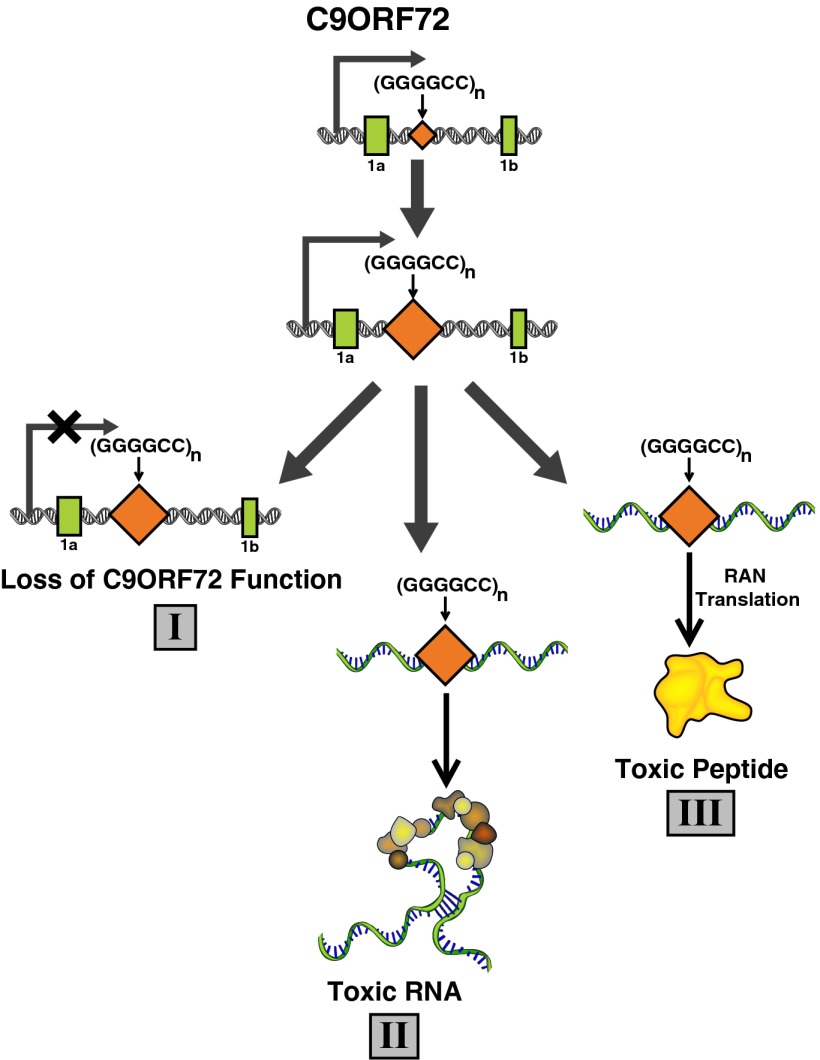
A recent and very stimulating finding in the field of neurodegenerative disease is the discovery that expansion of a hexanucleotide microsatellite DNA repeat (GGGGCC) in an intron of the C90RF72 gene is a common cause of two devastating neurodegenerative diseases for which there are no cures: frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) (1, 2). With this observation arose a fascinating question—how does expansion of this repeat induce a disease that can manifest as ALS, a motor neuron disease, as well as FTD, a leading cause of presenile dementia? Adding to the mystery is the almost complete absence of information on the C9ORF72 gene or the function of the protein it encodes. However, initial characterization of C9ORF72-FTD/ALS combined with work on some other neurodegenerative diseases suggests three pathogenic mechanisms (Fig. 1). First is the evidence indicating that mRNA levels of C9ORF72 transcripts are decreased in C9ORF72-FTD/ALS (1–3). Thus, a loss of C9ORF72 function might contribute to disease. Second, accumulation of RNA transcripts containing the GGGGCC repeat within nuclear foci in frontal cortex and spinal cord material in C9ORF72-FTD/ALS patients suggests a toxic RNA gain of function (1). This pathogenic mechanism is patterned after other noncoding expansion disorders where RNA foci lead to the sequestration and altered activity of RNA-binding proteins (4). A third possible pathogenic mechanism is repeat-associated non-ATG translation (RAN translation). RAN translation, a mode of translation that occurs in the absence of an initiating ATG codon, was first reported to occur across expanded CAG repeats to produce potentially toxic homopolymeric peptides (5). It is among these competing possibilities that now enters the work of Xu et al. in PNAS (6), showing that expression of the expanded C9ORF72 repeat is sufficient to cause neurodegeneration and that its ability to sequester the RNA-binding protein Pur α likely has a role in repeat-mediated neurodegeneration.
Fig. 1.
Scheme depicting the three major putative mechanisms underlying expanded C9ORF72 repeat ALS/FTD. Pathway I involves a loss of function of C9ORF72 and/or other nearby genes. Both mechanisms II and III involve a toxic gain of function, with pathway II consisting of a toxic RNA and in III, toxicity being attributable to a protein or peptide.
Using both mammalian neuronal cells and the fly Drosophila melanogaster, Xu et al. (6) first show that expression of an expanded GGGGCC repeat is sufficient to cause toxicity. This is a seminal observation because it directs one’s attention to the GGGGCC repeat itself regarding pathogenesis (i.e., pathways II and III, Fig. 1). In the case of the neuronal cell study, either a control (GGGGCC3) or an expanded (GGGGCC30) repeat was inserted into an EGFP expression vector between the start site of transcription and the translational ATG start codon. Only the neuronal Neuro-2a cells expressing the vector containing the expanded repeat exhibited a significant reduction in viability. To examine toxicity in vivo, Xu et al. used a well-established Drosophila genetic approach whereby expression of either a control or expanded GGGGCC repeat was directed to a variety of neuronal tissues. Flies expressing an expanded repeat showed significant signs of neuronal death. In both a cellular model and an animal model, expression of an expanded GGGGCC repeat was sufficient to cause damage. Two additional experiments using the Drosophila C9ORF72 disease model reveal that damage to retinal photoreceptors is more pronounced in older flies and that motor neurons, the primary disease target in ALS, undergo a progressive degeneration upon expression of an expanded GGGGCC repeat. These latter two results indicate that two seminal features of human C9ORF72 disease, age dependence of disease and susceptibility of motor neurons, are replicated with expression of expanded GGGGCC repeat in flies.
Therefore, how might an expanded GGGGCC repeat alter cellular function to cause disease? Building on this group’s experience with another DNA repeat-based disorder Fragile X tremor/ataxia syndrome (FXTAS), Xu et al. (6) looked at whether the capability of repeat containing RNA to bind normal RNA-binding proteins was enhanced; i.e., could C9ORF72 disease be one of the disorders where RNA foci lead to the sequestration and altered activity of RNA-binding proteins? Consistent with a RNA-mediated pathogenic mechanism, the authors show that synthesized GGGGCC repeat-containing RNAs are able to bind proteins in extracts prepared from mouse spinal cord. To identify proteins binding to GGGGCC repeat-containing RNA, they separated the bound proteins using polyacrylamide gel electrophoresis. Individual proteins were excised and identified using mass spectrometry. Intriguingly, the most prevalent C9ORF72 GGGGCC-binding proteins found were several members of the Pur family of RNA-binding proteins, with Pur α being the most abundant. Intriguingly, Pur α was previously found to interact with expanded CGG repeat-containing FXTAS RNA and modulated disease in a Drosophila model of FXTAS (7).
Binding of Pur α to GGGGCC repeats was examined in vitro using GST-tagged Pur α and radiolabeled GGGGCC repeats (6). Both mouse and Drosophila Pur α bound with similar affinities, Kd of 14.8 and 5.1 nM, respectively. Xu et al. further show that a tagged version of C9ORF GGGGCC-repeat RNA could interact with endogenous Pur α in Neuro-2a cells and in mouse and human brain extracts. The brain extract assay found no evidence that TAR DNA-binding protein (TDP)-43, another RNA-binding protein implicated in ALS/FTD, was able to bind C9ORF GGGGCC-repeat RNA. The heterogeneous nuclear ribonucleoprotein RNA-binding protein hnRNP A2/B1, which does bind to FXTAS CGG repeat-containing RNA, failed to bind to C9ORF72 GGGGCC-repeat RNA using either the in vitro or the in vivo assay. Xu et al. also failed to find evidence of C9ORF72 GGGGCC-repeat RNA binding to another hnRNP, hnRNP A3. However, the significance of their failure to detect C9ORF72 GGGGCC-repeat RNA to any of the hnRNPs needs to be taken with caution because another study found that C9ORF72 GGGGCC-repeat RNA bound to all three of these hnRNPs, with hnRNP A3 being detectable in the p62-positive, TDP-43–negative inclusions seen in the brains of patients with C9ORF disease (8). Xu et al. did find Pur α-positive inclusions in flies expressing expanded C9ORF72 GGGGCC repeat. Importantly, Pur α was found in inclusions in mutant C9ORF72 repeat carriers and in noncarriers with FTD-TDP.
Lastly, and perhaps one of the more informative results, is the authors’ demonstration that overexpression of Pur α is able to rescue expanded C9ORF72 repeat-induced neurodegeneration in the mammalian Neuro-2a neuronal cell system and Drosophila model of disease (6). Decreasing expression of Pur α in Neuro-2a cells in itself caused cell death. Moreover, overexpression of hnRNP A2/B1 (that, in this study, did not bind to the C9ORF72 repeat) failed to rescue expanded GGGGCC-induced toxicity. These results provide strong evidence that, at least in these model systems, manipulating the binding of Pur α levels impacts toxicity in a manner consistent with the hypothesis that sequestration of Pur α by mutant C9ORF72 repeats contributes to pathogenesis.
Collectively, Xu et al. (6) provide two major insights into understanding how an expanded C9ORF72 GGGGCC repeat alters neuronal function and induces disease. First, this study nicely demonstrates in two model systems that expression of the C9ORF72 expanded repeat itself, in the absence of other
The work of Xu et al. exemplifies what is becoming an effective strategy for identifying the events that have a substantive contribution to disease.
components of the C9ORF72 gene, is sufficient to be toxic. This finding strongly argues in favor of pathogenic models based on expression of the expanded repeat. Xu et al. further show that the sequestration of the RNA-binding protein Pur α by C9ORF72 RNA containing an expanded repeat contributes to pathogenesis in both the mammalian neuronal cell and Drosophila models of C9ORF72 disease, thus providing support for toxic RNA contributing to disease (pathway II, Fig. 1). This latter point is particularly noteworthy because it provides direct evidence linking a putative pathogenic pathway to C9ORF72 repeat-induced disease.
Although the work of Xu et al. (6) provides several important steps toward understanding the mechanism by which C9ORF72 disease develops, there are several key points remaining to be clarified. It remains a mystery why expansion of the C9ORF72 repeat leads to a neurodegenerative disease that presents as ALS and/or FTD. Another more specific issue that Xu et al. note is that this work used models of C9ORF72 disease where the size of the expanded repeats, 30 GGGGCC repeats, was limited by what was readily isolated and manipulated experimentally. Although this length is above the two- to eight-repeat-length tracts typically seen in unaffected individuals, it is far less than the 100–1,000 of hexanucleotide repeats seen in ALS/FTD patients (1, 2). One needs to remain open to the idea that expanded repeats so much longer than 30 GGGGCCs either might not sequester RNA-binding proteins and/or trigger another mechanism such as gene silencing at the C9ORF72 locus or translation of a toxic peptide. In this regard, it is worth noting that two groups recently reported immunostaining for GGGGCC repeat encoded insoluble peptides in the brains of patients who succumbed to C9ORF72 disease (9, 10). Whether these peptides actually contribute to C9ORF72 remains to be shown. As seen for many of the other unstable DNA repeat/microsatellite disorders (11), C9ORF72 GGGGCC expansion very likely will have several molecular outcomes. Importantly, the work of Xu et al. exemplifies what is becoming an effective strategy for identifying the events that have a substantive contribution to disease (i.e., the application of a cross-species spectrum of model systems for testing pathways fundamental to pathogenesis). It is through the identification of such pathways that the likelihood of developing effective therapeutic approaches will be enhanced.
Footnotes
The author declares no conflict of interest.
See companion article on page 7778.
References
- 1.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in non-coding region of C9ORF72 causes chromosome 9p-linked frontotemporal dementia and amyotrophic lateral sclerosis. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton AE, et al. ITALSGEN Consortium A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gijselinck I, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: A gene identification study. Lancet Neurol. 2012;11(1):54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 4.Renoux AJ, Todd PK. Neurodegeneration the RNA way. Prog Neurobiol. 2012;97(2):173–189. doi: 10.1016/j.pneurobio.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zu T, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci USA. 2011;108(1):260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Z, et al. Expanded hexanucleotide GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci USA. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin P, et al. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55(4):556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mori K, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013;125(3):413–423. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- 9.Ash PEA, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77(4):639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori K, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339(6125):1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 11.Nelson DL, Orr HT, Warren ST. The unstable repeats-three evolving faces of neurological disease. Neuron. 2013;77(5):825–843. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]