Background
Metformin is a first-line therapy for type 2 diabetes mellitus (T2DM, formerly ‘non-insulin-dependent diabetes mellitus’), and is one of the most commonly prescribed drugs worldwide. As a biguanide agent, metformin lowers both basal and postprandial plasma glucose (PPG) [1,2]. It can be used as a monotherapy or in combination with other antidiabetic agents including sulfonylureas, α-glucosidase inhibitors, insulin, thiazolidinediones, DPP-4 inhibitors as well as GLP-1 agonists. Metformin works by inhibiting the production of hepatic glucose, reducing intestinal glucose absorption, and improving glucose uptake and utilization. Besides lowering the blood glucose level, metformin may have additional health benefits, including weight reduction, lowering plasma lipid levels, and prevention of some vascular complications [3]. As the prevalence of obesity in the USA increases, the use of metformin is also increasing. Metformin is also used for other indications such as polycystic ovary syndrome (PCOS) [1]. Metformin is well tolerated by the majority of patients. However, the glycemic response to metformin is quite variable. Some patients respond extremely well, whereas others show no benefit [4]. This summary briefly reviews the pharmacokinetics of metformin (Fig. 1) and highlights genes mediating the diverse pharmacological responses to metformin treatment (Fig. 2). Knowledge of these pathways may help identify the genetic markers to predict variations in response as well as aid the tailoring of metformin therapy.
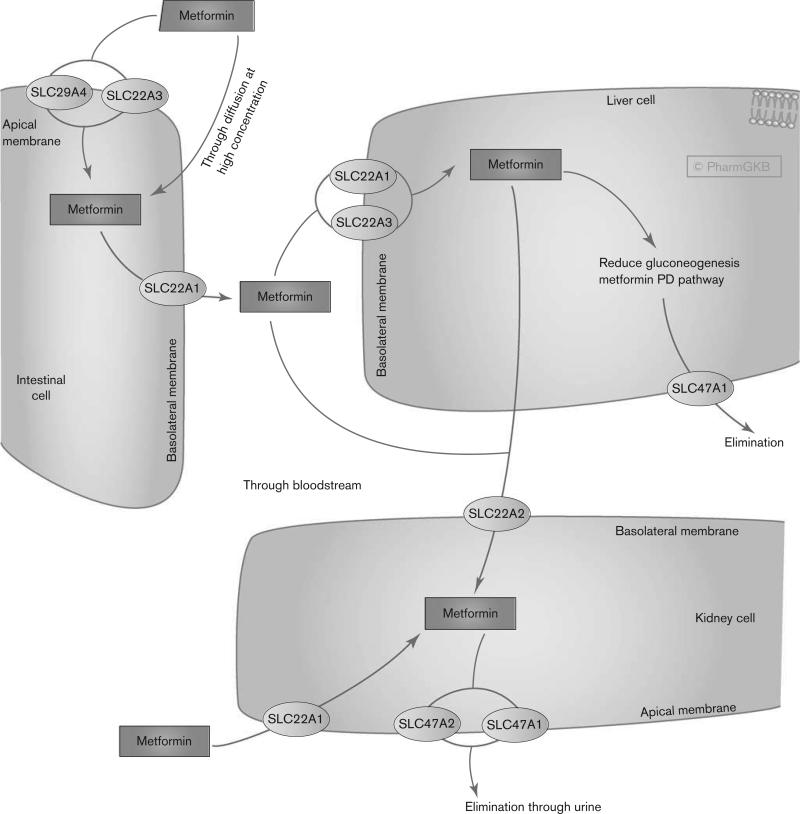
Fig. 1.
Pharmacokinetics pathway of metformin. Stylized cells depicting genes involved in the transport and clearance of metformin. A fully interactive version is available online at http://www.pharmgkb.org/pathway/PA165948259. PD, pharmacodynamics.
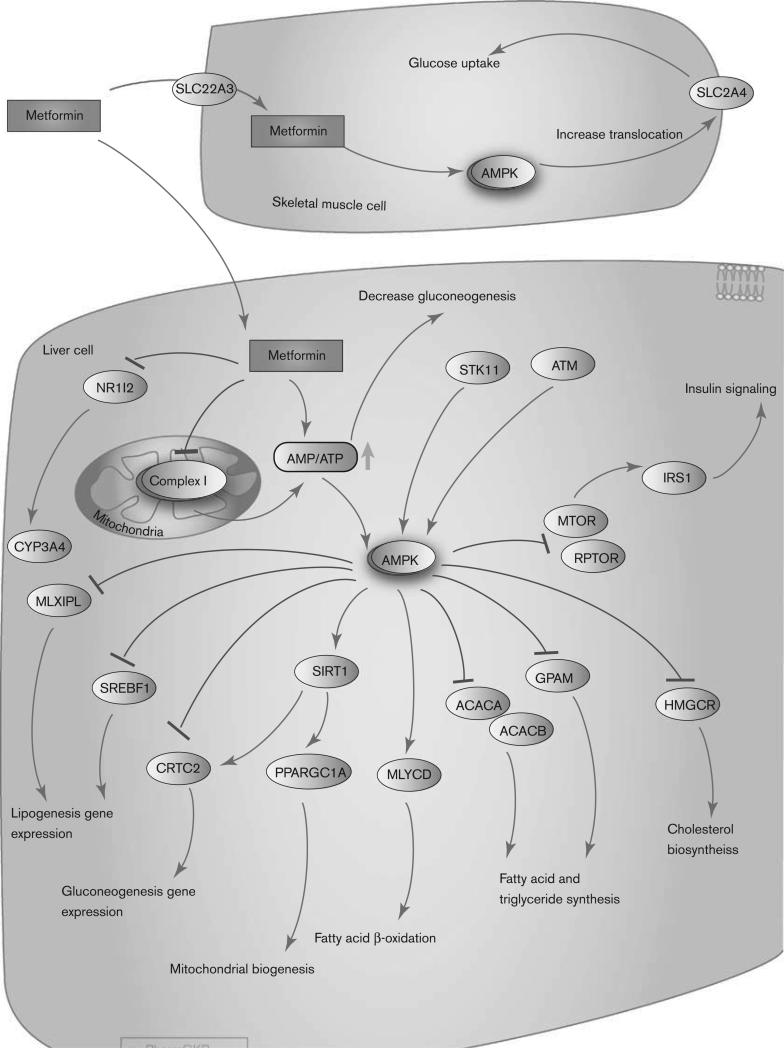
Fig. 2.
Pharmacodynamics pathway of metformin. Stylized cells depicting the mechanism of action of metformin. A fully interactive version is available online at http://www.pharmgkb.org/pathway/PA165948566.
Pharmacokinetics
Metformin is not metabolized [5] and is excreted unchanged in the urine, with a half-life of ~5 h [6]. The population mean for renal clearance (CLr) is 510±120 ml/min. Active tubular secretion in the kidney is the principal route of metformin elimination. The drug is widely distributed into body tissues including the intestine, liver, and kidney by organic cation transporters [6]. There is a large interindividual variability in metformin pharmacokinetics as measured by differences in trough steady-state metformin plasma concentration ranging from 54 to 4133 ng/ml [7].
The intestinal absorption of metformin may be primarily mediated by plasma membrane monoamine transporter (PMAT, encoded by gene SLC29A4), which is expressed on the luminal side of enterocytes [8] (Fig. 1). However, there are currently no in-vivo data on the role of PMAT in the disposition and pharmacological effect of metformin. OCT3 (gene SLC22A3) is also expressed on the brush border of the enterocytes and may contribute to metformin uptake [6,9]. In addition, OCT1 (gene SLC22A1), which is expressed on the basolateral membrane and cytoplasm of the enterocytes, may facilitate the transfer of metformin into the interstitial fluid [9]. The role of OCT1 and OCT3 in the intestinal transport of metformin remains to be defined.
The hepatic uptake of metformin is mediated primarily by OCT1 (SLC22A1) and possibly by OCT3 (SLC22A3). Both transporters are expressed on the basolateral membrane of hepatocytes [6,10–12]. In Oct1-deficient mice, the hepatic metformin concentration in the liver is significantly lower than that in control mice, suggesting that OCT1 is essential for the hepatic uptake of metformin [13]. In addition, the glucose-lowering effects of metformin were completely abolished in the Oct1-deficient mice. Metformin is also a good substrate for human multidrug and toxin extrusion 1 (MATE1, encoded by the gene SLC47A1) and MATE2-K (gene SLC47A2) [10,14–16]. MATE1 (SLC47A1) is highly expressed in the liver, kidney, and skeletal muscle [17], and may contribute toward the excretion of metformin from both the liver and the kidney. However, the role of MATE1 in hepatic secretion has been questioned, as biliary excretion of metformin seems to be insignificant in humans [6]. Data from a Mate1 knockout mouse study suggest that, at least in rodents, biliary excretion of metformin occurs [18].
The uptake of metformin from circulation into renal epithelial cells is primarily facilitated by OCT2 (gene SLC22A2) [10], which is expressed predominantly at the basolateral membrane in the renal tubules. Renal excretion of metformin from the tubule cell to the lumen is mediated through MATE1 (SLC47A1) and MATE2-K (SLC47A2) [14,15,19,20]. MATE1 and MATE2-K are expressed in the apical membrane of the renal proximal tubule cells, and studies in healthy individuals suggest that they contribute to the renal excretion of metformin [21]. OCT1 also appears to be expressed on the apical and subapical domain side of both the proximal and the distal tubules in the kidney, and may play an important role in metformin reabsorption in kidney tubules [22]. PMAT (gene SLC29A4) is expressed on the apical membrane of renal epithelial cells, and may play a role in the renal reabsorption of metformin [23]. However, there are no in-vivo data as yet supporting this role. In addition, P-gp (gene ABCB1) and BCRP (gene ABCG2) are involved in the efflux of metformin across placental apical membranes [24].
As metformin is not metabolized in the liver, drug–drug interactions through the inhibition of metformin transporters (OCTs and MATEs) are clinically relevant. Genetic polymorphisms in these transporter genes are also likely to have a direct impact on metformin pharmacokinetics and variability in drug responses (see the Pharmacogenomics section). Recent drug–drug interaction studies suggest that proton-pump inhibitors inhibit metformin uptake in vitro by inhibiting OCT1, OCT2, and OCT3 [25]. Oral antidiabetic drugs repaglinide and rosiglitazone also inhibited OCT1-mediated metformin transport in vitro [26]. The H2 blocker cimetidine is associated with reduced renal tubular secretion and increased systemic exposure to metformin when both drugs are coadministered [27]. Inhibition of MATEs, but not OCT2 [28], is the likely mechanism underlying the drug–drug interactions with cimetidine in renal elimination [20]. A recent study suggests the potential for a transporter-mediated drug–drug interaction between metformin and specific tyrosine kinase inhibitors (e.g. imatinib, nilotinib, gefitinib, and erlotinib), which may have clinical implications in the disposition, efficacy, and toxicity of metformin [29].
Pharmacodynamics
Metformin lowers both basal and PPG. It works mainly by suppressing excessive hepatic glucose production, through a reduction in gluconeogenesis [30]. Other potential effects of metformin include an increase in glucose uptake, an increase in insulin signaling, a decrease in fatty acid and triglyceride synthesis, and an increase in fatty acid β-oxidation. Metformin may also increase glucose utilization in peripheral tissues, and possibly reduce food intake and intestinal glucose absorption. As metformin does not stimulate endogenous insulin secretion, it does not cause hypoglycemia or hyperinsulinemia, which are common side effects associated with other antidiabetic drugs.
The molecular mechanisms underlying metformin action appear to be complex and remain a topic of considerable debate. However, there is general agreement that the administration of metformin results in the phosphorylation and activation of AMP-activated protein kinase (AMPK) in the liver, which in turn may lead to diverse pharmacologic effects, including inhibition of glucose and lipid synthesis [2,31]. Although the specific route of AMPK phosphorylation is not yet clear, the molecular components LKB1/STK11 and ATM have been shown to play a role in the phosphorylation of AMPK in the presence of metformin [31] (Fig. 2). However, ATM, LKB1, and AMPK are not the direct targets of metformin [32]. A recent study using liver-specific AMPK-knockout mice has shown that inhibition of hepatic glucose production by metformin is preserved, suggesting that metformin may inhibit hepatic gluconeogenesis in an LKB1-independent and AMPK-independent manner [33]. The findings from this study are yet to be replicated, and therefore, the role of AMP kinase in the inhibition of gluconeogenesis can still be considered. In a separate study in Oct-1-knockout mice, metformin both activated AMPK and reduced gluconeogenesis [13]. A separate group has also concluded that metformin inhibits hepatic gluconeogenesis through AMPK-dependent regulation of SHP [34]. Therefore, a reduction in gluconeogenesis may occur both ways, in an AMPK-dependent and an AMPK-independent manner. Although the direct target is not fully elucidated, metformin specifically inhibits complex I of the mitochondrial respiratory chain, suggesting that this inhibition may activate AMPK by increasing the cellular AMP: ATP ratio [32,35–37]. AMPK is a major cellular regulator of lipid and glucose metabolism. The activated AMPK phosphorylates and inactivates HMG-CoA reductase (encoded by gene HMGCR), MTOR (target of rapamycin); ACC-2 (encoded by gene ACACB); ACC (encoded by gene ACACA), glycerol-3-phosphate acyltransferase (encoded by gene GPAM); and carbohydrate response element-binding protein [31,38]. The activation of AMPK by metformin also suppresses the expression of SREBP-1 (encoded by gene SREBF1), a key lipogenic transcription factor [39]. Phosphorylated AMPK also activates SiRT1 and increases Pgc-1a (encoded by gene PPARGC1A) expression in the nucleus, leading to the downstream activation of mitochondrial biogenesis. Metformin disrupts the coactivation of PXR with SRC1, resulting in the downregulation of CYP3A4 gene expression [40]. Finally, activated AMPK results in an increase in glucose uptake in skeletal muscle by increasing the GLUT4 (encoded by gene SLC2A4) translocation activity [13]. The overall pharmacological effect of AMPK activation in the liver includes the stimulation of fatty acid oxidation with inhibition of cholesterol and triglyceride synthesis. Peripheral effects include the stimulation of fatty acid oxidation and glucose uptake in skeletal muscle as well as a systemic increase in insulin sensitivity [35]. However, the role of metformin in insulin-mediated glucose uptake has been debated [41].
Given the increased risk of cancer in T2DM patients, metformin has also been evaluated for its tumor suppression ability and its potential to protect from cancer [42]. Population studies have shown that metformin is associated with a significant reduction of neoplasia in multiple cancer types (cancer of the breast and prostate, in particular) [43]. Metformin may also inhibit the growth of cancer cells. The mechanisms underlying this protective effect are not well understood and may involve the activation of multiple pathways [2,42]. The cell cycle arrest in metformin-treated breast cancer cells seems to involve the activation of AMPK and down-regulation of cyclin D1, and requires p27Kip1 or p21Cip1 [44,45]. Metformin was reported to suppress HER2 (ERBB2) oncoprotein overexpression through inhibition of the mTOR effector p70S6K1(RPS6KB1) in human breast carcinoma cells [46].
Pharmacogenomics
The role of genetic factors in predicting response variations to metformin has been the subject of many investigations. Multiple studies have reported associations between genomic variations of metformin transporters and its pharmacokinetics and response, and a few have explored the role of pharmacodynamic genes/variants in drug efficacy (Table 1). However, the clinical relevance of these variants remains to be established in large-scale studies. Currently, no validated genetic predictor is being used in the clinic.
Table 1.
Summary of the genes and variants involved in metformin pharmacogenomics
| Genes | Variant | Associated phenotype |
|---|---|---|
| SLC22A1 (OCT1) | Reduced function alleles: R61C (rs12208357), G401S (rs34130495), 420del (rs142448543 or rs34305973 or rs35191146), and G465R (rs34059508) | High AUC, higher Cmax, and lower oral volume of distribution (V/F) [47] |
| Impaired response to a glucose tolerance test [47] | ||
| Increased CLr and decreased hepatic uptake, no exposure changes (AUC) [22] | ||
| Reduced lipid response (total CHO and triglycerides) and insulin responses to metformin in women with PCOS [48] | ||
| rs72552763 deletion, rs34130495 and other reduced function alleles | Reduced trough metformin steady-state concentration, and association with the initial absolute decrease in HbA1c [7] | |
| SLC22A2 (OCT2) | 596C>T, 602C>T, and 808G>T (rs316019) | Increase in AUC and Cmax and a decrease in CLr [49] |
| rs316019 (808G>T) | Reduced CLr but no effect on overall drug exposure [28] | |
| rs316019 (808G>T) | Higher metformin CLrs [50] | |
| SLC22A3 (OCT3) | T400I (rs8187725), V423F and T44M (rs8187715) | Impacted metformin uptake and AMPK activation in skeletal muscle cells [11] |
| SLC47A1 (MATE1) | rs2289669 | Reduction in HbA1c [51] |
| No observed association with metformin CLr or other pharmacokinetic parameters [51] | ||
| rs8065082 | Reduced diabetes incidence [52] | |
| SLC47A2 (MATE2K) | rs12943590 (–130G>A) | Poorer response to metformin treatment, assessed by the relative change in glycated hemoglobin [53] |
| SRR | rs391300 | Associated with levels of FPG, PPG, and CHO [54] |
| ATM | rs11212617 | Associated with metformin treatment success (Hba1c<7%) [55] |
| LKB/STK11 | rs8111699 | C allele associated with a significantly decreased chance of ovulation in PCOS women [56,57] |
AMPK, AMP-activated protein kinase; AUC, area under the concentration–time curve; Cmax, maximal plasma concentration; CHO, cholesterol; CLr, renal clearance; FPG, fasting plasma glucose; HbAlc, hemoglobin A1c; PCOS, polycystic ovary syndrome; PPG, postprandial plasma glucose.
Over the past few years, considerable progress has been made in understanding the effect of common genetic polymorphisms in transporter genes on the modulation of metformin pharmacokinetics. Considerable work has been carried out with the organic cation transporter family (SLC22A family) (reviewed by Nies et al. [58]). OCT1 (gene SLC22A1) is essential for the hepatic uptake of metformin [6]. In one study with 20 healthy volunteers, several genetic variants of OCT1: R61C (rs12208357), G401S (rs34130495), 420del (rs142448543 or rs34305973 or rs35191146), and G465R (rs34059508) exerted a significant effect on the pharmacokinetics of metformin after oral administration. Individuals carrying any of the reduced function OCT1 alleles showed a higher area under the concentration–time curve (AUC), higher Cmax, and a lower Vz compared with individuals carrying wild-type alleles [47]. A subsequent study in 103 healthy Caucasian men showed the impact of these low-activity alleles on pharmacokinetics, with increased CLrs and decreased hepatic uptake. However, unlike the earlier study, the reduced function allele did not lead to differences in metformin exposure (AUC) [22]. A recent study by Christensen et al. [7] has shown that reduced functional alleles of OCT1 were associated with decreased trough steady-state concentration of metformin and reduction in the absolute decrease in HbA1c during the initiation as well as maintenance period of the treatment. Overall, replication of OCT1 low-activity alleles in governing metformin disposition highlights the importance of these genetic variants in pharmacokinetics and may be taken into consideration for metformin therapy.
Studies in healthy volunteers have tested the effect of genetic variants in OCT2 (gene SLC22A2) on metformin pharmacokinetics. Genetic variants of OCT2 [c.596C > T, c.602C > T, and c.808G > T (rs316019)] were associated with differences in pharmacokinetics, compared with the reference genotype, with an increase in AUC and Cmax and a decrease in CLr [49]. A follow-up study in 15 healthy Chinese participants showed that rs316019 (808G > T) is associated with a reduced CLr, but not overall drug exposure [28]. Interestingly, in a separate healthy volunteer study including White and African-Americans, individuals heterozygous for the variant 808G/T had higher metformin CLrs than the reference group [50]. Similar to OCT1, the impact of OCT2 genetic variants is replicated, strongly suggesting the importance of these alleles in determining metformin exposure. Genetic variants of MATE1 or MATE2K have not yet been clinically associated with differences in metformin pharmacokinetics. However, in one healthy volunteer study, the administration of a MATE inhibitor, pyrimethamine, caused significant increases in metformin Cmax and AUC [21]. In-vivo studies have also shown the importance of the rodent Mate1 in modulating the pharmacokinetics of metformin through gene knockout [19].
In addition to pharmacokinetics, a number of studies have been carried out examining the role of genetic variants in metformin pharmacodynamics and response. Despite having an effect on CLr, well-established genetic polymorphisms of OCT1 and OCT2 that alter metformin disposition do not sufficiently explain the broad variation in clinical efficacy [59,60]. A pharmacogenetic study in healthy volunteers showed significant clinical effects of reduced function OCT1 variants (R61C, G401S, 420del, and G465R), causing an impaired response to a glucose tolerance test [13]. A prospective study carried out in patients with PCOS concluded that genetic variations in OCT1 may be associated with heterogeneity in the metabolic response to metformin [48]. Interestingly, the minor allele of an intronic variant of MATE1/SLC47A1, rs2289669 G > A, was significantly associated with a greater reduction in hemoglobin A1c (HbA1c), in a cohort of 116 metformin users, despite the lack of an association between the polymorphism and metformin CLr or other pharmacokinetic parameters [51]. In a meta-analysis by Jablonski et al. [52], the minor allele of rs8065082, a single-nucleotide polymorphism (SNP) in LD with MATE1 intronic SNP rs2289669, was associated with a reduced incidence of diabetes in patients taking metformin. A recent study by Choi et al. [53] also observed the association between this MATE1 intronic variant with a change in the HbA1c level, almost at the level of statistical significance. This evidence, along with the relatively large sample sizes, lends strong support for the functional impact of this MATE1 intronic variant. However, the missing link between pharmacokinetics and a reduction in HbA1c requires further research on this SNP and its mechanistic role.
Recently, a study by Choi et al. [53] showed that diabetic patients who were homozygous for g.-130G > A (rs12943590) in MATE2-K showed a significantly poorer response to metformin treatment, assessed by the relative change in glycated hemoglobin. In addition to the aforementioned transporters, the effect of variations in OCT3 has also been investigated. An in-vitro study showed that OCT3 (gene SLC22A3) may also play a role in the therapeutic action of metformin [11]. OCT inhibitors such as OCT3-specific short hairpin RNA significantly reduced the activating effect of metformin on AMPK in skeletal muscle cells. Also, genetic variants of OCT3 [T400I (rs8187725), V423F, and T44M (rs8187715)] significantly impacted metformin uptake and kinetics. In addition to transporters, an SNP in serene racemase (SRR), rs391300, showed an association with serum fasting plasma glucose, PPG, and cholesterol in 402 Chinese patients and 171 healthy controls taking metformin [54]. This discovery, although yet to be replicated, is promising for the discovery of other genetic variants that may affect clinical outcome. Finally, the exploration of gene–gene interaction may be a promising new area of research. In a study by Becker et al. [61], an interaction between two polymorphisms, rs622342 in OCT1 and rs2289669 in MATE1, was reported, suggesting that interactions between genes in the metformin pathway may impact metformin response. However, this study is small and the importance of the epistatic mechanism remains to be replicated.
The first genome-wide association study on metformin response by GoDARTs and UKPDS and WTCCC2 investigated 1024 Scottish individuals with T2DM, and was replicated in two cohorts including 1783 Scottish individuals and 1113 individuals from a UK prospective study [55]. The study found that common variants near the ATM (ataxia telangiectasia mutated) locus were associated with a glycemic response to metformin. The genes near this locus include CUL5, NPAT, C11org65, EXPH5, ACAT1, and KDELC2. The minor allele (C) of the most strongly associated SNP, rs11212617, had a population frequency of 44% and was associated with treatment success (achieving HbA1c < 7%). In the meta-analysis, SNP rs11212617 was significantly associated with treatment success with an odds ratio of 1.35. Despite the strong association, the SNP only accounts for 2.5% of the observed variability in glycemic response. ATM was selected as a causative gene because of its role in insulin resistance, increased risk of diabetes, and its role in AMPK activation. Furthermore, in-vitro functional studies performed by this group showed that inhibition of ATM by a chemical inhibitor (KU-55933) attenuated the metformin-induced phosphorylation and activation of AMPK. However, recent data suggest that this ATM inhibitor also inhibits OCT1 and may have acted through inhibition of metformin uptake rather than inhibition of ATM [62]. Overall, this recent finding suggests that the effect of ATM on activating AMPK and altering pharmacological outcomes is not conclusive.
In addition to the treatment of diabetes, metformin is used in the treatment of insulin resistance in individuals with PCOS. A small study has shown that a polymorphism in LKB/STK11 (rs8111699) is associated with ovulatory response to treatment with metformin alone in a prospective randomized trial, with the C allele associated with a significantly decreased chance of ovulation in PCOS women treated with metformin [56,57].
Conclusion
Although many new drugs have been developed for T2DM, metformin is still widely accepted as the first-line therapy because of its low incidence of microvascular and macrovascular events and its beneficial effects on plasma lipids and body weight. There is no validated genetic predictor to metformin response or pharmacokinetics, and epistatic mechanisms (gene–gene interactions) may be important [61]. Investigation of genetic variants in specific patient populations (e.g. stratification by ethnicity) as well as the investigation of gene–gene and gene–environment interactions may elucidate important polymorphisms. Furthermore, with advancements in sequencing platforms, whole exome or genome sequencing will facilitate the investigation of rare variants, copy number variants, insertions, deletions, and other genetic variants that may play a significant role in metformin pharmacokinetics and response.
Acknowledgements
PharmGKB is supported by the NIH/NIGMS (R24GM61374). Srijib Goswami and Kathleen M. Giacomini are supported in part by NIH Training Grant T32 GM007175 and NIH grant GM61390.
Footnotes
Li Gong and Srijib Goswami contributed equally to the writing of this article.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Scarpello JH, Howlett HC. Metformin therapy and clinical uses. Diab Vasc Dis Res. 2008;5:157–167. doi: 10.3132/dvdr.2008.027. [DOI] [PubMed] [Google Scholar]
- 2.Viollet B, Guigas B, Sanz Garcia N, Leclerc J, Foretz M, Andreelli F. Cellular and molecular mechanisms of metformin: an overview. Clin Sci (Lond) 2012;122:253–270. doi: 10.1042/CS20110386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeFronzo RA, Goodman AM. Efficacy of metformin in patients with non-insulin-dependent diabetes mellitus. The Multicenter Metformin Study Group. N Engl J Med. 1995;333:541–549. doi: 10.1056/NEJM199508313330902. [DOI] [PubMed] [Google Scholar]
- 4.Reitman ML, Schadt EE. Pharmacogenetics of metformin response: a step in the path toward personalized medicine. J Clin Invest. 2007;117:1226–1229. doi: 10.1172/JCI32133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardie DG. AMP-activated protein kinase as a drug target. Annu Rev Pharmacol Toxicol. 2007;47:185–210. doi: 10.1146/annurev.pharmtox.47.120505.105304. [DOI] [PubMed] [Google Scholar]
- 6.Graham GG, Punt J, Arora M, Day RO, Doogue MP, Duong JK, et al. Clinical pharmacokinetics of metformin. Clin Pharmacokinet. 2011;50:81–98. doi: 10.2165/11534750-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 7.Christensen MM, Brasch-Andersen C, Green H, Nielsen F, Damkier P, Beck-Nielsen H, Brosen K. The pharmacogenetics of metformin and its impact on plasma metformin steady-state levels and glycosylated hemoglobin A1c. Pharmacogenet Genomics. 2011;21:837–850. doi: 10.1097/FPC.0b013e32834c0010. [DOI] [PubMed] [Google Scholar]
- 8.Zhou M, Xia L, Wang J. Metformin transport by a newly cloned proton-stimulated organic cation transporter (plasma membrane monoamine transporter) expressed in human intestine. Drug Metab Dispos. 2007;35:1956–1962. doi: 10.1124/dmd.107.015495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller J, Lips KS, Metzner L, Neubert RH, Koepsell H, Brandsch M. Drug specificity and intestinal membrane localization of human organic cation transporters (OCT). Biochem Pharmacol. 2005;70:1851–1860. doi: 10.1016/j.bcp.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 10.Takane H, Shikata E, Otsubo K, Higuchi S, Ieiri I. Polymorphism in human organic cation transporters and metformin action. Pharmacogenomics. 2008;9:415–422. doi: 10.2217/14622416.9.4.415. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Pawlikowski B, Schlessinger A, More SS, Stryke D, Johns SJ, et al. Role of organic cation transporter 3 (SLC22A3) and its missense variants in the pharmacologic action of metformin. Pharmacogenet Genomics. 2010;20:687–699. doi: 10.1097/FPC.0b013e32833fe789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nies AT, Koepsell H, Winter S, Burk O, Klein K, Kerb R, et al. Expression of organic cation transporters OCT1 (SLC22A1) and OCT3 (SLC22A3) is affected by genetic factors and cholestasis in human liver. Hepatology. 2009;50:1227–1240. doi: 10.1002/hep.23103. [DOI] [PubMed] [Google Scholar]
- 13.Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest. 2007;117:1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsuda M, Terada T, Ueba M, Sato T, Masuda S, Katsura T, Inui K. Involvement of human multidrug and toxin extrusion 1 in the drug interaction between cimetidine and metformin in renal epithelial cells. J Pharmacol Exp Ther. 2009;329:185–191. doi: 10.1124/jpet.108.147918. [DOI] [PubMed] [Google Scholar]
- 15.Sato T, Masuda S, Yonezawa A, Tanihara Y, Katsura T, Inui K. Transcellular transport of organic cations in double-transfected MDCK cells expressing human organic cation transporters hOCT1/hMATE1 and hOCT2/hMATE1. Biochem Pharmacol. 2008;76:894–903. doi: 10.1016/j.bcp.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Tanihara Y, Masuda S, Sato T, Katsura T, Ogawa O, Inui K. Substrate specificity of MATE1 and MATE2-K, human multidrug and toxin extrusions/H(+)-organic cation antiporters. Biochem Pharmacol. 2007;74:359–371. doi: 10.1016/j.bcp.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Otsuka M, Matsumoto T, Morimoto R, Arioka S, Omote H, Moriyama Y. A human transporter protein that mediates the final excretion step for toxic organic cations. Proc Natl Acad Sci USA. 2005;102:17923–17928. doi: 10.1073/pnas.0506483102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito S, Kusuhara H, Kuroiwa Y, Wu C, Moriyama Y, Inoue K, et al. Potent and specific inhibition of mMate1-mediated efflux of type I organic cations in the liver and kidney by pyrimethamine. J Pharmacol Exp Ther. 2010;333:341–350. doi: 10.1124/jpet.109.163642. [DOI] [PubMed] [Google Scholar]
- 19.Tsuda M, Terada T, Mizuno T, Katsura T, Shimakura J, Inui K. Targeted disruption of the multidrug and toxin extrusion 1 (mate1) gene in mice reduces renal secretion of metformin. Mol Pharmacol. 2009;75:1280–1286. doi: 10.1124/mol.109.056242. [DOI] [PubMed] [Google Scholar]
- 20.Ito S, Kusuhara H, Yokochi M, Toyoshima J, Inoue K, Yuasa H, Sugiyama Y. Competitive inhibition of the luminal efflux by multidrug and toxin extrusions, but not basolateral uptake by organic cation transporter 2, is the likely mechanism underlying the pharmacokinetic drug-drug interactions caused by cimetidine in the kidney. J Pharmacol Exp Ther. 2012;340:393–403. doi: 10.1124/jpet.111.184986. [DOI] [PubMed] [Google Scholar]
- 21.Kusuhara H, Ito S, Kumagai Y, Jiang M, Shiroshita T, Moriyama Y, et al. Effects of a MATE protein inhibitor, pyrimethamine, on the renal elimination of metformin at oral microdose and at therapeutic dose in healthy subjects. Clin Pharmacol Ther. 2011;89:837–844. doi: 10.1038/clpt.2011.36. [DOI] [PubMed] [Google Scholar]
- 22.Tzvetkov MV, Vormfelde SV, Balen D, Meineke I, Schmidt T, Sehrt D, et al. The effects of genetic polymorphisms in the organic cation transporters OCT1, OCT2, and OCT3 on the renal clearance of metformin. Clin Pharmacol Ther. 2009;86:299–306. doi: 10.1038/clpt.2009.92. [DOI] [PubMed] [Google Scholar]
- 23.Xia L, Engel K, Zhou M, Wang J. Membrane localization and pH-dependent transport of a newly cloned organic cation transporter (PMAT) in kidney cells. Am J Physiol Renal Physiol. 2007;292:F682–F690. doi: 10.1152/ajprenal.00302.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemauer SJ, Patrikeeva SL, Nanovskaya TN, Hankins GD, Ahmed MS. Role of human placental apical membrane transporters in the efflux of glyburide, rosiglitazone, and metformin. Am J Obstet Gynecol. 2010;202:e381–e387. doi: 10.1016/j.ajog.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nies AT, Hofmann U, Resch C, Schaeffeler E, Rius M, Schwab M. Proton pump inhibitors inhibit metformin uptake by organic cation transporters (OCTs). PLoS One. 2011;6:e22163. doi: 10.1371/journal.pone.0022163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachmakov I, Glaeser H, Fromm MF, Konig J. Interaction of oral antidiabetic drugs with hepatic uptake transporters: focus on organic anion transporting polypeptides and organic cation transporter 1. Diabetes. 2008;57:1463–1469. doi: 10.2337/db07-1515. [DOI] [PubMed] [Google Scholar]
- 27.Somogyi A, Stockley C, Keal J, Rolan P, Bochner F. Reduction of metformin renal tubular secretion by cimetidine in man. Br J Clin Pharmacol. 1987;23:545–551. doi: 10.1111/j.1365-2125.1987.tb03090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang ZJ, Yin OQ, Tomlinson B, Chow MS. OCT2 polymorphisms and invivo renal functional consequence: studies with metformin and cimetidine. Pharmacogenet Genomics. 2008;18:637–645. doi: 10.1097/FPC.0b013e328302cd41. [DOI] [PubMed] [Google Scholar]
- 29.Minematsu T, Giacomini KM. Interactions of tyrosine kinase inhibitors with organic cation transporters and multidrug and toxic compound extrusion proteins. Mol Cancer Ther. 2011;10:531–539. doi: 10.1158/1535-7163.MCT-10-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hundal RS, Krssak M, Dufour S, Laurent D, Lebon V, Chandramouli V, et al. Mechanism by which metformin reduces glucose production in type 2 diabetes. Diabetes. 2000;49:2063–2069. doi: 10.2337/diabetes.49.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foretz M, Viollet B. Regulation of hepatic metabolism by AMPK. J Hepatol. 2011;54:827–829. doi: 10.1016/j.jhep.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Hardie DG. Neither LKB1 nor AMPK are the direct targets of metformin. Gastroenterology. 2006;131:973. doi: 10.1053/j.gastro.2006.07.032. author reply 974–975. [DOI] [PubMed] [Google Scholar]
- 33.Foretz M, Hebrard S, Leclerc J, Zarrinpashneh E, Soty M, Mithieux G, et al. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J Clin Invest. 2010;120:2355–2369. doi: 10.1172/JCI40671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim YD, Park KG, Lee YS, Park YY, Kim DK, Nedumaran B, et al. Metformin inhibits hepatic gluconeogenesis through AMP-activated protein kinase-dependent regulation of the orphan nuclear receptor SHP. Diabetes. 2008;57:306–314. doi: 10.2337/db07-0381. [DOI] [PubMed] [Google Scholar]
- 35.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J. 2000;348(Pt 3):607–614. [PMC free article] [PubMed] [Google Scholar]
- 37.El-Mir MY, Nogueira V, Fontaine E, Averet N, Rigoulet M, Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem. 2000;275:223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 38.Kawaguchi T, Osatomi K, Yamashita H, Kabashima T, Uyeda K. Mechanism for fatty acid ‘sparing’ effect on glucose-induced transcription: regulation of carbohydrate-responsive element-binding protein by AMP-activated protein kinase. J Biol Chem. 2002;277:3829–3835. doi: 10.1074/jbc.M107895200. [DOI] [PubMed] [Google Scholar]
- 39.Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krausova L, Stejskalova L, Wang H, Vrzal R, Dvorak Z, Mani S, Pavek P. Metformin suppresses pregnane X receptor (PXR)-regulated transactivation of CYP3A4 gene. Biochem Pharmacol. 2011;82:1771–1780. doi: 10.1016/j.bcp.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Natali A, Ferrannini E. Effects of metformin and thiazolidinediones on suppression of hepatic glucose production and stimulation of glucose uptake in type 2 diabetes: a systematic review. Diabetologia. 2006;49:434–441. doi: 10.1007/s00125-006-0141-7. [DOI] [PubMed] [Google Scholar]
- 42.Papanas N, Maltezos E, Mikhailidis DP. Metformin and cancer: licence to heal? Expert Opin Investig Drugs. 2010;19:913–917. doi: 10.1517/13543784.2010.499122. [DOI] [PubMed] [Google Scholar]
- 43.Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, Habel LA, et al. Diabetes and cancer: a consensus report. Diabetes Care. 2010;33:1674–1685. doi: 10.2337/dc10-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhuang Y, Miskimins WK. Cell cycle arrest in metformin treated breast cancer cells involves activation of AMPK, downregulation of cyclin D1, and requires p27Kip1 or p21Cip1. J Mol Signal. 2008;3:18. doi: 10.1186/1750-2187-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ben Sahra I, Laurent K, Loubat A, Giorgetti-Peraldi S, Colosetti P, Auberger P, et al. The antidiabetic drug metformin exerts an antitumoral effect in vitro and in vivo through a decrease of cyclin D1 level. Oncogene. 2008;27:3576–3586. doi: 10.1038/sj.onc.1211024. [DOI] [PubMed] [Google Scholar]
- 46.Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. The antidiabetic drug metformin suppresses HER2 (erbB-2) oncoprotein overexpression via inhibition of the mTOR effector p70S6K1 in human breast carcinoma cells. Cell Cycle. 2009;8:88–96. doi: 10.4161/cc.8.1.7499. [DOI] [PubMed] [Google Scholar]
- 47.Shu Y, Brown C, Castro RA, Shi RJ, Lin ET, Owen RP, et al. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin Pharmacol Ther. 2008;83:273–280. doi: 10.1038/sj.clpt.6100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gambineri A, Tomassoni F, Gasparini DI, Di Rocco A, Mantovani V, Pagotto U, et al. Organic cation transporter 1 polymorphisms predict the metabolic response to metformin in women with the polycystic ovary syndrome. J Clin Endocrinol Metab. 2010;95:E204–E208. doi: 10.1210/jc.2010-0145. [DOI] [PubMed] [Google Scholar]
- 49.Song IS, Shin HJ, Shim EJ, Jung IS, Kim WY, Shon JH, Shin JG. Genetic variants of the organic cation transporter 2 influence the disposition of metformin. Clin Pharmacol Ther. 2008;84:559–562. doi: 10.1038/clpt.2008.61. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Li S, Brown C, Cheatham S, Castro RA, Leabman MK, et al. Effect of genetic variation in the organic cation transporter 2 on the renal elimination of metformin. Pharmacogenet Genomics. 2009;19:497–504. doi: 10.1097/FPC.0b013e32832cc7e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Stricker BH. Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: a preliminary study. Diabetes. 2009;58:745–749. doi: 10.2337/db08-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jablonski KA, McAteer JB, de Bakker PI, Franks PW, Pollin TI, Hanson RL, et al. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 2010;59:2672–2681. doi: 10.2337/db10-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi JH, Yee SW, Ramirez AH, Morrissey KM, Jang GH, Joski PJ, et al. A common 5′-UTR variant in MATE2-K is associated with poor response to metformin. Clin Pharmacol Ther. 2011;90:674–684. doi: 10.1038/clpt.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dong M, Gong ZC, Dai XP, Lei GH, Lu HB, Fan L, et al. Serine racemase rs391300 G/A polymorphism influences the therapeutic efficacy of metformin in Chinese patients with diabetes mellitus type 2. Clin Exp Pharmacol Physiol. 2011;38:824–829. doi: 10.1111/j.1440-1681.2011.05610.x. [DOI] [PubMed] [Google Scholar]
- 55.Zhou K, Bellenguez C, Spencer CC, Bennett AJ, Coleman RL, Tavendale R, et al. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet. 2011;43:117–120. doi: 10.1038/ng.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldenberg N, Glueck CJ. Is pharmacogenomics our future? Metformin, ovulation and polymorphism of the STK11 gene in polycystic ovary syndrome. Pharmacogenomics. 2008;9:1163–1165. doi: 10.2217/14622416.9.8.1163. [DOI] [PubMed] [Google Scholar]
- 57.Legro RS, Barnhart HX, Schlaff WD, Carr BR, Diamond MP, Carson SA, et al. Ovulatory response to treatment of polycystic ovary syndrome is associated with a polymorphism in the STK11 gene. J Clin Endocrinol Metab. 2008;93:792–800. doi: 10.1210/jc.2007-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nies AT, Koepsell H, Damme K, Schwab M. Organic cation transporters (OCTs, MATEs), in vitro and in vivo evidence for the importance in drug therapy. Handb Exp Pharmacol. 2011;201:105–167. doi: 10.1007/978-3-642-14541-4_3. [DOI] [PubMed] [Google Scholar]
- 59.Zhou K, Donnelly LA, Kimber CH, Donnan PT, Doney AS, Leese G, et al. Reduced-function SLC22A1 polymorphisms encoding organic cation transporter 1 and glycemic response to metformin: a GoDARTS study. Diabetes. 2009;58:1434–1439. doi: 10.2337/db08-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shikata E, Yamamoto R, Takane H, Shigemasa C, Ikeda T, Otsubo K, Ieiri I. Human organic cation transporter (OCT1 and OCT2) gene polymorphisms and therapeutic effects of metformin. J Hum Genet. 2007;52:117–122. doi: 10.1007/s10038-006-0087-0. [DOI] [PubMed] [Google Scholar]
- 61.Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Stricker BH. Interaction between polymorphisms in the OCT1 and MATE1 transporter and metformin response. Pharmacogenet Genomics. 2010;20:38–44. doi: 10.1097/FPC.0b013e328333bb11. [DOI] [PubMed] [Google Scholar]
- 62.Yee SW, Chen L, Giacomini KM. The role of ATM in response to metformin treatment and activation of AMPK. Nat Genet. 2012;44:359–360. doi: 10.1038/ng.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]