Abstract
The origin of pathogenic autoantibodies remains unknown. Idiopathic pulmonary alveolar proteinosis is caused by autoantibodies against granulocyte–macrophage colony-stimulating factor (GM-CSF). We generated 19 monoclonal autoantibodies against GM-CSF from six patients with idiopathic pulmonary alveolar proteinosis. The autoantibodies used multiple V genes, excluding preferred V-gene use as an etiology, and targeted at least four nonoverlapping epitopes on GM-CSF, suggesting that GM-CSF is driving the autoantibodies and not a B-cell epitope on a pathogen cross-reacting with GM-CSF. The number of somatic mutations in the autoantibodies suggests that the memory B cells have been helped by T cells and re-entered germinal centers. All autoantibodies neutralized GM-CSF bioactivity, with general correlations to affinity and off-rate. The binding of certain autoantibodies was changed by point mutations in GM-CSF that reduced binding to the GM-CSF receptor. Those monoclonal autoantibodies that potently neutralize GM-CSF may be useful in treating inflammatory disease, such as rheumatoid arthritis and multiple sclerosis, cancer, and pain.
Keywords: autoantibody, biotherapeutic, cytokine, autoimmune, three-dimensional model
Although autoantibodies that cause paroxysmal cold hemoglobinuria were demonstrated more than a century ago (1), the origin of pathogenic autoantibodies remains unknown, despite immense progress in understanding immunology and in the knowledge of genes that predispose to autoimmunity (2). Ehrlich proposed that mechanisms exist to prevent antibody production against components of self and consequent damage (“horror autotoxicus”) (1). Burnet showed five decades later that immunological tolerance was acquired during fetal development and developed the clonal selection theory of antibody formation. Burnet proposed that autoimmunity was caused by a “forbidden clone” (3). Antibodies are made up of a heavy (H) chain and a light (L) chain, which contribute to the antigen-binding site through six variable peptide loops, termed complementary determining regions (CDRH1–3 and CDRL1–3). The diversity of the antigen-binding sites is achieved partially by combinations of H and L chains. Each H chain gene is formed somatically and stochastically by recombination and joining of one each of the ∼55 immunoglobulin heavy chain variable (IGHV), ∼27 immunoglobulin heavy chain diversity (IGHD), and 6 immunoglobulin heavy chain joining (IGHJ) genes. Each L chain gene is similarly formed by using one of either ∼40 immunoglobulin kappa variable (IGKV) or ∼30 immunoglobulin lambda variable (IGLV) genes, recombined with one of either 5 immunoglobulin kappa joining (IGKJ) or 4 immunoglobulin lambda joining (IGLJ) genes (4). There is evidence that certain V genes are more frequently used in autoantibodies, such as IGHV1-69 (5), and certain families of V genes are overexpressed in particular diseases, e.g., Graves disease (IGHV1 family), Hashimoto disease (IGHV3 family), myasthenia gravis, chronic idiopathic thrombocytopenic purpura (IGHV3-30), and Sjogren disease (IGHV1-69 and IGHV3-7) (6). Thus, one notion for the origin of pathogenic autoantibodies is the propensity of certain V genes or alleles or preferred combinations of H and L chains to give rise to high-affinity autoantibodies against self-antigens (6). An elegant study showed that authentic human monoclonal autoantibodies (mAbs) against transglutinaminase 2 (TG2) in celiac disease used preferred V genes (7). There were few somatic mutations in the mAbs so that germ-line V genes could produce high-affinity autoantibodies against TG2, but whether the autoantibodies against TG2 are pathogenic for celiac disease is unclear (7). Conversely, an alternative origin of pathogenic autoantibodies is that the autoantibodies arise in response to a pathogen antigen that mimics a self-antigen (8).
One autoimmune disease that is clearly caused by pathogenic autoantibodies that neutralize the bioactivity of the hemopoietic growth factor or cytokine, granulocyte–macrophage colony-stimulating factor (GM-CSF), is idiopathic pulmonary alveolar proteinosis (IPAP) (9–11). IPAP is a lung disease, caused by the accumulation of surfactant protein in the alveoli, that leads to severe respiratory distress. The typical treatment is periodic bronchio-alveolar lavage (BAL) (12). GM-CSF is produced by many types of cells when stimulated with microbial products or inflammatory cytokines and is also produced by antigen-stimulated T lymphocytes (13). GM-CSF stimulates the proliferation and differentiation of committed progenitors that generate neutrophils, monocytes, macrophages and dendritic cells and also activates differentiated myeloid effector cells such as neutrophils, eosinophils, or monocytes to increase their activity or maturation or prolong their survival (14–17). Small amounts of GM-CSF produced in the lung are necessary for the maturation of alveolar macrophages, which normally catabolize surplus surfactant in the alveoli. Moreover, administration of GM-CSF ameliorates IPAP (18). Animals or humans that lack functional genes encoding either GM-CSF or the GM-CSF receptor have pulmonary alveolar proteinosis (19).
Affinity-purified autoantibodies against GM-CSF from patients suffering from IPAP transferred to nonhuman primates cause the development of the characteristic features of IPAP, including milky BAL, an increased concentration of surfactants in BAL and serum, and increased pulmonary leucocytosis (11). Although transfers of serum or IgG have recapitulated the disease in hemolytic anemia, pemphigus (20), and myasthenia gravis (21), IPAP is one of the few human autoimmune diseases in which affinity-purified autoantibodies against a specific protein have been demonstrated to be sufficient to cause the disease, thus warranting a detailed study of their pathogenic mechanism.
We have generated 19 human mAbs against GM-CSF from six patients with IPAP. We demonstrate that the autoantibody response is polyclonal, uses multiple Ig V genes, and targets at least four different nonoverlapping epitopes on GM-CSF, suggesting that GM-CSF itself is driving the antibody response and thus the disease. The high number of somatic mutations suggests that T cells are involved and that the memory B cells have re-entered germinal centers and undergone somatic mutation and affinity selection multiple times. All of the autoantibodies specifically neutralized GM-CSF bioactivity, and the affinity of the antibodies to GM-CSF generally correlated with the neutralization of bioactivity. Epitope mapping and molecular modeling demonstrated that the mAbs most likely block, by steric interference, the interaction of GM-CSF with the GM-CSF receptor, thus preventing an effective signaling complex. These mAbs that potently neutralize GM-CSF may be useful in treating diseases in which the bioactivity of GM-CSF is pathogenic.
Results
Generation of mAbs Against GM-CSF from Patients with IPAP.
We generated 19 mAbs against GM-CSF from six patients with IPAP (Table S1). All 19 mAbs were structurally and genetically unrelated. The only sign of the preferred use of Ig V genes was the greater use of IGHV1-2 or -3. However, some of these mAbs using the same IGHV gene bound to different epitopes (below). We noted that some mAbs had a high number of somatic mutations in the IGHV gene, the highest number being 52, meaning that almost one in every five nucleotides had been mutated (Table S1). The median number of somatic mutations in the IGHV gene was 30 (Fig. S1). In contrast, the average mutation rate in human memory B cells and germinal center B cells is 13.6 ± 4.8 (22).
Neutralization of Bioactivity of GM-CSF.
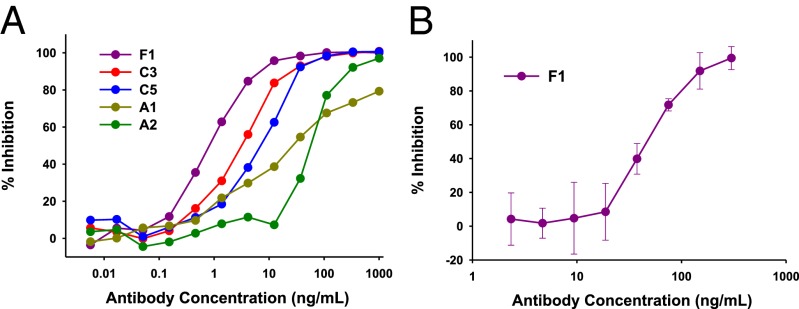
We used the human erythroleukemia cell line TF-1, which depends on growth factors such as GM-CSF or interleukin-3 (IL-3) to survive and proliferate, to compare the ability of the mAbs to neutralize the bioactivity of GM-CSF and IL-3 (10, 23). We found that all 19 mAbs had some capacity to neutralize the ability of GM-CSF to promote proliferation and survival of TF-1 cells (Table S1) and had no capacity to neutralize the bioactivity of IL-3 (Table S1 and Fig. S2A). Fig. 1A shows the dose–response of representative mAbs in inhibiting the bioactivity of glycosylated GM-CSF on TF-1 cells. Because high-affinity antibodies are more potent at neutralizing low concentrations of a growth factor (24), we used a low concentration of GM-CSF (200 pg/mL). From Fig. 1A, it can be seen that some mAbs were more potent than others at neutralizing the bioactivity of GM-CSF. The addition of mAb F1 at 100 ng/mL to TF-1 cells growing in GM-CSF could completely block cell survival in 6 d (Fig. S2B). We showed that selected mAbs could neutralize the bioactivity of naturally glycosylated GM-CSF produced in human cells on TF-1 cells (Fig. S2C). We also treated whole blood with GM-CSF for 30 min at 37 °C to stimulate primary neutrophils to express the integrin component, CD11b, on their cell-surface (23). Using flow cytometry, we measured the levels of CD11b on the surface of human neutrophils (Fig. S3) and observed in three experiments that the mAb F1 neutralized the ability of high concentrations of GM-CSF to induce CD11b expression on primary neutrophils (Fig. 1B). We also tested mAb C5 twice, and it inhibited the ability of GM-CSF to induce CD11b expression on primary neutrophils.
Fig. 1.
Neutralization by mAbs against GM-CSF of bioactivity of GM-CSF. (A) Washed TF-1 cells were incubated with titrations of the indicated mAbs and 200 pg/mL rhGM-CSF. After 4 d, metabolic activity of TF-1 cells was measured by the water-soluble tetrazolium assay. (B) mAb F1 inhibits CD11b expression level on primary neutrophils stimulated by GM-CSF. Titrations of F1 mAb were added to heparinized blood with a high concentration of GM-CSF (10 ng/mL). The levels of CD11b on neutrophils were subsequently determined by flow cytometry (Fig. S3).
Epitope Mapping.
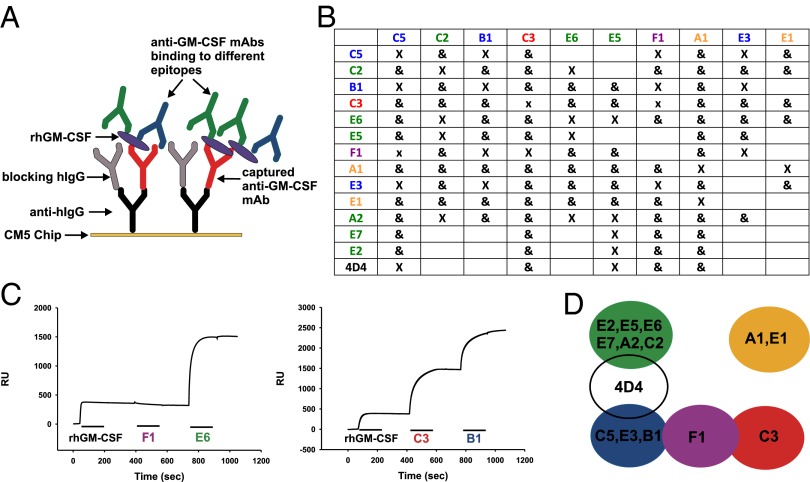
We used surface plasmon resonance (SPR) to map antibody epitopes and observed that our panel of mAbs bound to four nonoverlapping epitopes on GM-CSF (colored green, yellow, blue, and red) and an additional epitope (colored purple) that overlapped two of the epitopes (Fig. 2). In every case, when we obtained more than one mAb from a patient, they bound to at least two nonoverlapping epitopes. In two patients (C and E) from whom we generated six mAbs, they bound to at least three nonoverlapping epitopes. We also epitope-mapped a neutralizing murine mAb (4D4) against human GM-CSF (25) and observed that this epitope overlapped the green and blue epitopes (Fig. 2D). The epitopes bound by all 19 mAbs were conformational and dependent on disulfide bond formation, because none of the mAbs bound to GM-CSF on a reducing immunoblot (Table S1 and Fig. S4). Conversely, all mAbs bound to GM-CSF in a nonreducing immunoblot with the disulfide bonds intact.
Fig. 2.
Human mAbs against GM-CSF target multiple epitopes. SPR was used to map the binding of anti–GM-CSF mAbs to rhGM-CSF. (A) A schematic showing how an anti-hIgG is immobilized on a CM5 sensor chip to capture the first anti–GM-CSF mAb injected. After blocking of free sites with an excess of hIgG, rhGM-CSF is injected and bound by the first mAb. Subsequently, a second and third anti–GM-CSF mAb is injected, and the binding is monitored in real time by SPR. (B) A summary of the epitope-mapping experiments, with “X” meaning that the two mAbs cannot bind simultaneously and “&” meaning they can bind simultaneously to rhGM-CSF. mAbs in the leftmost column denote the captured mAb for each experiment. (C) Representative mapping experiments. (Left) After capture of C3 and binding of rhGM-CSF, only E6, and not F1, can bind. (Right) After capture of A1 and binding of rhGM-CSF, both C3 and B1 can bind simultaneously. (D) A map of the GM-CSF epitopes. Color coding is maintained throughout. The 4D4 is a mouse mAb raised against rhGM-CSF.
Amino Acid Mutations in GM-CSF to Map the Epitopes on GM-CSF.
We reasoned that antibodies that neutralized the bioactivity of GM-CSF might bind to the amino acids that GM-CSF uses to bind to its receptor subunits. We used single or double mutations of human GM-CSF, chosen for their effect on the interaction with the GM-CSF receptor alpha (GMR-α) or GM-CSF receptor beta common (GMR-βc) subunits of the GM-CSF receptor, to map where our mAbs in the different epitope groups bound to GM-CSF. Most GM-CSF mutants with altered binding to the GM-CSF receptor subunits had no obvious effect on the binding of most of the mAbs. However, the GM-CSF point mutation, D112R, drastically affected the binding of mAb B1 (Fig. S5A). We previously showed that residue D112 participates in a functionally important interaction with GMR-α (25) and is physically associated with GMR-α in the crystal structure of the GM-CSF receptor ternary complex (25). Thus, the mAbs that belong to the same nonoverlapping “blue” epitope group (B1, C5, and E3) probably neutralize GM-CSF by sterically inhibiting GM-CSF from binding to the GMR-α.
We detected effects of mAbs that bound to a different, nonoverlapping epitope group (the “green” group) in mutations of residues 14 and 15 that are located at the start of the first helix of GM-CSF, which binds to GMR-βc (25). As shown in Fig. S5 B and C, with mAb E7, we saw a decreased dissociation from GM-CSF mutated at either E14 or H15 to alanine. Conversely, the dissociation rate of mAb E5, another mAb from this epitope group, was increased by mutating residue H15 to alanine. H15 is a buried residue located on helix A of GM-CSF. Mutation of H15 to alanine will likely disrupt the interactions between H15 and helix C (via H83 and Y84) and helix D (via P118 and F119), perhaps altering the GM-CSF conformation and preventing the cytokine interacting with its receptor or mAbs. Thus, mAbs that belong to the green epitope group probably neutralize GM-CSF by binding to the surface of GM-CSF, which then sterically inhibits GM-CSF from binding to the βc.
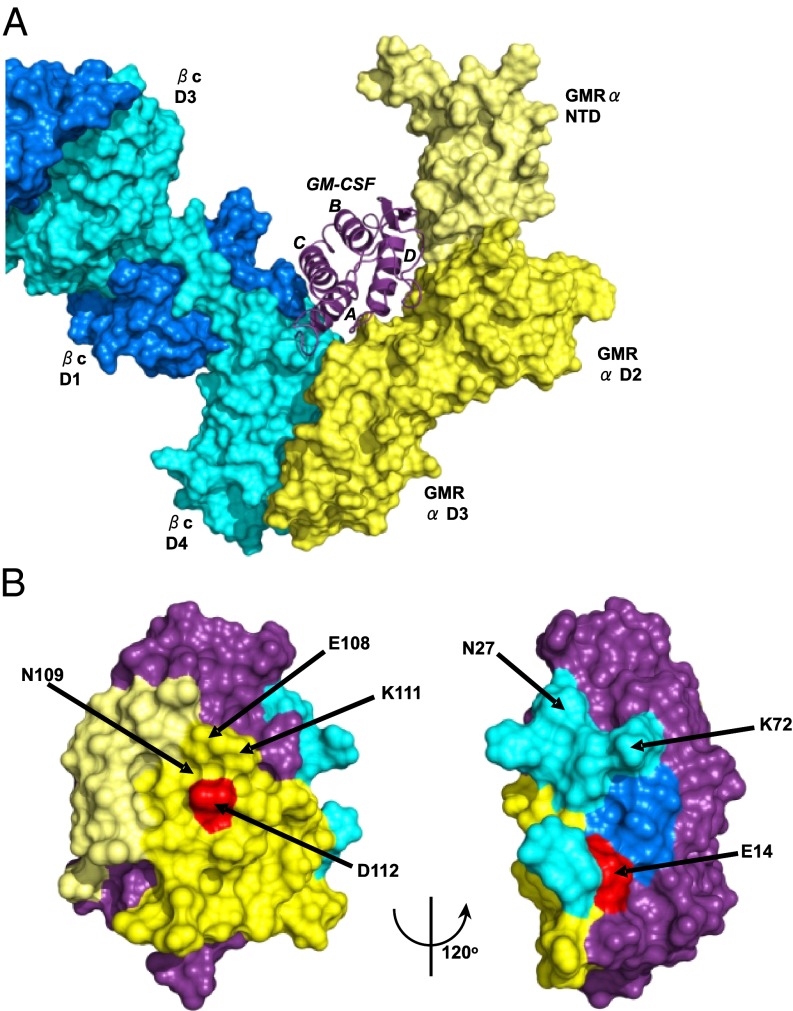
Fig. 3A depicts a model of the ternary complex, based on our ternary complex crystal structure (26), of a partially refined crystal structure of the GM-CSF:GMR-α binary complex and the crystal structure of the homologous IL-5 receptor alpha subunit (27). To visualize how anti–GM-CSF mAbs could interfere with the bioactivity of GM-CSF and its formation of a signaling complex with the GM-CSF receptor, we used Pisa (Version 1.37; www.ebi.ac.uk/msd-srv/prot_int/pistart.html) to map and quantify the interaction interface residues of the ternary GM-CSF:receptor complex model. Fig. 3B shows the location of the E14 and D112 mutations on the GM-CSF structure and the surface buried on GM-CSF by the two subunits of the GM-CSF receptor. Shown in Table S2 is the total solvent surface area of GM-CSF and the three parts of the GM-CSF receptor with which GM-CSF interacts, namely the GMR-α and two domains of different βc monomers. It can be seen that 30% of the surface area of GM-CSF is buried by interaction with the receptor: 19% by the interaction with the GMR-α, 7% with the D4 domain of βc (monomer 1), and 4% with the D1 domain of βc (monomer 2). A typical antiprotein antibody buries between 600 and 1,000 Å2 of the surface area with its antigen-binding site (28). If the binding sites of anti–GM-CSF mAbs overlap the area buried by interaction with the GM-CSF receptor, the mAbs could sterically interfere with the formation of the signaling complex.
Fig. 3.
Mutations on the surface of GM-CSF and area buried by the subunits of the GM-CSF receptor. (A) The GM-CSF:receptor ternary model. GM-CSF is shown in purple ribbon, the GMR-α in yellow molecular surface (with the NTD shown in lighter shade), and the βc shown in blue (different shades for the two monomer chains of the βc dimer). (B) Two views of the surface of GM-CSF using the coloring scheme in A. The surfaces that interact with the GMR-α are in yellow, with the GMR-βc in blue and the remainder of the cytokine colored purple. Some of the residues targeted for mutation are highlighted, and key residues, shown by mutation to markedly disrupt antibody binding, are shown in red.
Affinity and Kinetics of Binding to GM-CSF and Correlation with Neutralization of Bioactivity of GM-CSF.
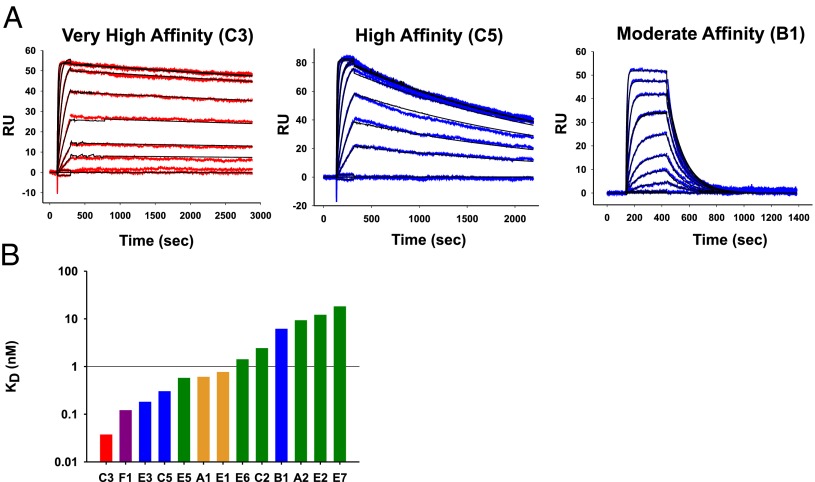
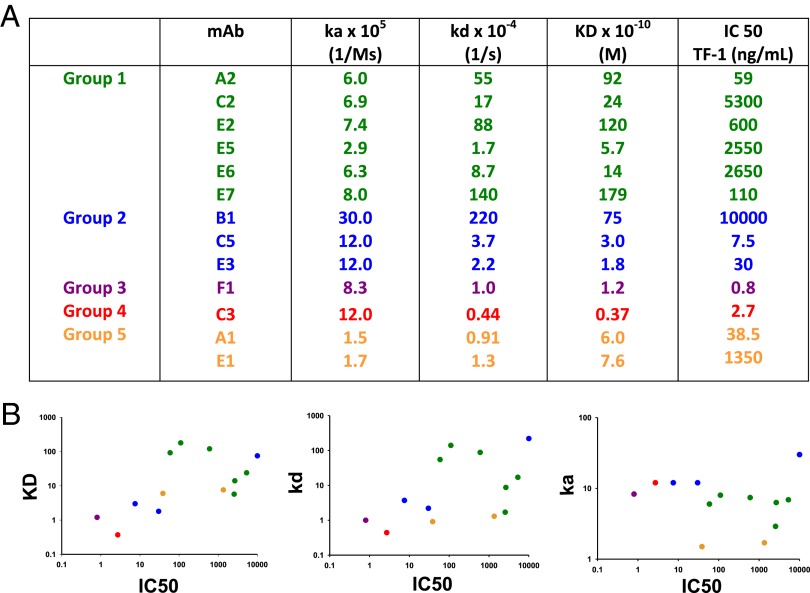
We used SPR to analyze the kinetics of binding of selected mAbs to GM-CSF. Shown in Fig. 4A are representative SPR data from a very high-affinity mAb, a high-affinity mAb, and a moderate-affinity mAb. In cases where we isolated multiple mAbs from the same patient, we observed a range of affinities (Fig. 4B). Fig. 5A shows the IC50 of representative mAbs from the TF-1 proliferation assay, in comparison with their binding kinetics. There was a general correlation of a decreased IC50 with a higher affinity and lower off-rate but there was no correlation with on-rate (Fig. 5B). However, we noted that a high affinity or a low off-rate did not strictly correlate with the ability to neutralize the bioactivity of GM-CSF. For example, mAb F1 has the best IC50 of 0.8 ng/mL, with mAb C3 the second best, with an IC50 of 2.7 ng/mL. However, mAb C3 has the highest affinity of all of the anti–GM-CSF mAbs (KD at 3.2 × 10−11 M), whereas mAb F1 has an almost fourfold lower affinity (KD at 1.2 × 10−10 M). The kinetics were not a factor because both the on-rate of mAb C3 was higher than mAb F1 and the off-rate of mAb C3 was lower. Even for mAbs against the same overlapping epitopes, the affinity did not correlate with the potency of neutralizing bioactivity of GM-CSF. For example, of the mAbs A2, E5, and E6 that bound to the green epitope, mAb A2 had the lowest IC50 of 59 ng/mL and the highest Kd of 9.2 × 10−9 M, whereas the mAb E5 and E6 had dramatically higher IC50 of 2,650 and 2,550 ng/mL and had 17- and 7-fold higher affinities (KD of 5.7 × 10−10 and 1.4 × 10−9 M), respectively. Likewise, mAbs E3 and C5 bound to the same blue epitope; mAb E3 had a higher affinity than mAb C5, but mAb C5 had a fourfold lower IC50.
Fig. 4.
Affinities of the human anti–GM-CSF mAbs. (A) Shown are examples of SPR data and fit for a representative very-high-affinity (C3), high-affinity (C5), and moderate-affinity (B1) mAb. (B) Relative affinities of the human anti–GM-CSF mAbs.
Fig. 5.
Relationship between kinetics of binding and potency at inhibiting the bioactivity of GM-CSF. (A) Shown are the equilibrium dissociation constants and rate constants for the rhGM-CSF and mAb complexes and comparison with their IC50 values from the TF-1 proliferation assay. (B) A graphical representation of the data from A analyzing the correlation between neutralization activity on the y axis vs. the KD (Left), kd (Center), and ka (Right) on the y axis. A nonparametric Spearman correlation test was used to calculate the P values reported for each comparison. ka, P = 0.4131; kd, P = 0.0436; KD, (P = 0.0315).
Discussion
The genetics and epitope specificity of the mAbs against GM-CSF generated from six IPAP patients (Table S1) makes several hypotheses about their pathogenesis unlikely. That all 19 mAbs generated were genetically unrelated and used multiple V genes in each patient shows that there was not preferred V-gene use like those seen in autoantibodies against TG2 (7). The fact that the autoantibodies were polyclonal excludes a single forbidden clonotype of pathogenic autoantibodies in each patient. The fact that there were at least three nonoverlapping conformational epitopes of mAbs against GM-CSF in two patients and a total of four nonoverlapping conformational epitopes in the six patients would appear to exclude a cross-reactive B-cell epitope on a pathogen (“molecular mimicry”) (8) for the following reasons. Because the area of a B-cell epitope on a protein is at least 600–935 Å2, the proper description of a B-cell epitope is through crystallography of an antigen/antigen-binding site complex (28). The probability that a pathogen antigen should have four nonoverlapping, conformational B-cell epitopes that are cross-reactive with GM-CSF is small. If another unrelated antigen initiates the antibody response and somatic mutations change the antigen-binding site so it binds to an epitope on GM-CSF, it is unlikely that chance somatic mutations would change the antigen-binding sites to bind four nonoverlapping, conformational epitopes on GM-CSF. Two crystallographic studies of affinity maturation of human antibodies (29, 30) establish that the increase in affinity is due to stabilization of the original antigen contacts and that the antigen-binding site of the original antibody has the same general conformation as the affinity-matured, high-affinity antibodies. If subsequent affinity maturation is driven by GM-CSF, the epitope on the mutated antibody should still overlap the original epitope on GM-CSF.
Our tentative conclusion is that GM-CSF most likely drove the original antibody response. There is no evidence for B-cell tolerance or anergy for an antigen like GM-CSF that is not detectable in the plasma (31); indeed, there is evidence to the contrary (32, 33). There is evidence of B-cell anergy for soluble antigens at serum concentration of >0.1 nM (34). Even in inflammatory conditions like rheumatoid arthritis, GM-CSF is present in synovial fluid at <35 pM (31, 35). One group has reported that the frequency of autoantibodies in normal subjects reaches 100% if cytokine–autoantibody complexes are dissociated at low pH before autoantibodies are measured against IL-2, granulocyte colony-stimulating factor (G-CSF), VEGF, TNF, IL-8 (32), and GM-CSF (33). However, the latter report caused some controversy, and Bazin et al. suggested that IgG, treated with low pH, can bind artifactually to GM-CSF (36). The question of the physiological production of autoantibodies against GM-CSF could be answered by measuring the frequency of memory B cells in healthy humans that produce autoantibodies against GM-CSF.
The involvement of T follicular helper cells in the pathogenic autoantibodies to GM-CSF is suggested by the high numbers of mutations in the IGHV genes of mAbs that we cloned from IPAP patients. We speculate that a pathogen, with a T-cell epitope that is cross-reactive with GM-CSF, will present pathogen-associated molecular patterns (like endotoxin or viral RNA) to the dendritic cells and activate T cells, breaking T-cell tolerance. If the pathogen stimulates the release of GM-CSF, the GM-CSF–specific B cells will endocytose GM-CSF and will present the cross-reactive T-cell epitope to T cells activated by the pathogen. The T-cell epitope on the pathogen need not correspond to the amino acid sequence of the T-cell epitope in GM-CSF, because the T-cell receptor (TCR) is polyreactive, and multiple peptides of different sequences on the MHC II can be recognized by the same TCR (37). In the future, it will be important to investigate the frequency of the T cells that are stimulated by GM-CSF in IPAP. The importance of T-cell central tolerance as a mechanism for preventing the development of autoantibodies against cytokines is suggested by the autoimmune polyendocrine syndrome type I, in which subjects have a mutation in the gene for the Autoimmune Regulator and make autoantibodies against IL-17A, IL-17F, and IL-22 (38, 39).
A paper that was published recently characterized pathogenic mAbs that cause pemphigus vulgaris (40). They reverted the somatic mutations from four mAbs against desmoglein-3 (DSG3) and found that the unmutated antibodies did not bind to DSG3. They concluded that autoantibodies against DSG3 were created by somatic mutations generated in the response to an antigen unrelated to DSG3. However, an antibody with a KD of 3 × 10−4 M could still activate B cells when expressed as a BCR in mice (41). In another study, although somatic mutations were reverted in three affinity-matured antibodies and the unmutated mAbs do not bind to the antigen, when the unmutated mAbs were expressed in a B-cell line as a BCR, they signaled when antigen was added (42). Furthermore, the probability that antibodies that bind to one unrelated antigen should have fortuitous somatic mutations that bind to two or three nonoverlapping epitopes of a DSG3 is small. The mechanism for pathogenic autoantibodies that cause pemphigus may be similar to the mechanism we propose for pathogenic autoantibodies in IPAP, namely that a pathogen that activates T cells cross-reactive with peptides of DSG3 and that humans exhibit no B-cell tolerance or anergy for DSG3 (43).
There is one report of four human mAbs against GM-CSF obtained from peripheral blood B cells from normal subjects or from patients with IPAP and with in vitro immunization of peptides from human GM-CSF (44). However, this publication does not provide information as to whether the four antibodies against GM-CSF were derived from IPAP patients or normal donors or include the results from in vitro immunization. Nor did it provide data on the autoantibodies against GM-CSF about V-gene use, somatic mutations, or epitopes.
All of the mAbs we examined neutralized the bioactivity of GM-CSF, although they did not neutralize the bioactivity of IL-3 (Table S1 and Fig. S2A). Each mAb when bound to GM-CSF could sterically interfere with the formation of the dodecamer signaling complex (26) (Fig. 3). The relationship between affinity and off-rate with the IC50 generally correlated, but the epitope where the mAb bound to GM-CSF also mattered (Fig. 5). Because we used GM-CSF binding to select specific memory B cells and the ELISA to screen, we would have not generated very low-affinity antibodies. Because these were long-time patients with IPAP, we expected to find high-affinity mAbs cloned from the GM-CSF–specific memory B cells. For example, in donors C and E, the affinities ranged, respectively, from a KD of 2.4 × 10−9 to 3.7 × 10−11 M and from a KD of 1.2 × 10−8 to 1.8 × 10−10 M. We speculate that most of the high-affinity memory B cells were retained in the lymphoid tissue and were not in the blood.
Those mAbs that potently neutralize the biological activity of GM-CSF may be useful in treating inflammatory diseases (45) like arthritis (46) and multiple sclerosis (47–49). Anti–GM-CSF mAbs may treat juvenile myelomonocytic leukemias (50) and epithelial cancers, which secrete GM-CSF that suppresses the immune response against cancer (51, 52). These mAbs may also be useful in treating pain (53, 54). The risk of causing IPAP with these therapeutic anti–GM-CSF mAbs is low because very small amounts of GM-CSF are necessary to mature alveolar macrophages, and IPAP is only caused by a critical threshold of autoantibodies against GM-CSF (33).
Materials and Methods
Generation of mAbs.
Ethical clearance for samples from patients was obtained from the ethics boards of the Universities of British Columbia, Melbourne, and Toronto. Blood samples were collected from individuals treated for IPAP. Individual memory B cells binding to GM-CSF were sorted into individual wells (55) (Fig. S6). After 1 wk, wells were screened for the production of GM-CSF–specific antibodies by ELISA. RNA was purified from positive clones, and cDNA from clones was synthesized by using constant region primers followed by PCR amplification with primers taken from the leader sequences of human V genes (Table S3). The International ImMunoGeneTics Information System (www.imgt.org) was used to determine Ig V gene use and somatic mutations.
Proliferation Assay Using TF-1 Cells.
mAbs were serially diluted, added to GM-CSF at a concentration of 400 pg/mL, and preincubated at 37 °C for 1 h. The mAb/GM-CSF mixtures were added to an equal volume of washed TF-1 cells, 1,000 cells per well, yielding a final concentration of 200 pg/mL GM-CSF. KE5, which is a human mAb IgG1 molecule against human cytomegalovirus, was used as a negative control (56). To control that the mAbs did not inhibit TF-1 cells, the GM-CSF was replaced by a final concentration of 1% (vol/vol) gibbon IL-3–conditioned medium. After incubation at 37 °C for 4 d, water-soluble tetrazolium-1 reagent was added, and incubation was continued for 4 h. The absorbance values at wavelength 450 nm, with reference wavelength of 690 nm values subtracted, were determined by using a plate reader.
Inhibition of CD11b Expression Level on Neutrophils Stimulated by GM-CSF.
Triplicate samples of heparinized human whole blood (100 μL) were incubated with GM-CSF (10 ng/mL) with 2.34–300 ng/mL mAb. Cells were stained with anti-human CD11b-PE Ab, and FACS was performed to evaluate CD11b levels (Fig. S3).
SPR Analysis.
The SPR experiments were performed at 25 °C on a Biacore 3000 (GE Healthcare) by using a carboxymethylated dextran 5 (CM5) sensor chip. An anti-human IgG (Fc) was immobilized similarly on the experimental and reference flow cells by using standard amine coupling. For affinity measurements, recombinant human GM-CSF (rhGM-CSF) was injected over the mAb-coated cell and reference cell at the following concentrations: 100, 50, 25, 12.5, 6.25, 3.12, 1.5, and 0.8 nM. The flow rate was 30 μL/min, and triplicate blank injections and replicate injections of the 12.5 nM rhGM-CSF were included to account for any drift and to ensure assay reproducibility. Data were fit to a 1:1 model with (for those with very fast on-rates) or without a mass transport term by using the Biacore 3000 evaluation software.
Statistical Analysis.
A nonparametric Spearman correlation test was used to assess the correlation between mAb potency and affinity for GM-CSF using Statview.
Modeling the GM-CSF Ternary Complex.
The ternary model shown in Fig. 3 is based partly on the crystal structure of the published ternary GM-CSF:receptor complex (Protein Data Bank ID code 3CXE). However, the GMR-α extracellular region in this complex was incomplete, with only the extracellular domain 3 (D3) of GMR-α well defined (26). Thus, extracellular domains 1 [N-terminal domain (NTD)] and 2 (D2) of GMR-α were modeled based on a partially refined crystal structure of the GM-CSF:GMR-α binary complex and the crystal structure of the homologous IL-5 receptor α subunit (27). Our GM-CSF:GMR-α binary complex data suggest that the GMR-α NTD–D2 linker is flexible and that the NTD may adopt a variety of orientations with respect to D2 and D3.
Supplementary Material
Acknowledgments
We thank the staff of the Royal Melbourne Hospital and the Toronto General Hospital; Dr. John Seymour; Dr. Christopher Brown; and the University of British Columbia Centre for Biothermodynamics. This study was supported by the Canadian Institutes of Health Research and the Canadian Arthritis Network (J.W.S.); by a National Health and Medical Research Council of Australia (NHMRC) Development Grant (to J.A.H., J.W.S., and G.P.A.) and Program Grant 565217 (to A.L.); and infrastructure support from the Victorian State Government Operational Infrastructure Support Program (M.W.P.). J.W.S. is a Canada Research Chair and M.W.P. and J.A.H. are Senior Principal Research Fellows of the NHMRC.
Footnotes
Conflict of interest statement: G.P.A. and J.A.H. have a relevant licensed patent, and JWS has a relevant patent application.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216011110/-/DCSupplemental.
References
- 1.Silverstein AM. Autoimmunity versus horror autotoxicus: The struggle for recognition. Nat Immunol. 2001;2(4):279–281. doi: 10.1038/86280. [DOI] [PubMed] [Google Scholar]
- 2.Zenewicz LA, Abraham C, Flavell RA, Cho JH. Unraveling the genetics of autoimmunity. Cell. 2010;140(6):791–797. doi: 10.1016/j.cell.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burnet FM. A reassessment of the forbidden clone hypothesis of autoimmune disease. Aust J Exp Biol Med Sci. 1972;50(1):1–9. doi: 10.1038/icb.1972.1. [DOI] [PubMed] [Google Scholar]
- 4.Watson CT, Breden F. The immunoglobulin heavy chain locus: Genetic variation, missing data, and implications for human disease. Genes Immun. 2012;13(5):363–373. doi: 10.1038/gene.2012.12. [DOI] [PubMed] [Google Scholar]
- 5.Breden F, et al. Comparison of antibody repertoires produced by HIV-1 infection, other chronic and acute infections, and systemic autoimmune disease. PLoS ONE. 2011;6(3):e16857. doi: 10.1371/journal.pone.0016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foreman AL, Van de Water J, Gougeon M-L, Gershwin ME. B cells in autoimmune diseases: Insights from analyses of immunoglobulin variable (Ig V) gene usage. Autoimmunity Rev. 2007;6(6):387–401. doi: 10.1016/j.autrev.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Niro R, et al. High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions. Nat Med. 2012;18(3):441–445. doi: 10.1038/nm.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowley D, Jenkin CR. Antigenic cross-reaction between host and parasite as a possible cause of pathogenicity. Nature. 1962;193:151–154. doi: 10.1038/193151a0. [DOI] [PubMed] [Google Scholar]
- 9.Kitamura T, et al. Idiopathic pulmonary alveolar proteinosis as an autoimmune disease with neutralizing antibody against granulocyte/macrophage colony-stimulating factor. J Exp Med. 1999;190(6):875–880. doi: 10.1084/jem.190.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uchida K, et al. High-affinity autoantibodies specifically eliminate granulocyte-macrophage colony-stimulating factor activity in the lungs of patients with idiopathic pulmonary alveolar proteinosis. Blood. 2004;103(3):1089–1098. doi: 10.1182/blood-2003-05-1565. [DOI] [PubMed] [Google Scholar]
- 11.Sakagami T, et al. Human GM-CSF autoantibodies and reproduction of pulmonary alveolar proteinosis. N Engl J Med. 2009;361(27):2679–2681. doi: 10.1056/NEJMc0904077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trapnell BC, Carey BC, Uchida K, Suzuki T. Pulmonary alveolar proteinosis, a primary immunodeficiency of impaired GM-CSF stimulation of macrophages. Curr Opin Immunol. 2009;21(5):514–521. doi: 10.1016/j.coi.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metcalf D. Hematopoietic cytokines. Blood. 2008;111(2):485–491. doi: 10.1182/blood-2007-03-079681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handman E, Burgess AW. Stimulation by granulocyte-macrophage colony-stimulating factor of Leishmania tropica killing by macrophages. J Immunol. 1979;122(3):1134–1137. [PubMed] [Google Scholar]
- 15.Hamilton JA, Stanley ER, Burgess AW, Shadduck RK. Stimulation of macrophage plasminogen activator activity by colony-stimulating factors. J Cell Physiol. 1980;103(3):435–445. doi: 10.1002/jcp.1041030309. [DOI] [PubMed] [Google Scholar]
- 16.Lopez AF, et al. Recombinant human granulocyte-macrophage colony-stimulating factor stimulates in vitro mature human neutrophil and eosinophil function, surface receptor expression, and survival. J Clin Invest. 1986;78(5):1220–1228. doi: 10.1172/JCI112705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hercus TR, et al. The granulocyte-macrophage colony-stimulating factor receptor: Linking its structure to cell signaling and its role in disease. Blood. 2009;114(7):1289–1298. doi: 10.1182/blood-2008-12-164004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonfield TL, Kavuru MS, Thomassen MJ. Anti-GM-CSF titer predicts response to GM-CSF therapy in pulmonary alveolar proteinosis. Clin Immunol. 2002;105(3):342–350. doi: 10.1006/clim.2002.5301. [DOI] [PubMed] [Google Scholar]
- 19.Carey B, Trapnell BC. The molecular basis of pulmonary alveolar proteinosis. Clin Immunol. 2010;135(2):223–235. doi: 10.1016/j.clim.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anhalt GJ, Labib RS, Voorhees JJ, Beals TF, Diaz LA. Induction of pemphigus in neonatal mice by passive transfer of IgG from patients with the disease. N Engl J Med. 1982;306(20):1189–1196. doi: 10.1056/NEJM198205203062001. [DOI] [PubMed] [Google Scholar]
- 21.Toyka KV, et al. Myasthenia gravis. Study of humoral immune mechanisms by passive transfer to mice. N Engl J Med. 1977;296(3):125–131. doi: 10.1056/NEJM197701202960301. [DOI] [PubMed] [Google Scholar]
- 22.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453(7195):667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uchida K, et al. GM-CSF autoantibodies and neutrophil dysfunction in pulmonary alveolar proteinosis. N Engl J Med. 2007;356(6):567–579. doi: 10.1056/NEJMoa062505. [DOI] [PubMed] [Google Scholar]
- 24.Rathanaswami P, et al. Demonstration of an in vivo generated sub-picomolar affinity fully human monoclonal antibody to interleukin-8. Biochem Biophys Res Commun. 2005;334(4):1004–1013. doi: 10.1016/j.bbrc.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Hercus TR, et al. Identification of residues in the first and fourth helices of human granulocyte-macrophage colony-stimulating factor involved in biologic activity and in binding to the alpha- and beta-chains of its receptor. Blood. 1994;83(12):3500–3508. [PubMed] [Google Scholar]
- 26.Hansen G, et al. The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell. 2008;134(3):496–507. doi: 10.1016/j.cell.2008.05.053. [DOI] [PubMed] [Google Scholar]
- 27.Patino E, et al. Structure analysis of the IL-5 ligand-receptor complex reveals a wrench-like architecture for IL-5Rα. Structure. 2011;19(12):1864–1875. doi: 10.1016/j.str.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 28.Laver WG, Air GM, Webster RG, Smith-Gill SJ. Epitopes on protein antigens: Misconceptions and realities. Cell. 1990;61(4):553–556. doi: 10.1016/0092-8674(90)90464-p. [DOI] [PubMed] [Google Scholar]
- 29.Thomson CA, et al. Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus. EMBO J. 2008;27(19):2592–2602. doi: 10.1038/emboj.2008.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt AG, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci USA. 2013;110(1):264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steidl S, Ratsch O, Brocks B, Dürr M, Thomassen-Wolf E. In vitro affinity maturation of human GM-CSF antibodies by targeted CDR-diversification. Mol Immunol. 2008;46(1):135–144. doi: 10.1016/j.molimm.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe M, et al. Anti-cytokine autoantibodies are ubiquitous in healthy individuals. FEBS Lett. 2007;581(10):2017–2021. doi: 10.1016/j.febslet.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 33.Uchida K, et al. Granulocyte/macrophage-colony-stimulating factor autoantibodies and myeloid cell immune functions in healthy subjects. Blood. 2009;113(11):2547–2556. doi: 10.1182/blood-2009-05-155689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adelstein S, et al. Induction of self-tolerance in T cells but not B cells of transgenic mice expressing little self antigen. Science. 1991;251(4998):1223–1225. doi: 10.1126/science.1900950. [DOI] [PubMed] [Google Scholar]
- 35.Ottonello L, et al. Synovial fluid from patients with rheumatoid arthritis inhibits neutrophil apoptosis: Role of adenosine and proinflammatory cytokines. Rheumatology (Oxford) 2002;41(11):1249–1260. doi: 10.1093/rheumatology/41.11.1249. [DOI] [PubMed] [Google Scholar]
- 36.Bazin R, St-Amour I, Laroche A, Lemieux R. Activated cryptic granulocyte-macrophage colony-stimulating factor autoantibodies in intravenous immunoglobulin preparations. Blood. 2010;115(2):431. doi: 10.1182/blood-2009-08-240309. [DOI] [PubMed] [Google Scholar]
- 37.Felix NJ, et al. Alloreactive T cells respond specifically to multiple distinct peptide-MHC complexes. Nat Immunol. 2007;8(4):388–397. doi: 10.1038/ni1446. [DOI] [PubMed] [Google Scholar]
- 38.Kisand K, et al. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J Exp Med. 2010;207(2):299–308. doi: 10.1084/jem.20091669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puel A, et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J Exp Med. 2010;207(2):291–297. doi: 10.1084/jem.20091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Zenzo G, et al. Pemphigus autoantibodies generated through somatic mutations target the desmoglein-3 cis-interface. J Clin Invest. 2012;122(10):3781–3790. doi: 10.1172/JCI64413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dal Porto JM, Haberman AM, Kelsoe G, Shlomchik MJ. Very low affinity B cells form germinal centers, become memory B cells, and participate in secondary immune responses when higher affinity competition is reduced. J Exp Med. 2002;195(9):1215–1221. doi: 10.1084/jem.20011550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lingwood D, et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489(7417):566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akkaraju S, Canaan K, Goodnow CC. Self-reactive B cells are not eliminated or inactivated by autoantigen expressed on thyroid epithelial cells. J Exp Med. 1997;186(12):2005–2012. doi: 10.1084/jem.186.12.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, et al. Human antibodies for immunotherapy development generated via a human B cell hybridoma technology. Proc Natl Acad Sci USA. 2006;103(10):3557–3562. doi: 10.1073/pnas.0511285103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol. 2008;8(7):533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 46.Cook AD, Braine EL, Campbell IK, Rich MJ, Hamilton JA. Blockade of collagen-induced arthritis post-onset by antibody to granulocyte-macrophage colony-stimulating factor (GM-CSF): Requirement for GM-CSF in the effector phase of disease. Arthritis Res. 2001;3(5):293–298. doi: 10.1186/ar318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McQualter JL, et al. Granulocyte macrophage colony-stimulating factor: A new putative therapeutic target in multiple sclerosis. J Exp Med. 2001;194(7):873–882. doi: 10.1084/jem.194.7.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Codarri L, et al. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12(6):560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 49.El-Behi M, et al. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol. 2011;12(6):568–575. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iversen PO, et al. Inhibition of granulocyte-macrophage colony-stimulating factor prevents dissemination and induces remission of juvenile myelomonocytic leukemia in engrafted immunodeficient mice. Blood. 1997;90(12):4910–4917. [PubMed] [Google Scholar]
- 51.Bayne LJ, et al. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 2012;21(6):822–835. doi: 10.1016/j.ccr.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell. 2012;21(6):836–847. doi: 10.1016/j.ccr.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schweizerhof M, et al. Hematopoietic colony-stimulating factors mediate tumor-nerve interactions and bone cancer pain. Nat Med. 2009;15(7):802–807. doi: 10.1038/nm.1976. [DOI] [PubMed] [Google Scholar]
- 54.Cook AD, et al. Granulocyte-macrophage colony-stimulating factor is a key mediator in inflammatory and arthritic pain. Ann Rheum Dis. 2013;72(2):265–270. doi: 10.1136/annrheumdis-2012-201703. [DOI] [PubMed] [Google Scholar]
- 55.Thomson CA, et al. Pandemic H1N1 influenza infection and vaccination in humans induces cross-protective antibodies that target the hemagglutinin stem. Front Immunol. 2012;3:87. doi: 10.3389/fimmu.2012.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McLean GR, et al. Recognition of human cytomegalovirus by human primary immunoglobulins identifies an innate foundation to an adaptive immune response. J Immunol. 2005;174(8):4768–4778. doi: 10.4049/jimmunol.174.8.4768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.