Abstract
Microhomology-mediated end joining (MMEJ) is a major pathway for Ku-independent alternative nonhomologous end joining, which contributes to chromosomal translocations and telomere fusions, but the underlying mechanism of MMEJ in mammalian cells is not well understood. In this study, we demonstrated that, distinct from Ku-dependent classical nonhomologous end joining, MMEJ—even with very limited end resection—requires cyclin-dependent kinase activities and increases significantly when cells enter S phase. We also showed that MMEJ shares the initial end resection step with homologous recombination (HR) by requiring meiotic recombination 11 homolog A (Mre11) nuclease activity, which is needed for subsequent recruitment of Bloom syndrome protein (BLM) and exonuclease 1 (Exo1) to DNA double-strand breaks (DSBs) to promote extended end resection and HR. MMEJ does not require S139-phosphorylated histone H2AX (γ-H2AX), suggesting that initial end resection likely occurs at DSB ends. Using a MMEJ and HR competition repair substrate, we demonstrated that MMEJ with short end resection is used in mammalian cells at the level of 10–20% of HR when both HR and nonhomologous end joining are available. Furthermore, MMEJ is used to repair DSBs generated at collapsed replication forks. These studies suggest that MMEJ not only is a backup repair pathway in mammalian cells, but also has important physiological roles in repairing DSBs to maintain cell viability, especially under genomic stress.
Keywords: BLM/Exo1, CtIP, DNA repair pathway, DNA damage, genome stability
DNA double-strand breaks (DSBs) can be repaired by multiple pathways. The classical nonhomologous end joining (C-NHEJ) pathway relies on Ku70/Ku80 and ligates DSB ends without a template (1). Homologous recombination (HR), an error-free pathway, uses a homologous template to repair DSBs (2) and is initiated by end resection from DSB ends to generate a long stretch of single-strand DNA (ssDNA) for strand invasion. Although C-NHEJ is active throughout the cell cycle, HR is used when cells enter S and G2 because cyclin-dependent kinases (CDKs) are needed for promoting end resection to activate HR (3–5).
In the absence of C-NHEJ factors such as Ku70, Ku80, or DNA ligase IV, robust alternative nonhomologous end joining (alt-NHEJ) activity is observed in various organisms including yeast and mammals (6, 7). Many alt-NHEJ events, classified as microhomology-mediated end joining (MMEJ), require end resection and join the ends by base pairing at microhomology sequences (5–25 nucleotides), resulting in deletions at the junctions (6). However, other alt-NHEJ pathways without using microhomology regions also exist.
Genetic analyses in yeast reveal that MMEJ is Rad52-independent, distinguishing it from HR and single-strand annealing (SSA) repair pathways, whereas the Mre11-Rad50-Xrs2 (MRX) complex and DNA ligase IV are needed for MMEJ (8–10). Further studies suggest that in yeast, Srs2 helicase, Sae2 nuclease [CtBP-interacting protein (CtIP) homologue], Tel1 [ataxia telangiectasia mutated (ATM) homologue], and DNA polymerases (Pol4, Rev3, and Pol32) are important for MMEJ (11). In mammalian cells, Mre11, CtIP, and DNA ligase III have critical roles in MMEJ (12–17).
MMEJ is error-prone and contributes to genome instabilities, such as chromosomal translocations and telomere fusions (18–20), but its use and regulation under normal physiological conditions in mammalian cells are unclear. In this study, we demonstrated that MMEJ is distinct from Ku-dependent C-NHEJ but shares the initial end resection step with HR in a CDK-dependent manner. The Mre11 nuclease activity is required for initial short-range end resection, even of fewer than 20 bp, at DSB ends to promote MMEJ, which is essential for subsequent recruitment of BLM and Exo1 to DSBs to launch extended end resection and activate HR. Using a MMEJ and HR competition repair substrate, we showed that MMEJ with short-range end resection is used at a substantial frequency to repair DSBs in normal cycling cells even when both C-NHEJ and HR pathways are available. Furthermore, MMEJ is activated when replication forks are collapsed, suggesting that MMEJ has critical biological functions to cope with genomic stress in mammalian cells.
Results
Mre11 Nuclease Activity Is Required for Initial End Resection of Fewer than 20 bp to Promote MMEJ, Even in the Absence of Ku70.
We constructed an EGFP-based MMEJ repair substrate, EGFP-MMEJ, which requires short-range end resection (14 or 18 bp on either side of the DSB) to reveal the 9-bp microhomology regions for annealing to create a functional EGFP after I-SceI cleavage (Fig. 1A and Fig. S1 A and B) (21). We designed this MMEJ substrate with short-range end resection for two reasons. First, C-NHEJ does not require end resection but often processes small deletions of 1–4 bp at noncompatible DSB ends (1, 5, 22). In comparison, the MMEJ construct was designed to produce the smallest possible deletion (27 bp) upon end resection and repair. Short-range end resection also distinguishes this MMEJ substrate from HR and SSA, which both need long-range end resection. Second, studies in yeast suggest that end resection is carried out by initial end resection (100–200 bp) followed by extended end resection (23). In mammalian cells, although replication protein A (RPA) foci formation and checkpoint kinase 1 (Chk1) phosphorylation are used to monitor extended end resection (24), no assays readily exist for examining initial short-range end resection. Our designed MMEJ substrate allows us to monitor DSB repair requiring fewer than 20 bp of end resection.
Fig. 1.
The nuclease activity of Mre11 is important for MMEJ. (A) EGFP-MMEJ DSB repair substrate. (B–E) U2OS cells with single integration of EGFP-MMEJ or EGFP-HR substrate and stably expressing shRNAs against Ku70 and/or Mre11 or No, with or without stably expressed Myc- or HA-tagged Mre11 (WT or H129N), as indicated (Fig. S4), were induced by I-SceI and assayed. For this figure and all of the following figures in the main text, data shown are mean of three independent experiments, with error bars as SD and P values as noted: *P ≤ 0.05; **P ≤ 0.01; and ***P ≤ 0.001.
We generated stable human osteosarcoma U2OS and glioblastoma T98G cell lines carrying a single integrated copy of EGFP-MMEJ at different genomic loci, and substantial levels of MMEJ (varying from 0.3% to 2.5%) were detected after I-SceI expression with unperturbed HR or NHEJ function (Fig. S1 C and D). Using our HR repair substrate (Fig. S2A) (21), we found that suppressing Ku70 increased HR and elevated MMEJ in U2OS single clone cell lines, and a more profound effect was observed in a Ku70-deficient mouse embryo fibroblast (MEF) cell population carrying EGFP-MMEJ (Fig. 1B and Fig. S2 B and C) (15). Thus, although suppressed by C-NHEJ, MMEJ is functionally active in mammalian cells even when both HR and NHEJ are available.
Both the Mre11-Rad50-Nbs1 complex (MRN) and CtIP are required for MMEJ in mammalian cells (12–15), and similar results were obtained using our EGFP-MMEJ substrate, where the relative reduction of MMEJ in the absence of MRN or CtIP was similar to that of HR (Fig. S3). The Mre11-H129N nuclease mutant (25) was also impaired in MMEJ to a similar extent as HR (Fig. 1C and Fig. S4A). These studies suggest that Mre11 nuclease activity not only is important for HR, but also is required for MMEJ involving short-range end resection.
In yeast, the need of MRX for end resection can be bypassed in the absence of Ku (26, 27). Interestingly, we found that elevated MMEJ due to Ku70 deficiency was greatly reduced by loss of Mre11 or Mre11 nuclease activity (Fig. 1 D and E and Fig. S4 B and C) This suggests that, in mammalian cells, Mre11 and its nuclease activity are indispensable for MMEJ and HR, even in the absence of Ku at the endonuclease-generated “clean” DSB ends.
MMEJ Is Increased When Cells Enter the S Phase.
To examine MMEJ activity during the cell cycle, we generated an inducible I-SceI construct, DD-HA-I-SceI-GR (Fig. 2A, Left). This construct contains a destabilization domain (DD) for rapid degradation of HA-I-SceI and is stabilized by the Shield1 ligand (Fig. 2A, Lower Left) (28). I-SceI is also fused to the glucocorticoid receptor (GR) ligand-binding domain, which leads to nuclear localization upon addition of triamcinolone acetonide (TA) (Fig. 2A, Right, and Fig. S5A) (29). After adding TA and Shield1, DD-HA-I-SceI-GR efficiently induces HR and MMEJ (Fig. S5B).
Fig. 2.
MMEJ increases when cells enter S/G2. (A) Inducible I-SceI construct, DD-HA-I-SceI-GR. Stabilization (immunoblot) and localization (immunostaining) after Shield1 (2 h) or TA (15 min). (B) T98G cells with single integration of MMEJ substrate were asynchronized, G0/G1-arrested, or G0/G1-arrested and released and assayed. (C) Experimental scheme and assay for repair in T98G (MMEJ) cells arrested at G0/G1 or released to Lov-, Thy-, or Noc-containing medium. (D) U2OS cells with single integration of MMEJ substrate treated ± Rosc for 24 h or expressing Cdk2 shRNAs were assayed.
T98G cells can be synchronized by serum starvation (Fig. S5C) (30). We expressed DD-HA-I-SceI-GR in T98G (MMEJ) single integration cell lines and assayed for MMEJ. MMEJ in G0/G1-arrested (serum-starved) cells was substantially low compared with asynchronous cells or cells released from serum starvation (Fig. 2B and Fig. S5D). We also induced I-SceI upon releasing to lovastatin (Lov)-containing medium to prevent cells from entering S phase (31) and showed that MMEJ was significantly lower than those cells released to thymidine (Thy)- and nocodazole (Noc)-containing medium, which were arrested in early S phase or had experienced G1, S, and G2 before being arrested in early mitosis (Fig. 2C and Fig. S5E). The expression level and nuclear localization of induced I-SceI were similar throughout the cell cycle (Fig. S6 A and B). These data suggest that MMEJ activity is low in G0 and G1 and is significantly elevated in cycling cells and when cells enter S and G2. Therefore, MMEJ with short-range end resection to remove fewer than 20 bp at DSBs is regulated during the cell cycle in mammalian cells.
Cdk2 Promotes MMEJ, and CtIP Is an Important Target for This Regulation.
Since MMEJ is increased upon S-phase entry, we probed whether CDK is needed to activate MMEJ. Roscovitine (Rosc) treatment (2 µM) suppressed MMEJ (Fig. 2D, Left). Significantly, expression of shRNAs for Cdk2 also reduced MMEJ (Fig. 2D, Right, and Fig. S6C). This suggests that MMEJ, even with very short end resection, is promoted by CDK activities.
CtIP is also important for MMEJ (13, 15), and CDK-mediated phosphorylation of CtIP at site S327 facilitates CtIP interaction with BRCA1 to promote HR but not MMEJ (32–34). We showed that mutating another conserved CDK site (T847) (35) reduced both MMEJ and HR (Fig. S6D), implicating CtIP as one critical target of CDKs to promote not only HR but also the initial short-range end resection to activate MMEJ.
MMEJ Is Used at the Levels of 10–20% of HR to Repair DSBs in Mammalian Cells.
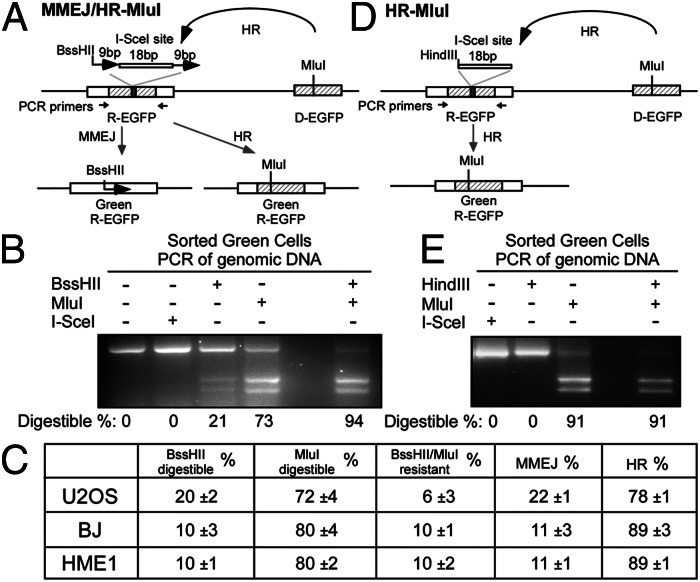
We observed MMEJ activity in cells with unperturbed HR and NHEJ pathways. To directly compare the relative frequency of MMEJ with HR, we generated an EGFP-based MMEJ and HR competition substrate, EGFP-MMEJ/HR-MluI (Fig. 3A), where the recipient EGFP (R-EGFP) cassette of EGFP-HR (Fig. S2A) was replaced with the EGFP-MMEJ cassette (Fig. 1A). A unique MluI site in the donor EGFP (D-EGFP) cassette was created via a silent mutation at the BssHII site.
Fig. 3.
MMEJ is used frequently to repair DSBs in mammalian cells. (A) EGFP-MMEJ/HR-MluI competition repair substrate. (B) U2OS cells with single integration of EGFP-MMEJ/HR-MluI substrate were induced with I-SceI and sorted for EGFP-positive cells, and PCR and digestion analysis was performed with percentages of digestible products shown. (C) U2OS (from B) and BJ and HME1 cell populations carrying EGFP-MMEJ/HR-MluI were assayed as in B, with repair frequencies of three independent experiments. (D and E) EGFP-HR-MluI repair substrate and assay.
Upon I-SceI cleavage, restoration of a functional EGFP cassette results in loss of the I-SceI site after cells undergo MMEJ or HR. PCR analysis of the sorted green cells (Fig. S7A) using primers specific for R-EGFP (Fig. 3A) was performed. Repair by MMEJ would retain one copy of the 9-bp duplication with the BssHII site, and the percentage of BssHII-digestible bands among total PCR products reflects the MMEJ frequency. Repair by HR transfers the MluI site from D-EGFP to R-EGFP, and thus the percentage of MluI-digestible bands reflects the HR frequency.
As shown in Fig. 3B, BssHII and MluI digestions yielded 21% and 73% digestible bands, respectively, among total PCR products. We also observed 6% (100% minus 94%) of the PCR products resistant to digestion by both BssHII and MluI, which we attributed to contamination of nongreen cells in the sorted green cell population (Fig. S7B). These contaminant cells that have lost the BssHII site without gaining the MluI site have likely used repair mechanisms other than the designed MMEJ and HR. When excluded, we estimate that the percentage of using MMEJ to HR in U2OS cells is about 22% versus 78% (Fig. 3C). Similar results were obtained when the EGFP-MMEJ/HR-MluI substrate was integrated at a different genomic locus (Fig. S7C).
To show the efficiency of MluI to mark HR-repaired events, we generated the EGFP-HR-MluI substrate, where the R-EGFP contains a HindIII site instead of 9-bp duplication, and the D-EGFP contains the MluI site (Fig. 3D and Fig. S7D). EGFP in this substrate can be restored only by HR, as assayed by gain of MluI site and loss of HindIII site (Fig. 3D and Fig. S7E). Ninety-one percent of the PCR products from sorted green cells were digested by MluI with no detectable HindIII-digested bands (Fig. 3E). The remaining 9% HindIII- and MluI-resistant band was due to contaminant nongreen cells. This suggests that virtually all HR-repaired, EGFP-positive products were marked by MluI within the detectable resolution.
We further examined the use of MMEJ and HR in primary cells including telomerase-immortalized human primary fibroblast (BJ) cells and human mammary epithelium (HME1) cells. We generated cell populations carrying EGFP-MMEJ/HR-MluI and found that the ratio of MMEJ to HR was ∼11% versus 89% (Fig. 3C), which is relatively lower than in the U2OS tumor cell line but still at a substantial level. These studies suggest that MMEJ is used in mammalian cells at the levels of 10–20% of HR when both HR and NHEJ mechanisms are also available.
BLM and Exo1 Are Not Required for Initial End Resection.
Using the EGFP-HR/MMEJ-MluI competition substrate, we showed that the relative ratio of MMEJ to HR was largely unaffected despite the overall increased repair frequency in Ku70-deficient cells and decreased repair frequency in Mre11-, CtIP-, or Cdk2-deficient cells (Fig. 4A and Fig. S8A). In addition, the ratio of MMEJ to HR in the thymidine-arrested early S-phase cells was similar to asynchronous cells, suggesting that MMEJ and HR are increased proportionally when cells enter the S phase (Fig. S8 B and C). Thus, HR and MMEJ are similarly regulated by Ku70, Mre11, CtIP, and Cdk2, and MMEJ and HR likely share the same initial end resection step in a CDK-dependent manner, which is promoted by Mre11 and CtIP but suppressed by the Ku complex.
Fig. 4.
The role of various repair proteins and Cdk2 in MMEJ and HR. (A and C) U2OS cells with single integration of EGFP-MMEJ/HR-MluI substrate and stably expressing Ku70, Mre11, CtIP, Cdk2, BLM, or Exo1 shRNAs or control were assayed as in Fig. 3. (B and D) U2OS (EGFP-HR) or U2OS (EGFP-MMEJ) cells with BLM, Exo1, or H2AX shRNAs or control were induced with I-SceI and assayed.
Like Mre11 and CtIP, BLM and Exo1 are required for extended end resection and HR in mammalian cells (Fig. 4B, Left) (36). Depletion of BLM or Exo1 did not reduce but increased MMEJ activity (Fig. 4B, Right, and Fig. S8D) in U2OS cells and in Exo1 knockout MEFs, which was suppressed with exogenous HA-Exo1 expression (Fig. S8E). Consistently, when BLM or Exo1 was inactivated in U2OS (EGFP-MMEJ/HR-MluI) cells, the MMEJ-to-HR ratio was increased (Fig. 4C, Lower table), with a slight decrease of overall repair frequency (Fig. 4C, Upper). These data support that end resection in mammalian cells is carried out by two distinct steps. While the nuclease activity of Mre11 is required for the initial end resection to promote MMEJ, BLM and Exo1 are dispensable for this process and instead promote extended end resection to activate HR.
MMEJ Occurs at DSB Ends and Is Independent of γH2AX.
Upon DSB formation, ATM-mediated phosphorylation of H2AX recruits DSB repair proteins to DSB-flanking chromatin regions (37). H2AX is important for HR-mediated DSB repair (Fig. 4D, Left) (38). However, we found that MMEJ was not reduced but elevated when H2AX was inactivated (Fig. 4D, Right). These data suggest that, in contrast to HR, H2AX is dispensable for MMEJ, and thus the initial end resection at the DSB ends required for MMEJ is independent of γH2AX chromatin recruitment activity.
Initial End Resection Is Important for Recruiting BLM and Exo1 to DSBs.
In yeast, Exo1 and Dna2 endonuclease/helicase binding to DSBs depends on Mre11 but not on its nuclease activity (27). We monitored the recruitment of BLM and Exo1 to laser-generated DSBs in live mammalian cells. Interestingly, BLM and Exo1 recruitment to the damage sites was impaired not only in Mre11 and CtIP knockdown cells (Fig. 5A and Fig. S9A) but also in the Mre11-H129N nuclease mutant cell line (Fig. 5B and Fig. S9B). These studies suggest that the initial end resection activity mediated by the nuclease activity of Mre11 is important for subsequent recruitment of BLM and Exo1 to chromosomal DSBs to promote extended end resection to activate HR (Fig. 5C).
Fig. 5.

Recruitment of BLM and Exo1 to laser-induced DSBs depends on Mre11 nuclease activity. U2OS cells stably expressing enhanced blue fluorescent protein-marked Mre11 or CtIP shRNAs or control (A), or HA-Mre11 (WT or H129N) with endogenous Mre11 silenced (B), were transfected with monomeric red fluorescent protein-marked BLM or Exo1. Laser microirradiation, live-cell imaging, and recruitment analyses were performed as described in SI Materials and Methods. (C) A proposed two-step model for the regulation of MMEJ and HR in mammalian cells.
MMEJ Is Used to Repair DSBs Occurring at Collapsed Replication Forks.
U2OS cells with EGFP-HR or EGFP-MMEJ were sorted to remove accumulated green cells and cultured. Spontaneous recombination was readily observed in U2OS (EGFP-HR) cells (Fig. 6A, Left, and Fig. S10A), possibly due to replication restart by short-track gene conversion from stalled replication forks (Fig. 6F, Top) (39), but relatively low spontaneous MMEJ was detected (Fig. 6A, Right). However, when U2OS (EGFP-HR) or U2OS (EGFP-MMEJ) cells were treated with a high dose of hydroxyurea (HU) (2 mM for 24 or 34 h), fork collapse was induced (39) and led to a higher frequency of MMEJ repair compared with HR (Fig. 6B). Ataxia-telangiectasia and Rad3-related (ATR) and the Claspin/Timeless (Tim)/Tipin complex are important for protecting stalled forks from collapse (40, 41). We inactivated ATR or Tim by shRNAs, which would cause fork collapse even without HU treatment, and observed increased MMEJ with and without HU (Fig. 6C). Inactivation of Tim resulted in DSB formation, as revealed by H2AX phosphorylation (Fig. S10B) (42), but did not influence HR and MMEJ to repair I-SceI–induced DSBs (Fig. S10C). Thus, increased MMEJ in Tim-deficient cells observed without HU treatment (Fig. 6C, Right) is induced by accumulated DSBs at collapsed replication forks due to Tim deficiency in the absence of exogenous DNA-damaging agents.
Fig. 6.
MMEJ is used frequently to repair DSBs at collapsed replication forks. (A and B) U2OS cells with single integration of EGFP-HR and EGFP-MMEJ substrate were sorted for EGFP-negative cells and (in A) assayed for EGFP-positive events or (in B) treated with 2 mM HU, recovered for 14 h and assayed. (C) U2OS (EGFP-MMEJ) with ATR- or Tim-shRNAs or control were treated with HU (as in B) and assayed. (D and E) U2OS (EGFP-MMEJ) cells with LigIII shRNA or control were induced with I-SceI (in D) or HU (in E, treated as in C) and assayed. (F) Model for replication restart and DSB repair at collapsed replication forks (Discussion).
DNA ligase III is required for MMEJ (Fig. 6D and Fig. S10D) (16, 17). Its inactivation reduced HU-induced MMEJ, suggesting that DNA ligase III is needed for MMEJ at collapsed forks (Fig. 6E), and further supporting that MMEJ is actively used to repair DSBs at collapsed replication forks.
Discussion
MMEJ is Ku-independent and often considered as a backup alt-NHEJ mechanism (7), yet some evidence also suggests that it might be active when C-NHEJ is available. In yeast, MMEJ is used at a comparable level to C-NHEJ in response to IR (8). In mammals, MMEJ was observed in the presence of normal C-NHEJ function when using truncation mutants of V(D)J recombinase proteins Rag1 and Rag2 during recombination and when forming telomere fusions (18, 20). In this study, we observed substantial MMEJ activity in mammalian cells when C-NHEJ and HR repair pathways were unperturbed and when DSBs were generated by I-SceI–producing 4-bp complementary overhangs, which are optimal substrates for C-NHEJ. Using a MMEJ and HR competition repair substrate, we found that MMEJ is used at a level ∼20% of HR in U2OS cells and about 10% of HR in primary nontumor cells. Thus, MMEJ does not merely serve as a backup repair mechanism but exhibits substantial activities in mammalian cycling cells. Differences of MMEJ frequency in U2OS and other cell lines may be due to the tumor background or cell-type differences, which will be investigated further.
Because MMEJ is used during the normal cell cycle and in the presence of Ku-dependent C-NHEJ, we define MMEJ as a “Ku-independent” rather than as a “Ku-alternative” (in the absence of Ku) end-joining mechanism, which uses microhomology sequences (5–25 bp) for end joining (6). The concept of MMEJ is not interchangeable with Ku-independent NHEJ or alt-NHEJ, as MMEJ constitutes only one end-joining repair mechanism, although it could be a major one, of probably multiple Ku-independent NHEJ or alt-NHEJ repair pathways.
MMEJ requires end resection to expose microhomology for annealing (6), and this resection can be very limited, as in our MMEJ substrate with fewer than 20 bp processed, compared with HR that requires extended end resection. Interestingly, we found that MMEJ, even with short-range end resection is cell-cycle regulated and is significantly elevated when cells enter S phase. This is different from C-NHEJ, which is active throughout the cell cycle (1, 5). We also found that CDK activity is important for promoting MMEJ like HR, and CtIP is an important CDK target for both MMEJ and HR. We thus propose that MMEJ shares the initial end resection step with HR. This is also supported by the MMEJ/HR competition experiments in which Cdk2, MRN, and CtIP as well as the Mre11 nuclease activity are equally required for both MMEJ and HR, and the Ku complex suppresses MMEJ and HR to a similar extent.
Our studies support the two-step end resection model proposed in yeast (23). We found that whereas the Mre11 complex and CtIP are required for the initial short-range end resection to promote MMEJ, BLM and Exo1 are dispensable for this process but are needed for extended end resection and HR (Fig. 5C) (36, 43), thus revealing different genetic requirements for the two distinct end resection steps in mammalian cells. Notably, the need for Mre11 nuclease activity in end resection and HR is different in yeast and mammals. First, Mre11-H129N knock-in mice are embryonic lethal (25), whereas the yeast Mre11 nuclease-defective mutants are viable with only mild sensitivity to IR (44). Second, the yeast Mre11 complex and Sae2 participate in initial end resection, followed by long-range end resection mediated by Sgs1 (helicase)/Dna2 and Exo1, but loss of Mre11 or Sae2 can be compensated by Sgs1 and Exo1 activities, although with slower resection kinetics (23). We showed that the nuclease activity of human Mre11 is required for initial end resection and cannot be substituted by other nucleases such as BLM and Exo1. Third, loss of Ku70 in yeast can bypass the requirement of MRN for end resection (26, 27), whereas, in mammalian cells, elevated MMEJ and HR in Ku70- or Ku80-deficient cells are still dependent on the Mre11 complex and Mre11 nuclease activity, even with DSB ends generated by the I-SceI endonuclease in our study. This finding is supported by the in vitro analysis using Xenopus extracts where depletion of Mre11 blocks end resection from restriction enzyme-generated DSBs from the very initial step at the first nucleotide (45). Therefore, Mre11-nuclease activity is necessary in mammalian cells for initial processing of the 5′ strand to generate 3′ ssDNA even from the “clean” DSB ends free of Ku complex binding.
In yeast, Mre11 interacts with and recruits Sgs1 to DSB ends (46, 47). In mammalian cells, the physical interaction of MRN with BLM/Exo1 may also promote binding of BLM and Exo1 to DSB ends and facilitate end resection (48). We further demonstrated that the Mre11 nuclease activity is required for BLM and Exo1 recruitment to DSBs. Thus, initial end resection occurring at DSB ends is needed for further loading of BLM and Exo1 to DSBs, where BLM and Exo1 may favor binding to initially processed DSB ends or to proteins that bind to resected DNA ends. Notably, biochemical analysis has shown that preresected DNA ends are better substrates for yeast Exo1 (49). We propose that, although initial end resection may facilitate BLM/Sgs1 and Exo1 recruitment to DSBs in both yeast and mammals, Mre11 nuclease activity is essential for initial end resection only in mammalian cells and thus is indispensable for BLM/Exo1 recruitment for extended end resection. These studies reveal an important underlying mechanism for requiring Mre11 nuclease activity for HR in mammals wherein the physical interactions of MRN with BLM/Exo1 may still contribute to promoting and stabilizing localization of BLM/Exo1 to DSBs. We also showed that, although H2AX is important for HR (38), it is dispensable for MMEJ. Thus, the initial end resection required for MMEJ likely occurs at DSB ends independently of H2AX chromatin recruitment function. Although the exact role of H2AX in HR needs to be further investigated, our study suggests that H2AX is involved in promoting HR after the initial step of limited end resection at DSBs. Impaired function of HR may lead to increased use of MMEJ.
MMEJ is active in normal cycling cells, and we further showed that MMEJ is actively used to repair DSBs occurring at collapsed replication forks. Observed spontaneous HR (Fig. 6A) is likely due to replication restart in a Rad51-dependent manner at stalled replication forks without fork breakage (Fig. 6F, Top) (39). In mammalian cells, it was suggested that replication does not restart at collapsed forks but is resumed upon new origin firing from adjacent origins (Fig. 6F, Left), which leads to two-ended DSBs when new forks encounter collapsed forks (39). Based on our findings of I-SceI–induced DSB repair, HR would be used eight- to ninefold more frequently than MMEJ at two-ended DSBs. However, upon fork collapse, we observed a significant increase of MMEJ, but not of HR, suggesting that other mechanisms may be involved. Due to replication stalling, the breaks at collapsed forks are surrounded by ssDNAs (Fig. 6F, Left) and cannot be repaired by C-NHEJ because the Ku complexes do not bind to DSB ends with ssDNA tails. Mus81 has been implicated in cleaving the single-strand and double-strand junctions at blocked leading strands of stalled forks to produce one-ended DSB breaks (50, 51) and, once generated, may be immediately repaired by MMEJ before HR is engaged (Fig. 6F, Right Inset). After short-range end resection at one break end, a helicase activity would be needed at the other break end to expose the microhomology for MMEJ (Fig. 6F, Right Inset). A role of helicases for MMEJ in mammalian cells has yet to be uncovered, but Srs2 is important for MMEJ in yeast (11). Intriguingly, we did not observe substantial increase of HR when replication forks are collapsed by HU treatment, suggesting that HR may not be a major pathway to repair fork collapse-induced DSBs under this circumstance. However, we cannot completely rule out the possibility that failure in observing HR on collapsed forks is because the EGFP cassettes in the artificial HR substrate need to be mis-aligned for gene conversion, which may cause constraint on chromatin leading to unfavorable use, although the EGFP-HR substrate readily reveals spontaneous HR events.
HR requires relatively time-consuming extended end resection, whereas MMEJ needs only limited end resection. Thus, under critical situations, MMEJ may be an advantageous choice for cells to quickly repair DSBs at collapsed replication forks to avoid cell death. A switch from classical break-induced replication (BIR) to microhomology-mediated BIR under cellular stress has been described (52). Although MMEJ is error-prone and by its nature causes deletions, mammalian cells may afford to tolerate this to some extent, as intron sequences are abundant in the genome. It should be noted that our observations are based on using a MMEJ substrate to process a 20-bp end resection and to anneal at a 9-bp microhomology sequence. It remains to be seen whether MMEJ requiring longer end resection or using other sizes of microhomology may be regulated differently.
Even though MMEJ is error-prone, our studies suggest that MMEJ with short-range end resection plays an important role in repairing DSBs in normal cycling cells, especially under conditions critical to cell viability. Thus, MMEJ is a double-edged sword and its precise regulation is surely critical for the balance of its function in preventing or causing genome instability.
Materials and Methods
Cell culture, RNA interference, cell lysis, immunoblotting, immunostaining, flow cytometry, and laser microirradiation are described in refs. 21 and 32. Additional details are provided in SI Materials and Methods. Analysis of EGFP-based DSB repair substrates are described in ref. 21 and in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Steven S. Reed and Leonardo Teixeira (The Scripps Research Institute), Frederick Alt (Harvard Medical School), Winfried Edelmann (Albert Einstein College of Medicine), Maria Jasin (Memorial Sloan-Kettering Cancer Center), Kum Kum Khanna (Queensland Institute of Medical Research), Marek Rusin (Maria Sklodowska-Curie Memorial Institute), and Jeremy Stark (City of Hope) for kindly providing valuable reagents, and James E. Haber (Brandeis University, Waltham, MA) and Sang Eun Lee (University of Texas Health Science Center at San Antonio, San Antonio, TX) for valuable comments. This work was supported by National Institutes of Health (NIH) Grants CA102361, GM080677, CA140972, and CA102361-07S1 (to X.W.); the Beckman Laser Institute Foundation (to M.W.B.); and by NIH Training Grant DK007022-30 (to L.N.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213431110/-/DCSupplemental.
References
- 1.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11(3):196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431(7011):1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23(24):4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothkamm K, Krüger I, Thompson LH, Löbrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23(16):5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008;24(11):529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131(2):223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol. 2003;23(23):8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Paull TT. The Mre11/Rad50/Xrs2 complex and non-homologous end-joining of incompatible ends in S. cerevisiae. DNA Repair (Amst) 2005;4(11):1281–1294. doi: 10.1016/j.dnarep.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Wilson TE, Grawunder U, Lieber MR. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388(6641):495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 11.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176(4):2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16(8):814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rass E, et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16(8):819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 14.Dinkelmann M, et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol. 2009;16(8):808–813. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4(6):e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279(53):55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65(10):4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 18.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449(7161):478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 19.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17(4):410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rai R, et al. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J. 2010;29(15):2598–2610. doi: 10.1038/emboj.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, et al. CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded breaks. J Biol Chem. 2012;287(25):21471–21480. doi: 10.1074/jbc.M112.355354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guirouilh-Barbat J, et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14(5):611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134(6):981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jazayeri A, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8(1):37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 25.Buis J, et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135(1):85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29(19):3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shim EY, et al. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010;29(19):3370–3380. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126(5):995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soutoglou E, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9(6):675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee AY, et al. Dbf4 is direct downstream target of ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) protein to regulate intra-S-phase checkpoint. J Biol Chem. 2012;287(4):2531–2543. doi: 10.1074/jbc.M111.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu JR, Gilbert DM. Lovastatin arrests CHO cells between the origin decision point and the restriction point. FEBS Lett. 2000;484(2):108–112. doi: 10.1016/s0014-5793(00)02135-9. [DOI] [PubMed] [Google Scholar]
- 32.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283(12):7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 33.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24(21):9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459(7245):460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284(14):9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22(20):2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 2011;25(5):409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie A, et al. Control of sister chromatid recombination by histone H2AX. Mol Cell. 2004;16(6):1017–1025. doi: 10.1016/j.molcel.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37(4):492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulsen RD, Cimprich KA. The ATR pathway: Fine-tuning the fork. DNA Repair (Amst) 2007;6(7):953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 41.Errico A, Costanzo V. Mechanisms of replication fork protection: A safeguard for genome stability. Crit Rev Biochem Mol Biol. 2012;47(3):222–235. doi: 10.3109/10409238.2012.655374. [DOI] [PubMed] [Google Scholar]
- 42.Smith KD, Fu MA, Brown EJ. Tim-Tipin dysfunction creates an indispensible reliance on the ATR-Chk1 pathway for continued DNA synthesis. J Cell Biol. 2009;187(1):15–23. doi: 10.1083/jcb.200905006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci USA. 2008;105(44):16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19(1):556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao S, Guay C, Toczylowski T, Yan H. 2012. Analysis of MRE11’s function in the 5′→3′ processing of DNA double-strand breaks. Nucleic Acids Res 40(10):4496–4506. [DOI] [PMC free article] [PubMed]
- 46.Niu H, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467(7311):108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiolo I, et al. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol Cell Biol. 2005;25(13):5738–5751. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nimonkar AV, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25(4):350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicolette ML, et al. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17(12):1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanada K, et al. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol. 2007;14(11):1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 51.Whitby MC, Osman F, Dixon J. Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J Biol Chem. 2003;278(9):6928–6935. doi: 10.1074/jbc.M210006200. [DOI] [PubMed] [Google Scholar]
- 52.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10(8):551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.