue. To optimize 2D crystal contacts, the C terminus was extended by five amino acids (highlighted in orange). The IT c-subunit, known to provide sufficient 2D crystal contacts (23), was used as a template. An amino acid alignment of the C-terminal transmembrane helix from IT with OF4 WT and OF4 extWT is given below. (B) Comparison of c-ring/c-ring interaction with and without a c-subunit C-terminal extension. Crystal contacts in 2D crystals are established by the soluble regions of the proteins; the extensions allow formation of new crystal contacts (red circles).
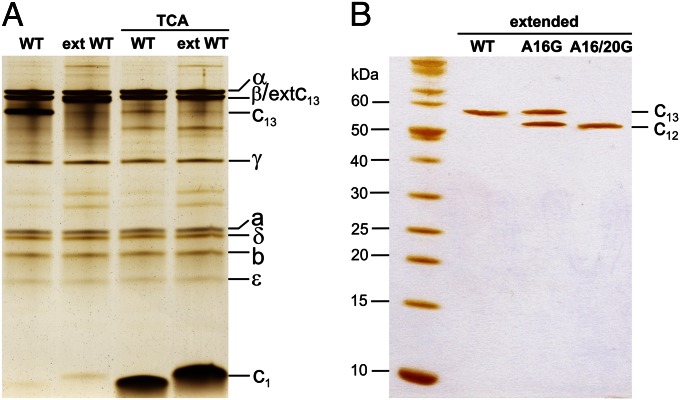
We next purified three different extended c-ring mutants—extWT, extA16G, and extA16/20G—from complete B. pseudofirmus OF4 F1Fo-ATP synthase, and compared their migration profiles using SDS/PAGE (Fig. 3). These constructs showed SDS-stable c-rings ∼50 kDa in size, which was independent of the pH (7.5 and 10.5) of the growth medium (Fig. S2). The stability with respect to high temperature (85 °C) and a range of pH values (2–11.5) was still remarkably high, as in the WT (Fig. S3). In contrast to the nonextended WT c-ring, the extWT c-ring ran slightly slower, owing to the C-terminal extension, but the WT c13 stoichiometry was retained in the extWT, as demonstrated by AFM (Fig. 4). The purified extA16G mutant showed two gel bands, one at the exact level of the extWT c13 ring and the other, slightly faster, below the extWT band (Fig. 3B). The faster-migrating band ran at a size expected for a c12 ring. These results suggest that the extA16G produces a mix of two SDS stable c-rings, c12 and c13, or it may make a c12 ring that sometimes has an additional c-monomer attached (30).
Fig. 3.
SDS/PAGE of purified B. pseudofirmus OF4 WT and alanine mutant ATP synthase and c-rings. (A) Isolated ATP synthases. F1Fo-ATP synthase with extended WT c-ring and WT c-ring before and after TCA treatment, which monomerizes the c-ring. The nonextended WT c13 and c1 (indicated) run faster than the extWT. (B) Isolated c-rings. The extWT c13 ring runs as a single band at ∼58 kDa; a molecular weight marker (in kDa) is on the left. The extA16G mutant shows two bands, one at the WT c13 level and the other at the c12 level. The double mutant extA16/20G runs as a single band, faster than extWT, at the c12 level. The samples were separated on 11% and 13.2% acrylamide gels in A and B, respectively, and stained with silver.
Fig. 4.
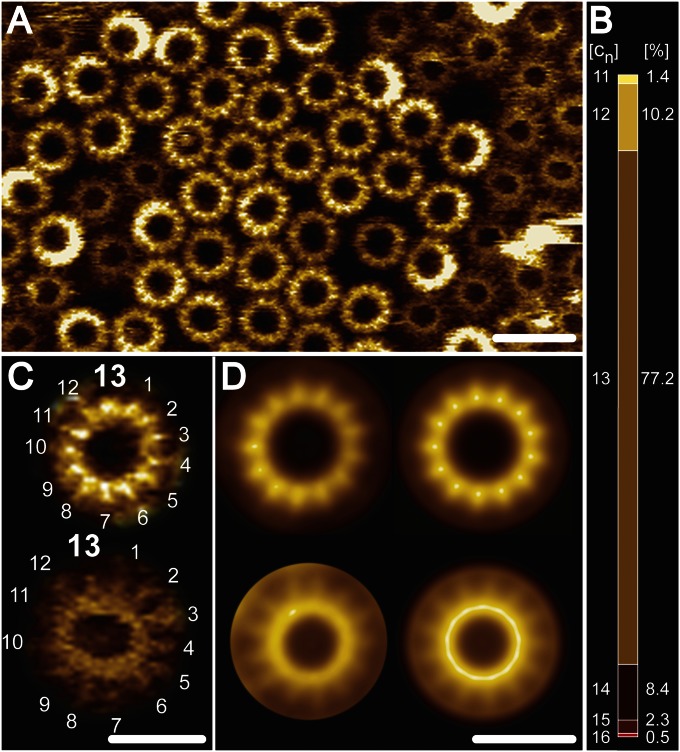
High-resolution AFM topographs of reconstituted B. pseudofirmus OF4 extWT c-rings. An isolated extWT B. pseudofirmus OF4 c13 ring sample was densely reconstituted in lipid vesicles and high-resolution topographic images were recorded in quasi 2D-crystalline regions. (A) Overview. Higher-protruding (lighter) and lower-protruding (darker) rings become visible, showing the cytoplasmic and periplasmic ends of the extWT c-rings, respectively (20, 23, 27). No central lipid plugs are visible (28). (B) extWT c-ring stoichiometry statistics ranging from c11 to c16 (n = 270), with a majority of 77.2% c13 stoichiometry. [cn] represents the stoichiometry, and [%] is the percentage of the c-rings. (C) Unprocessed images of cytoplasmic (Upper) and periplasmic (Lower) sides with countable 13 subunits. (D) Reference-free single-particle averages of the cytoplasmic (Upper; n = 350) and periplasmic (Lower; n = 390) sides of the c13 rings. Nonsymmetrized (Left) and symmetrized (Right) averages confirm 13-fold symmetry of the complex. The average diameter of the cytoplasmic side of the c13 ring is 6.7 ± 0.3 nm (n = 30). (Scale bars: 10 nm in A; 5 nm in C and D.)
In contrast to the extA16G mutant, the double mutant extA16/20G showed only the lower, putative c12 ring band in the c-oligomeric region of the SDS gel (Fig. 3B). Thus, the two AxAxAxA motif mutants contain c-rings, which are incorporated in the B. pseudofirmus OF4 ATP synthase and appear to contain an altered stoichiometry, that is, c12 instead of WT c13.
We next reconstituted the extWT and the mutants extA16G and extA16G/A20G in MGDG lipid bilayer vesicles and analyzed them by AFM. Compared with the nonextended WT (Fig. S1), all extended versions of the c-rings formed remarkably large and densely protein-packed vesicles up to 10 μm in size, which were more stable and suitable for topographic imaging. Figs. 4 and 5 show images of the extWT, extA16G, and extA16/20G c-rings. In extWT, higher and lower protruding c-rings were visible (Fig. 4A), corresponding to the cytoplasmic- and periplasmic-facing sides, respectively (23, 27, 28). This interpretation is also supported by the fact that the N- and C-termini of Bacillus c-subunits are particularly short compared with those of other species (19, 20). In contrast to the WT c13, no central mass was observed in these c-rings. In addition, the single c-subunits were well resolved because of better packing. They were clearly visible and countable directly by eye in the unprocessed images. This finding demonstrates the positive effect of the C-terminal extension for AFM imaging. Moreover, the lower protruding rings were highest at the inner rings’ rims and lower at their outer rims, another effect of the C-terminal extension (Fig. 2).
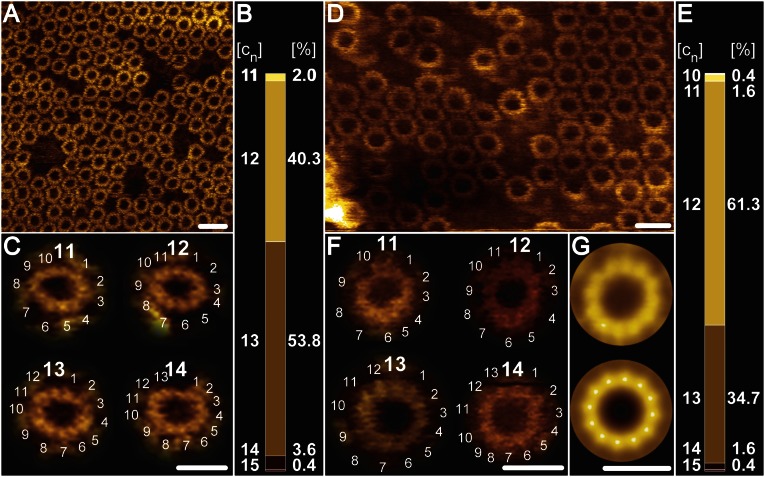
Fig. 5.
High-resolution AFM topographs of reconstituted B. pseudofirmus OF4 extA16G and extA16/20G c-ring mutants. Each of the two mutant B. pseudofirmus OF4 c-ring samples was densely reconstituted in lipid vesicles, and high-resolution topographic images were recorded in quasi 2D-crystalline regions. (A) Overview: extA16G c-rings viewed from the cytoplasmic side (high-protruding c-rings). (B) extA16G stoichiometry statistics ranging from c11 to c15 (n = 253) with a majority of c13 (53.8%) and c12 (40.3%) rings. (C) Gallery showing extA16G c-rings of variable stoichiometries. (D) Overview: extA16/20G c-rings viewed from the cytoplasmic side (high-protruding c-rings). (E) extA16/20G stoichiometry statistics ranging from c10 to c15 (n = 248), with a majority of c12 (61.3%) and c12 (34.7%) rings. (F) Gallery showing extA16/20G c-rings of variable stoichiometries. (G) Nonsymmetrized (Upper) and symmetrized (Lower) reference-free single-particle averages of the extA16/20G mutant, cytoplasmic side (n = 100). The average diameters of the c13 rings are 6.6 ± 0.2 nm (extA16G; n = 30) and 6.3 ± 0.3 nm (extA16/20G; n = 30), respectively. [cn] represents stoichiometry and [%] percentage of the c-rings. (Scale bars: 10 nm in A and D; 5 nm in C, F, and G.)
The high resolution of the extWT c-ring facilitated evaluation of the c-ring stoichiometries from 270 individual c-rings (Fig. 4B). Among these, c13 was the most abundant (77.2%), but other c-ring stoichiometries (c11–c16) were apparent as well. In the extA16G and extA16/20G mutants (Fig. 5), the higher protruding (cytoplasmic side) c-rings were clearly visible and the c-subunits were countable, but no c-rings closer to the membrane level (extracellular side) could be imaged at a sufficiently high resolution to view the individual c-subunits in the rings. In contrast to the extWT and extA16G, the extA16/20G mutant had a protruding phospholipid mass present on the extracellular side of the ring. Despite some lateral distortions during AFM imaging of the mutated c-rings, individual c-subunits remained distinguishable on the cytoplasmic c-ring side. The stoichiometries of the extA16G mutant varied between 11 and 15 c-subunits, with a preponderance of c13 and c12 rings (53.8% and 40.3%, respectively) (Fig. 5B). This distribution detected by AFM analysis closely mirrored that inferred by the SDS/PAGE migration profile. The extA16/20G mutant c-rings also varied between 10 and 15 c-subunits, with a preponderance of c12 and c13 rings (61.3% and 34.7%, respectively; Fig. 5E), but only the c12 species was visible on the gel, implying that this was the most stable species of this mutant (Fig. 3B). Taken together, the AFM data reveal variable stoichiometry in all extended c-rings, which could not be visualized by SDS/PAGE because some of the rings are either susceptible to the SDS treatment or below the detection limits of the silver-stained SDS gel.
Structure of the A16/20G c12 Ring from B. pseudofirmus OF4 Determined by X-Ray Crystallography.
To learn more about the structural factors that led to the change in stoichiometry from c13 to c12, we solved the A16/20G c12 ring by X-ray crystallography at 4.1 Å (Table S1). Despite the low resolution of the data, the electron density map allowed us to unambiguously trace the Cα backbone of the subunits and some of the larger side chains (Fig. 6). The model thus obtained confirmed that the A16/20G mutant forms a c12 ring. We then measured a set of distances in the mutant c12 and WT c13 rings (Fig. S4). The chosen residues were 16 and 20, describing the c/c-subunit distance at the inner ring of α-helices; residue 56, as a c/c-subunit distance marker on the outer ring of α-helices; and residues 16 and 56, as the (intra) c-subunit distance marker from the inner helix to the outer helix. Whereas the distances 16–56 and 56–56 remained very close to the WT c13 distance, the inner ring of the α-helices was more tightly packed in the mutant c12, resulting in significantly decreased 16–16 and 20–20 c/c-subunit distances (Fig. S4). This effect was even more evident when comparing the circumferences of a theoretically calculated WT c12 ring and the experimentally obtained mutant c12 ring. The latter, 97.1 Å (on the level of the A20G mutation), was significantly smaller than the calculated WT c12 circumference of 100.5 Å, apparently caused by the A16/20G mutations.
Fig. 6.
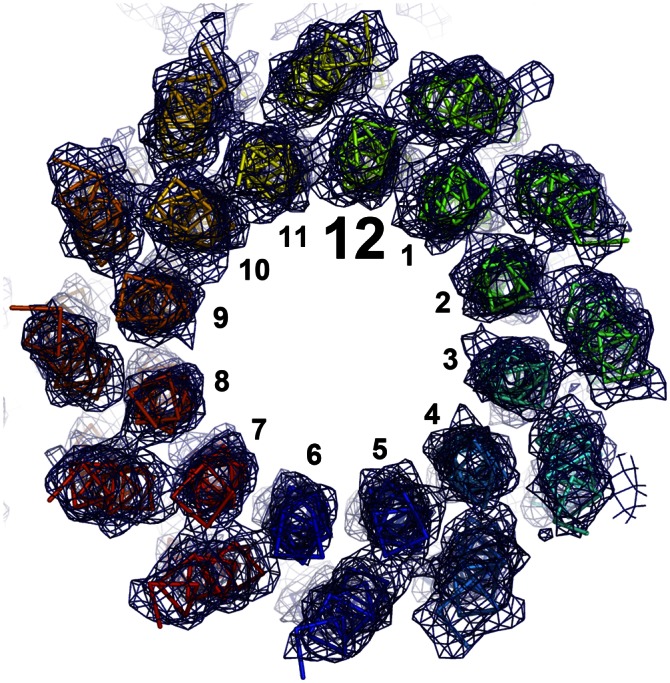
Structure of the c12 A16/20G mutant c-ring from B. pseudofirmus OF4. View of the c12 ring from the cytoplasmic side of the membrane. The backbone of the individual subunits is shown in ribbon representation. The electron densities (2Fobs − Fcalc) are shown at 1.7σ as a blue mesh. The individual helices were unambiguously traceable, confirming that the A16/20G mutant c-ring consists of 12 c-subunits (see also Figs. S7 and S8).
Growth Studies of c-Ring Mutants.
Our investigation of alanine mutants demonstrated that the stoichiometry of the rotor rings can be altered by site-directed mutagenesis. These c-rings were incorporated into the complete F1Fo-ATP synthases of B. pseudofirmus OF4 (Fig. 3A). To determine the effect on growth of extending the c-ring with or without accompanying alanine mutations, we carried out growth curves using concentrations of malate or glucose (50 mM) that are not growth-limiting as the carbon and energy source at pH 7.5 or 10.5. The extWT strain had a longer lag phase growing on malate (Fig. S5) than the WT strain (nonextended), but both strains grew at comparable rates. The extA16G mutant strain grew like the nonextended A16G mutant on malate at pH 10.5 and slightly better at pH 7.5. The A16/20G mutants exhibited the most pronounced phenotype of the alanine mutants. The double mutant defect, described previously, was more severe in a screening assay involving a single 14-h A600 measurement than in a growth curve experiment conducted under different conditions (24). Interestingly, the extA16/20G double mutant grew somewhat better compared with its nonextended homolog on malate at both pH values. The extension might have served as a stabilizing factor for the c-ring in vivo for the double mutant, resulting in improved growth.
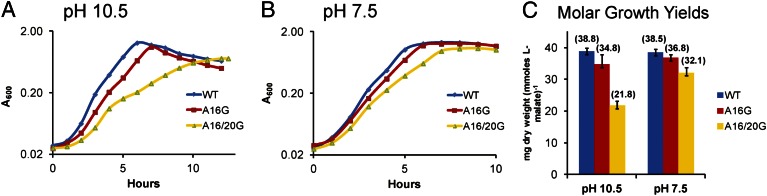
Assays of molar growth yields provide rigorous assessments of the comparative ability of two strains to convert limiting substrate into cell mass. Given the similar overall growth patterns of strains bearing nonextended c-rings and strains bearing extended c-rings, we performed growth curves on the strains with nonextended c-rings. We used a limiting concentration of the nonfermentable substrate malate, 5 mM, instead of the usual nonlimiting concentration of 50 mM. At time points corresponding to the growth maximum, samples were obtained for assays of malate consumption, and the dry weight was calculated from a standard curve relating turbidity at 600 nm to dry weight. Compared with the WT, the A16/20G mutant grew slower and to a lower final density at pH 10.5, exhibiting a smaller deficit at pH 7.5 (Fig. 7). The molar growth yield of the A16/20G mutant was only 56% of that of the WT (21.8 mg vs. 38.8 mg dry weight per mmol L-malate consumed) at pH 10.5, but was 83% of that of the WT at pH 7.5. The A16G mutant was somewhat less affected relative to WT than the double mutant, in a manner apparently consistent with the finding that the majority of its c-ring content was of the WT c13 stoichiometry. In summary, reducing the c-ring stoichiometry appeared to specifically lower the rate of ATP synthesis and molar growth yield during oxidative phosphorylation at very alkaline pH, and had a more modest impact at near-neutral pH.
Fig. 7.
Growth curves and molar growth yields of the WT and A16G and A16/20G mutants (nonextended) on limiting malate at pH 10.5 and 7.5. Strains were grown on both 0.1% yeast extract alone and 5 mM L-malate (in the presence of 0.1% yeast extract). The maximum A600 value observed on yeast extract alone was subtracted from the maximum A600 value on malate for the calculation of dry weight. The growth curves are the average of between five and eight independent experiments. (A) Growth curves at pH 10.5 on 5 mM L-malate. The A600max values (without yeast extract correction) for the three strains were 1.293 ± 0.067, 1.108 ± 0.055, and 0.724 ± 0.021, respectively. (B) Growth curves at pH 7.5. The A600max values (without yeast extract correction) for the three strains were 1.350 ± 0.027, 1.280 ± 0.029, and 1.076 ± 0.038, respectively. (C) Molar growth yields at both pH values.
Discussion
This work has addressed the question of whether the AxAxAxA motif in the alkaliphilic B. pseudofirmus OF4 c-subunit is an adaptation to alkaline pH resulting in higher c-ring stoichiometries, which enables the synthesis of ATP by oxidative phosphorylation at low proton motive force. A previously observed gel shift of the alanine mutant c-rings (24) can now be visually confirmed as a change in the stoichiometry. Whereas the extWT contains 13 subunits per c-ring almost exclusively, the A16G and A16/20G mutants exhibit altered stoichiometries, consisting mainly (but not exclusively) of c12 + c13 and c12, respectively. In agreement with previous reports (24, 26), growth studies on nonfermentable medium at pH 7.5 support the finding that these altered c-rings are part of functional ATP synthases and coexist within a single cell. A shift of the growth medium to pH 10.5 causes major growth deficits, including reduced molar growth yields on malate. Our data indicate that B. pseudofirmus OF4 can assemble and operate ATP synthases with different stoichiometries of c-rings in the range of c11 to c15, but robust growth at high pH is restricted to strains with a majority of c-rings with at least the c13 stoichiometry.
Alanine Motif Influences c-Ring Stoichiometry.
The alanine motif in the c13 ring from B. pseudofirmus OF4 is the area of tightest packing and, with its involvement in the c/c-subunit interface, contributes to a high complex stability. In particular, the Ala16Cα atom contributes notably to the stabilizing interactions within this motif. Indeed, the A16G mutant demonstrated the strongest effect on the c-ring stoichiometry, and this effect was enhanced by a second mutation, A20G, sharing the side with A16G on the α-helix. The double mutant lost a total of seven hydrophobic c/c-subunit interactions per subunit; phenotypically, it changed 63% of the c-rings from c13 to c12. Also intuitively correct, A-to-G mutations (this work) reduce the c-ring stoichiometry and circumference, whereas G-to-A mutations increase it (25).
The X-ray crystallography structure of the mutant c12 ring showed a reduced c12 ring circumference relative to the WT c13, that is, reduced single c/c-subunit distances and slightly closer packing at the level of the mutations. These subtle changes in distances disturb the native preferred c13 conformation and lead to an energetically more preferred, and hence statistically more prevalent, c12 ring of the double mutant. Our data support the idea that the motif is a hotspot for the c-ring stoichiometry determination; selected mutations cause subtle but pivotal steric effects within the c/c-subunit packing (Fig. 1C), changing the stoichiometry.
Our results correspond to the observation that the alanines (or serines) found in the motif lead to an extended ring diameter and higher stoichiometry in alkaliphilic Bacillus species (20). The changes in the tertiary structure of these mutants have no influence on complex stability with respect to pH, temperature, or detergent, but further mutations finally destabilize the c-ring (24, 26). In full agreement with the findings of a study on I. tartaricus rotor rings (25), some residues in the motif served as hotspots for the stoichiometry. In contrast, other parts of the structure, such as the C-terminal extension (“ext” in this work) and other mutations in the c-subunit (31) did not influence the complex stability in such a profound way. The determinants of c-ring stoichiometry seem to point toward multiple factors that in combination affect the final stoichiometry, with the alanine/glycine motif a particularly important factor.
Conclusion.
Alkaliphilic Bacillus species have evolved several adaptations to cope with the bioenergetic challenge of ATP synthesis at a low pmf. One of these adaptations is the stretch of AxAxAxA instead of GxGxGxG in the c-subunit sequence encoding the inner ring of α-helices of the c-ring. This alteration was proposed to enhance both the c-ring stoichiometry and its size in alkaliphilic Bacillus species (19, 20, 26). Our observations here concur with the expectation that a higher c-ring stoichiometry (i.e, increased i) would be advantageous at a low overall pmf in alkaliphiles (21, 32), because the phosphorylation potential for ATP synthesis (ΔGp) at thermodynamic equilibrium equals i × pmf. The common theme of the enlarged c-ring stoichiometries found in alkaliphilic Bacillus species (c13) and the contrasting smaller c10 rings found in neutralophilic Bacillus species [e.g., Bacillus PS3 (33), Geobacillus kaustophilus (Fig. S6)] further support this proposed evolutionary adaptation mechanism. The alanine motif is a necessary, but insufficient, adaptation of alkaliphilic Bacillus bacteria. It has a direct influence on the c-ring stoichiometry and its indigenous property to determine the ATP synthase i value, and thus directly modulates the cell’s physiology and bioenergetics, facilitating growth at pH >10. Remarkably, and in agreement with previous work (25), this observation also suggests that i can be adapted by just one or two selected mutations. This property enables adaptation to new environmental challenges, a process that can occur within a rather short evolutionary time frame.
Materials and Methods
Cloning and Purification of Mutant c-Rings from B. pseudofirmus OF4 Cells.
The WT strain is B. pseudofirmus OF4 811M, a marked strain derived from B. pseudofirmus OF4, for brevity referred to as B. pseudofirmus OF4 herein. Cloning of extended c-ring mutants was done as described in SI Materials and Methods. Purification of the ATP synthase and extraction of the mutant c-rings were performed as described previously (19) but using 0.5% (wt/vol) n-decyl-β-D-maltoside (DM) instead of 0.05% (wt/vol) n-dodecyl-β-D-maltoside (DDM) for hydroxyapatite chromatography (30). Stability tests of WT and mutant B. pseudofirmus OF4 c-rings and purification of G. kaustophilus c-rings from native cells were performed as described in SI Materials and Methods.
Reconstitution and 2D Crystallization of B. pseudofirmus OF4 Rotor Rings.
For 2D crystallization of the WT and the mutant c-rings, samples of decylmaltoside-solubilized c-rings (1 mg/mL) were mixed with MGDG (Larodan Fine Chemicals) in 2% DM at lipid-to-protein ratios of 0.65, 0.75, 0.85, and 0.95. The setups were dialyzed in 30-µL buttons (Hampton Research) against 250 mL of 100 mM Tris⋅HCl (pH 7.5–8.0), 200 mM NaCl, 2–5 mM MgCl2, and 3 mM NaN3 buffer with 12,000–14,000-kDa cutoff membranes (Spectra/Por 2, Spectrumlabs) for 1 wk at 25 °C, then shifted to 37 °C until turbidity was visible. The samples were removed from the dialysis buttons and stored at 25 °C until further use. The most successful reconstitution conditions were as follows: (i) extWT: 0.1 M Tris⋅HCl (pH 8.0), 0.2 M NaCl (buffer D) plus MGDG with a lipid-to-protein ratio (LPR) of 0.75, with dialysis at 25 °C for 7 d, then at 37 °C for 10 d; (ii) extA16G: buffer D plus 2 mM MgCl2 and MGDG (LPR = 0.75), with dialysis at 25 °C for 14 d; and (iii) extA16/20G: buffer D plus 5 mM MgCl2 and MGDG (LPR = 0.85), with dialysis for 10 d at 25 °C and then for 10 d at 37 °C.
AFM.
The contact-mode atomic force microscope was equipped with a 100-μm X–Y piezo scanner (NanoScope IIIa; Veeco) and 200-µm 0.05 N/m cantilevers (OMCL RC-800-PS; Olympus) (SI Materials and Methods). The c-ring images from series of topographs were pooled to ensure more than 200 particle datasets for averaging for WT, more than 1,900 datasets for extWT, and more than 700 datasets for extA16G and extA16/20G. All molecules had the same in-plane orientation in the membrane of tightly packed vesicles, and averaging was performed by crystallographic methods with no reference bias. A color-converted gray-level scale corresponds to 2.0 nm in all AFM images (Figs. 4 and 5 and Fig. S1).
Structure Determination of the Nonextended A16/20G Mutant c-Ring by X-Ray Crystallography.
The mutant protein was purified as described previously (19). Crystals were grown in hanging drops by vapor diffusion. For this, 1 μL of protein was supplied with 2% (wt/vol) α-nonyl-maltoside (NM) (Anatrace; Affymetrix) and mixed with 1 μL of crystallization buffer [0.1 M Tris⋅HCl (pH 9.0) and 23% (vol/vol) polyethylene glycol (PEG) 400]. After 24 h, the crystals appeared. Before the crystals were flash-frozen in liquid nitrogen, they were soaked for 3 min in a cryoprotective solution containing 0.1 M Tris⋅HCl (pH 9.0), 30% (vol/vol) PEG 400, and 2% (wt/vol) NM. Data collection, structure determination, and refinement were done as described in SI Materials and Methods. The atomic coordinates and structure factors of the B. pseudofirmus OF4 c12 ring have been deposited in the Protein Data Bank (PDB ID code 3zo6).
Supplementary Material
Acknowledgments
We thank Petra Schwille for granting access to the atomic force microscopy equipment and laboratory space, and Werner Kühlbrandt for his generous support of T.M.’s laboratory. This project was supported by the Collaborative Research Center 807 of the German Research Foundation, the Cluster of Excellence Frankfurt Macromolecular Complexes at Goethe University (Project EXC 115), and the National Institute of General Medical Sciences (Research Grant R01 GM28454, to T.A.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3zo6).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303333110/-/DCSupplemental.
References
- 1.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370(6491):621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 2.Boyer PD. The ATP synthase—a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 3.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386(6622):299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 4.Junge W, Sielaff H, Engelbrecht S. Torque generation and elastic power transmission in the rotary F(O)F(1)-ATPase. Nature. 2009;459(7245):364–370. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- 5.Schemidt RA, Qu J, Williams JR, Brusilow WSA. Effects of carbon source on expression of F0 genes and on the stoichiometry of the c subunit in the F1F0 ATPase of Escherichia coli. J Bacteriol. 1998;180(12):3205–3208. doi: 10.1128/jb.180.12.3205-3208.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watt IN, Montgomery MG, Runswick MJ, Leslie AG, Walker JE. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci USA. 2010;107(39):16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pogoryelov D, et al. The c15 ring of the Spirulina platensis F-ATP synthase: F1/F0 symmetry mismatch is not obligatory. EMBO Rep. 2005;6(11):1040–1044. doi: 10.1038/sj.embor.7400517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meier T, Faraldo-Gómez JD, Börsch M. Molecular Machines in Biology. Cambridge, UK: Cambridge Univ Press; 2011. ATP synthase, a paradigmatic molecular machine; pp. 208–238. [Google Scholar]
- 9.Murata T, Yamato I, Kakinuma Y, Leslie AG, Walker JE. Structure of the rotor of the V-type Na+-ATPase from Enterococcus hirae. Science. 2005;308(5722):654–659. doi: 10.1126/science.1110064. [DOI] [PubMed] [Google Scholar]
- 10.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science. 2005;308(5722):659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson SJ. ATP synthase: From sequence to ring size to the P/O ratio. Proc Natl Acad Sci USA. 2010;107(39):16755–16756. doi: 10.1073/pnas.1012260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hicks DB, Liu J, Fujisawa M, Krulwich TA. F1F0-ATP synthases of alkaliphilic bacteria: Lessons from their adaptations. Biochim Biophys Acta. 2010;1797(8):1362–1377. doi: 10.1016/j.bbabio.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol. 2011;9(5):330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heberle J, Riesle J, Thiedemann G, Oesterhelt D, Dencher NA. Proton migration along the membrane surface and retarded surface to bulk transfer. Nature. 1994;370(6488):379–382. doi: 10.1038/370379a0. [DOI] [PubMed] [Google Scholar]
- 15.Mulkidjanian AY, Cherepanov DA, Heberle J, Junge W. Proton transfer dynamics at membrane/water interface and mechanism of biological energy conversion. Biochemistry (Mosc) 2005;70(2):251–256. doi: 10.1007/s10541-005-0108-1. [DOI] [PubMed] [Google Scholar]
- 16.Sandén T, Salomonsson L, Brzezinski P, Widengren J. Surface-coupled proton exchange of a membrane-bound proton acceptor. Proc Natl Acad Sci USA. 2010;107(9):4129–4134. doi: 10.1073/pnas.0908671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimroth P, Cook GM. Bacterial Na+- or H+-coupled ATP synthases operating at low electrochemical potential. Adv Microb Physiol. 2004;49:175–218. doi: 10.1016/S0065-2911(04)49004-3. [DOI] [PubMed] [Google Scholar]
- 18.Janto B, et al. Genome of alkaliphilic Bacillus pseudofirmus OF4 reveals adaptations that support the ability to grow in an external pH range from 7.5 to 11.4. Environ Microbiol. 2011;13(12):3289–3309. doi: 10.1111/j.1462-2920.2011.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preiss L, Yildiz Ö, Hicks DB, Krulwich TA, Meier T. A new type of proton coordination in an F1Fo-ATP synthase rotor ring. PLoS Biol. 2010;8:e000443. doi: 10.1371/journal.pbio.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthies D, et al. The c13 ring from a thermoalkaliphilic ATP synthase reveals an extended diameter due to a special structural region. J Mol Biol. 2009;388(3):611–618. doi: 10.1016/j.jmb.2009.03.052. [DOI] [PubMed] [Google Scholar]
- 21.Meier T, et al. A tridecameric c ring of the adenosine triphosphate (ATP) synthase from the thermoalkaliphilic Bacillus sp. strain TA2.A1 facilitates ATP synthesis at low electrochemical proton potential. Mol Microbiol. 2007;65(5):1181–1192. doi: 10.1111/j.1365-2958.2007.05857.x. [DOI] [PubMed] [Google Scholar]
- 22.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: Structure and implications. Science. 1997;276(5309):131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 23.Vonck J, et al. Molecular architecture of the undecameric rotor of a bacterial Na+-ATP synthase. J Mol Biol. 2002;321(2):307–316. doi: 10.1016/s0022-2836(02)00597-1. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, et al. Mutations in a helix-1 motif of the ATP synthase c-subunit of Bacillus pseudofirmus OF4 cause functional deficits and changes in the c-ring stability and mobility on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochemistry. 2011;50(24):5497–5506. doi: 10.1021/bi2005009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pogoryelov D, et al. Engineering rotor ring stoichiometries in the ATP synthase. Proc Natl Acad Sci USA. 2012;109(25):E1599–E1608. doi: 10.1073/pnas.1120027109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Fujisawa M, Hicks DB, Krulwich TA. Characterization of the functionally critical AXAXAXA and PXXEXXP motifs of the ATP synthase c-subunit from an alkaliphilic Bacillus. J Biol Chem. 2009;284(13):8714–8725. doi: 10.1074/jbc.M808738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stahlberg H, et al. Bacterial Na(+)-ATP synthase has an undecameric rotor. EMBO Rep. 2001;2(3):229–233. doi: 10.1093/embo-reports/kve047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meier T, Matthey U, Henzen F, Dimroth P, Müller DJ. The central plug in the reconstituted undecameric c cylinder of a bacterial ATP synthase consists of phospholipids. FEBS Lett. 2001;505(3):353–356. doi: 10.1016/s0014-5793(01)02837-x. [DOI] [PubMed] [Google Scholar]
- 29.Meier T, et al. Evidence for structural integrity in the undecameric c-rings isolated from sodium ATP synthases. J Mol Biol. 2003;325(2):389–397. doi: 10.1016/s0022-2836(02)01204-4. [DOI] [PubMed] [Google Scholar]
- 30.Meier T, Ferguson SA, Cook GM, Dimroth P, Vonck J. Structural investigations of the membrane-embedded rotor ring of the F-ATPase from Clostridium paradoxum. J Bacteriol. 2006;188(22):7759–7764. doi: 10.1128/JB.00934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Hicks DB, Guffanti AA, Baldwin K, Krulwich TA. Replacement of amino acid sequence features of a- and c-subunits of ATP synthases of alkaliphilic Bacillus with the Bacillus consensus sequence results in defective oxidative phosphorylation and non-fermentative growth at pH 10.5. J Biol Chem. 2004;279(25):26546–26554. doi: 10.1074/jbc.M401206200. [DOI] [PubMed] [Google Scholar]
- 32.von Ballmoos C, Cook GM, Dimroth P. Unique rotary ATP synthase and its biological diversity. Annu Rev Biophys. 2008;37:43–64. doi: 10.1146/annurev.biophys.37.032807.130018. [DOI] [PubMed] [Google Scholar]
- 33.Mitome N, Suzuki T, Hayashi S, Yoshida M. Thermophilic ATP synthase has a decamer c-ring: Indication of noninteger 10:3 H+/ATP ratio and permissive elastic coupling. Proc Natl Acad Sci USA. 2004;101(33):12159–12164. doi: 10.1073/pnas.0403545101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.