Life Emerges From Functions of a System of Synchronized Clocks
A hierarchical system of clocks within cells throughout the body creates and synchronizes rhythmic functions from which life emerges. Biological circadian rhythms, linked to light and darkness in an organism's environment, are generated by transcription-translation feedback mechanisms on universal “core” clock genes, e.g., Clock, Bmal1, Per1, Per2, Cry1 and Cry2 (16). Clocks within the brain broadcast signals to all tissues (5). Many cell types also exhibit their “local” circadian clocks, as well as infradian and ultradian clocks that rhythmically regulate cell type-specific functions on time scales commensurate with the functions. Functional regulation by clocks range from activation-inactivation-reactivation cycles of molecules (e.g., ion channels) at millisecond scales, the heart's beating at cycles of hundreds of milliseconds, respiration at cycle of seconds, low-frequency autonomic signaling at cycles of tens of seconds, to diurnal and infradian variations of numerous body functions.
The Hierarchy of Clocks Within the Heart
Cardiovascular function is regulated by a complex hierarchical clock system. Circadian regulation contributes to normal heart function. Disruption of circadian clocks within mice leads to cardiac pathology that includes reductions in heart rate, beating rate variability (1), arrhythmias, altered substrate metabolism and contractile function (2), and altered adaptations of the heart in response to hypertrophic stimuli (6). Circadian patterns of regulation of Ca2+ channel subunits and Ca2+ current densities and phosphorylation of key signaling molecules, including ERK, p38, Akt, and GSK, have been identified (7).
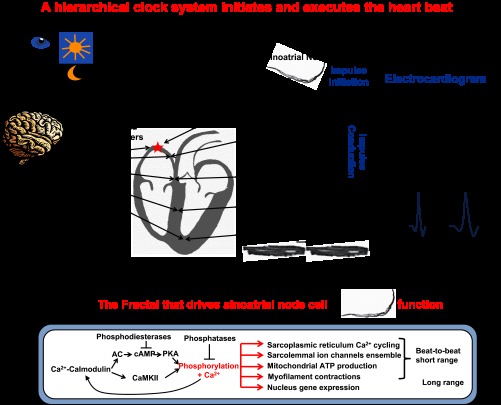
A hierarchy of ultradian clocks within the heart regulates beat-to-beat cardiac function (12). Synchronization of these “local” clocks over short time scales in different parts of the heart ensures the rhythmic generation and execution of each heartbeat (Fig. 1): The “period” of clocks within the sinoatrial node (SAN) pacemaker cell from which impulses emerge that initiate each heart beat is synchronized with the periods of the other parts of the electrical system that conduct the impulse to the ventricular myocardium to elicit a synchronized contraction of ventricular myocytes that is required to eject blood from the heart. Regulation of heart rate and rhythm by the hierarchical clock system is modulated by autonomic signaling from the brain via neurotransmitter release from the vagus and sympathetic nerves (Fig. 1). Desynchronization of brain-heart clock functions or of any clock periods within and among different cardiac cell types leads to abnormalities in impulse initiation by SAN cells, in conduction of the impulse, and to abnormalities of cardiac contraction. Interaction and spread of electrical excitation across cell types of the hierarchical clock system (brain to ventricular myocytes) generates a body surface potential captured in the electrogram (ECG). Malfunctions within the clock hierarchy produce abnormal ECGs (Fig. 1, right).
Fig. 1.
Rhythmic initiation (electrical component) and execution (contractile component) of each heartbeat by a hierarchical clock system (brain-heart) that synchronizes functions of cells within and among other tissues. Impulse is initiated in the sinoatrial node (SAN) and conducted to ventricular myocytes. Bottom: SAN pacemaker cells generate rhythmic electrical impulses that emerge from the SAN. Right panels schematically illustrate that heart-specific disruption of circadian clock (Bmal1-/-) results in abnormal rhythm and rate (ECG) and impulse conduction (QRS).
There Is a Formidable Gap in Knowledge About How Functions of Cardiac Circadian Clock Link to Those of Cardiac Ultradian Clocks
In this issue of American Journal of Physiology-Cell Physiology, Schroder et al. (15) bridged this gap by investigating cardiac clock mechanisms in vivo and in vitro in adult ventricular cardiomyocytes by 1) a deletion of the cardiac clock gene Bmal1 in adult mice and 2) assessing the effect of this deletion on heart rate and rhythm in mice and in isolated cardiomyocytes. The diurnal pattern of heart rate observed in wild-type (WT) mice was suppressed in Bal1-/- mice. In addition to this altered circadian heart rhythmicity, Shroder et al. observed changes in ultradian clocks that regulate beat-to-beat function: 1) a reduction in the rate and disruption of the rhythm at which impulses emanate from the SAN, manifested by prolonged and dysrhythmic R-R intervals on the ECG (Fig. 1); and 2) abnormal spread of impulse conduction across the ventricle, i.e.,prolonged QRS complex on the ECG (Fig. 1).
Schroder et al. screened circadian expression profiles of genes known to be involved in regulation of heart beating rate and rhythm. They observed that Scn5a, which codes Nav1.5, the major subunit of cardiac-type Na+ channels (i.e., those that generate Na+ current, INa), exhibited diurnal expression in WT hearts, but this pattern was suppressed in hearts of Bmal1-/- mice. But how is the altered diurnal expression of Scn5a, i.e., a long oscillatory period, in Bmal1-/- mice coupled to abnormalities in ultradian clock phenotypes of the cardiac electrical impulse initiation and conduction? A clue to the link between circadian and ultradian abnormalities produced by the cardiac-specific disruption of the Bmal1-/- gene is that Schroder et al. observed that not only is the cardiac diurnal expression of pattern of Scn5a (i.e., Nav1.5 transcript) disturbed in Bmal1-/-, but also Bmal1-/- hearts have reduced 1) Nav1.5 protein expression and 2) ad hoc peak INa amplitude density in their ventricular myocytes. The authors note that the Bmal1-/- phenotype is strikingly similar to that of mice with targeted disruption of Scn5a, in which INa was reduced by 50% (13). This suggests that one mechanism that links circadian to ultradian cardiac clocks is rhythmic change in transcription, translation of proteins involved in the regulation of long- and short-range cardiac rhythmicity.
How Do Na Channels Participate in Regulation of the Heart Rate and Rhythm?
Cells within each component of the clock hierarchy (Fig. 1) have Na+ channels. In mice, Nav1.5 is absent from the center but present in the periphery of the SAN, whereas Nav1.1 (neuronal isoform) is present throughout the SAN (9). Both Na+ channel isoforms are likely important for SAN function in mice: the two isoforms are involved in impulse initiation, but only the cardiac Nav1.5 isoform participates in propagation of the action potential from the SAN to the surrounding atrial muscle. HCN4-positive-Cx43-negative cells within atrioventricular (AV) node or His bundle, and Purkinje fibers in mice have high expression levels of Nav1.5 (14). Moreover, a late Na+ current via Na+ channels that operate in slow gating modes [as opposed to transient INa that generates the action potential (AP) upstroke] is found in ventricular myocytes. In heart failure, transient INa is reduced, but late INa increases, and is involved in arrhythmias, due to effects to disperse AP repolarization, and create cell Ca2+ overload, with attendant increase in diastolic Ca2+ levels and in likelihood for the occurrence of spontaneous Ca2+ release (see review in Ref. 11).
Regulation of the Cardiac Impulse beyond the Na+ Channels
But Na+ channels are only one component of the functions of chemical and electrical clocks within components of the clock hierarchy that generates, conducts, and executes cardiac impulse. Recent discoveries have led to the idea that cardiac impulse initiation and conduction involves the coupling of chemical to electrical clocks within cardiac pacemaker cells (3, 8, 17). Numerous molecular clock functions, present within cells of each tissue component of the hierarchical system in mice (10), become synchronized to create an intracellular clock system that delivers the “payload” of that cell type in a rhythmic manner with a specified period. For example, ultradian clock functions embedded in node cells (Fig. 1, bottom) include sarcoplasmic reticulum (SR) rhythmic spontaneous Ca2+ cycling, rhythmic ion channel current activation and inactivation, rhythmic oscillatory mitochondria ATP production, and oscillatory contractile displacement and force production. Gene expression patterns within the nucleus and their epigenetic modulation regulate transcription and translation of proteins that control the ultradian SAN cell (SANC) clock functions (Fig. 1). But the kinetics of spontaneous rhythms that drive ultradian functions in SANC is regulated by posttranslational protein modification, i.e., phosphorylation of proteins that are involved in rhythmic AP firing of SANC. Remarkably, a constitutively activated Ca2+-calmodulin-activated adenylyl cyclase (AC)/cAMP/PKA-CaMKII signaling system drives the kinetics of SR Ca2+ cycling, surface membrane channel activation and inactivation, mitochondria ATP production, contractile displacement, and force production (8, 10, 18, 19) (Fig. 1). Moreover, this signaling also synchronizes the function that drives the rhythm of AP generation. When this signaling cascade within SANC becomes disabled and Ca2+, cAMP, and phosphorylation levels are reduced, the rhythmic spontaneous AP firing by these cells become dys-rhythmic. Thus, this Ca2+-calmodulin-activated AC/cAMP/PKA-CaMKII signaling system within nodal cells is analogous to a fractal, not only regulating numerous functions, but also the synchronization from which rhythmicity emerges.
There is evidence that intracellular fractals very similar to that of nodal cells (Fig. 1, bottom) also exist within cells of other tissues of the hierarchical clock cascade and may contribute to regulation of similar rhythmic functions within these cells. Furthermore, when neurotransmitters bind to their receptors, the Ca2+-calmodulin-activated AC/cAMP/PKA-CaMKII signaling system becomes engaged in cells of each component of the system depicted in Fig. 1. Therefore, one may envision that a fractal common to each of these cell types is involved in initiating and synchronizing processes not only within but also among components of the hierarchy that initiate and execute each heartbeat. That each oscillator within the hierarchy has a slightly different autonomous period (i.e., when not driven by the impulses emanating from the SAN) suggests that the kinetic parameters that drive these fractal elements within cells of each component are set at slightly different rates. The periodicity of the fractal-generated functions would be the shortest in SANC, due to higher basal Ca2+-AC-cAMP/PKA/CaMKII-mediated phosphorylation of Ca2+ cycling proteins. At the other extreme, the periodicity driven by the fractal in ventricular myocytes is suppressed, due to a shift in balance of cAMP production and its degradation by phosphodiesterases that dampens basal fractal signaling. This suppression of fractal signaling is removed when β-adrenergic receptors are stimulated by an increase in neurotransmitter release above the basal level. Thus, each tissue component that generates the cardiac impulse and cells within each component can be conceived as a fractal of a system that ensures timely impulse initiation and culminates in robust heart muscle contractions during heartbeat. It may also be envisioned that common fractals drive periodic functions within different parts of the brain to synchronize processes that become synchronized regulate its neurotransmitter release that modulate cardiac functions. Of note, a strikingly similar fractal to that depicted in Fig. 1 within SANC is operative within the retina (4) to sense photons and this fractal may be implicated in the rhythmic diurnal regulation of the heartbeat.
Future Directions
In summary, the study by Schroder et al., in demonstrating that Scn5a is regulated by a cardiomyocyte circadian clock, uncovers what is likely to be “a tip of the iceberg” of complex processes distributed among fractals that control the function within and among different cardiac cell types on a broad time scale (milliseconds to hours to days and beyond). Future studies are required to test the hypothesis that clock genes regulate ultradian fractal-like clock regulation and synchronization of functions within cells (as depicted for SANC in Fig. 1) of other tissues components of the depicted hierarchy and that these same fractals link ultradian rhythms to circadian rhythms.
GRANTS
This research was supported entirely by the Intermural Research Program of the National Institutes of Health, National Institute on Aging.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
E.G.L., Y.Y., and V.A.M. drafted the manuscript; edited and revised the manuscript; approved the final version of the manuscript.
REFERENCES
- 1.Binah O, Weissman A, Itskovitz-Eldor J, Rosen MR. Integrating beat rate variability: from single cells to hearts. Heart Rhythm. doi:10.1016/j.hrthm.2013.02.013 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray MS, Shaw CA, Moore MW, Garcia RA, Zanquetta MM, Durgan DJ, Jeong WJ, Tsai JY, Bugger H, Zhang D, Rohrwasser A, Rennison JH, Dyck JR, Litwin SE, Hardin PE, Chow CW, Chandler MP, Abel ED, Young ME. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am J Physiol Heart Circ Physiol 294: H1036–H1047, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Cheng H, Smith GL, Hancox JC, Orchard CH. Inhibition of spontaneous activity of rabbit atrioventricular node cells by KB-R7943 and inhibitors of sarcoplasmic reticulum Ca2+ ATPase. Cell Calcium 49: 56–65, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dessauer CW, Posner BA, Gilman AG. Visualizing signal transduction: receptors, G-proteins, and adenylate cyclases. Clin Sci (Lond) 91: 527–537, 1996 [DOI] [PubMed] [Google Scholar]
- 5.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72: 517–549, 2010 [DOI] [PubMed] [Google Scholar]
- 6.Durgan DJ, Tsai JY, Grenett MH, Pat BM, Ratcliffe WF, Villegas-Montoya C, Garvey ME, Nagendran J, Dyck JR, Bray MS, Gamble KL, Gimble JM, Young ME. Evidence suggesting that the cardiomyocyte circadian clock modulates responsiveness of the heart to hypertrophic stimuli in mice. Chronobiol Int 28: 187–203, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko ML, Shi L, Grushin K, Nigussie F, Ko GY. Circadian profiles in the embryonic chick heart: L-type voltage-gated calcium channels and signaling pathways. Chronobiol Int 27: 1673–1696, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lakatta EG, Maltsev VA, Vinogradova TM. A coupled SYSTEM of intracellular Ca2+ clocks and surface membrane voltage clocks controls the timekeeping mechanism of the heart's pacemaker. Circ Res 106: 659–673, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei M, Jones SA, Liu J, Lancaster MK, Fung SS, Dobrzynski H, Camelliti P, Maier SK, Noble D, Boyett MR. Requirement of neuronal- and cardiac-type sodium channels for murine sinoatrial node pacemaking. J Physiol 559: 835–848, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Sirenko S, Juhaszova M, Ziman B, Shetty V, Rain S, Shukla S, Spurgeon HA, Vinogradova TM, Maltsev VA, Lakatta EG. A full range of mouse sinoatrial node AP firing rates requires protein kinase A-dependent calcium signaling. J Mol Cell Cardiol 51: 730–739, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maltsev VA, Undrovinas A. Late sodium current in failing heart: friend or foe? Prog Biophys Mol Biol 96: 421–451, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandel Y, Weissman A, Schick R, Barad L, Novak A, Meiry G, Goldberg S, Lorber A, Rosen MR, Itskovitz-Eldor J, Binah O. Human embryonic and induced pluripotent stem cell-derived cardiomyocytes exhibit beat rate variability and power-law behavior. Circulation 125: 883–893, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papadatos GA, Wallerstein PM, Head CE, Ratcliff R, Brady PA, Benndorf K, Saumarez RC, Trezise AE, Huang CL, Vandenberg JI, Colledge WH, Grace AA. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc Natl Acad Sci USA 99: 6210–6215, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Remme CA, Verkerk AO, Hoogaars WM, Aanhaanen WT, Scicluna BP, Annink C, van den Hoff MJ, Wilde AA, van Veen TA, Veldkamp MW, de Bakker JM, Christoffels VM, Bezzina CR. The cardiac sodium channel displays differential distribution in the conduction system and transmural heterogeneity in the murine ventricular myocardium. Basic Res Cardiol 104: 511–522, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroder EA, Lefta M, Zhang X, Bartos D, Feng HZ, Zhao Y, Patwardhan A, Jin JP, Esser KA, Delisle BP. The cardiomyocyte molecular clock, regulation of Scn5a, and arrhythmia susceptibility. Am J Physiol Cell Physiol (January 30, 2013). doi:10.1152/ajpcell.00383.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siepka SM, Yoo SH, Park J, Lee C, Takahashi JS. Genetics and neurobiology of circadian clocks in mammals. Cold Spring Harb Symp Quant Biol 72: 251–259, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ter Keurs HE, Boyden PA. Calcium and arrhythmogenesis. Physiol Rev 87: 457–506, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang D, Lyashkov AE, Li Y, Ziman BD, Lakatta EG. RGS2 overexpression or Gi inhibition rescues the impaired PKA signaling and slow AP firing of cultured adult rabbit pacemaker cells. J Mol Cell Cardiol 53: 687–694, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yaniv Y, Juhaszova M, Lyashkov AE, Spurgeon HA, Sollott SJ, Lakatta EG. Ca2+-regulated-cAMP/PKA signaling in cardiac pacemaker cells links ATP supply to demand. J Mol Cell Cardiol 51: 740–748, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]