Abstract
To investigate the association of survivin −31G/C, −141G/C, and −241T/C polymorphisms with colorectal cancer (CRC) susceptibility and explore the mechanisms of the survivin polymorphism in CRC development. A case–control study was conducted of 275 CRC cases and 270 healthy controls. Polymorphisms of survivin −31G/C, −141G/C, and −241T/C were genotyped by polymerase chain reaction–restriction fragment length polymorphism. Survivin and Ki-67 expression was analyzed by immunohistochemistry by the Envision technique for the paraffin sections of 152 CRC. It showed that the −31G/C genotype and allele distribution were significantly different between the CRC cases and controls. The −31CC genotype and −31C allele were over-represented among the CRC cases. Compared with the CC genotype, the GC and GG genotypes had a significantly decreased risk of CRC (p=0.015). Survivin and Ki-67 expression of patients with the CC genotype was significantly higher than the patients with the GC and GG genotypes. In addition, a significantly positive correlation was found between expression of Survivin and Ki-67. There were no significant difference of the −141G/C and −241T/C polymorphism distributions among cases and controls. Survivin 31G/C may adjust the Survivin expression, and it might contribute to a risk of developing CRC.
Three polymorphisms of the survivin gene were examined in normal individuals and colorectal cancer patients. Tissues were studied for expression of survivin and Ki-67 expression by immunohistochemistry. 31G/C altered protein expression and may contribute to the risk of colorectal cancer.
Introduction
Defects in the apoptosis signaling pathway that are common in cancer cells are thought to play an important role in tumor initiation, progression, and metastasis. The inhibitor of apoptosis proteins (IAPs) is a family of antiapoptotic proteins that inhibit initiator (caspase-9) and effector caspases (caspase-3 and caspase-7) to prevent apoptosis (Thompson, 1995; Raff, 1998). Survivin, a member of the IAP family, plays a key role in modulating processes, such as cell division, tissue patterning, cytokine activation, and activation of various cell signaling pathways (Altieri, 2003; Pennati et al., 2008). Experimental data have implicated the role of survivin in promotion of cell proliferation as well as inhibition of apoptosis in cancer cells (Ambrosini et al., 1998). Highly expressed Survivin has been detected in many types of human cancers, including colorectal cancer (CRC) (Gazouli et al., 2009; Srivastava et al., 2012). Survivin overexpression was associated with neoplasia and metastasis, suggesting that it might be used as one of the most important diagnostic and prognosis markers for many types of cancers (Altieri, 2003; Montorsi et al., 2007; Li et al., 2008). Polymorphisms in the survivin may influence Survivin production or activity, thereby modulating susceptibility to cancer (Jang et al., 2008). As of yet, around 199 single-nucleotide polymorphisms (SNPs) have been identified in the survivin gene in humans (Srivastava et al., 2012).
CRC, one of the frequent cancers worldwide, arises as a result of the accumulation of genetic errors, many of which affect the control of apoptosis. Effective chemoprevention strategies for CRC must rectify these genetic defects. Ki-67, a nuclear antigen, is a marker of cell growth. It is expressed during all phases of the cell cycle (G1, S, G2, and mitosis), except phase G0. In recent studies, important correlation was found between Ki-67 and the growth, invasion, metastasis, and prognosis of cancer (Gzy Zew Ska et al., 2004). In this study, our aim was to investigate whether the −31G/C, −141G/C, and −241T/C SNPs of the human survivin could represent a risk factor for CRC development and explore the expression of Survivin and Ki-67 in CRC patients with different genotypes.
Materials and Methods
Study population
This case–control study consisted of 275 CRC patients and 270 healthy controls. All subjects were unrelated Han nationality living in Luzhou City, Sichuan Province, and surrounding regions. CRC patients were found with newly diagnosed disease and were recruited from Department of Oncology, the First Affiliated Hospital of Luzhou Medical College, between July 2009 and March 2011. The patients were histological confirmed by the local diagnosing pathologists. Controls were randomly selected from 3200 individuals who participated in a healthy checkup during the same period as the cases were recruited. These control subjects were matched with the patients in terms of age and sex, excluding those with medical history of surgery or chronic disease. Approval of the study was given by the ethics committee of Chinese Human Genome. Each subject was personally face-to-face interviewed by trained interviewers for information on demographic data as well as related risk factors such as tobacco smoking, alcohol drinking, and medical history of viral hepatitis type. After interview, ∼5-mL venous blood sample was collected. DNA was extracted from the peripheral blood with the phenolic chloroform method and frozen at −20°C until tested. Paraffin blocks were collected from the CRC patient postoperative sample.
Table 1 summarizes the characteristics of the 275 CRC patients and 270 healthy control subjects included in this study. The median age of the patients was 54 years (range 35–82 years) and for the controls was 56 years (range 32–80 years). No significant differences were identified between the cases and controls for the age, sex, smoking, and alcohol consumption status. This suggested that the matching based on these two variables was adequate. The criteria for exclusion of the study subjects were as follows: the subjects with recurring CRC, history of cancers, other serious diseases, genetic relationship, or the subjects refusing to inquisition, and no detailed personal and clinicopathological data.
Table 1.
Characteristics of the Study Population
| Variables | CRC patients, n=275(%) | Controls, n=270(%) | χ2 | p |
|---|---|---|---|---|
| Age | ||||
| ≥60 | 130 (47.3) | 125 (46.3) | 0.052 | 0.819 |
| <60 | 145 (52.7) | 145 (53.7) | ||
| Sex | ||||
| Male | 152 (55.3) | 143 (53.0) | 0.293 | 0.588 |
| Female | 123 (44.7) | 127 (47.0) | ||
| Smoking status | ||||
| Smokers | 107 (38.9) | 126 (46.7) | 3.350 | 0.067 |
| Nonsmokers | 168 (61.1) | 144 (53.3) | ||
| Alcohol use | ||||
| Drinkers | 132 (48.0) | 122 (45.2) | 0.434 | 0.510 |
| Nondrinkers | 143 (52.0) | 148 (54.8) | ||
CRC, colorectal cancer.
Genotypes
The −31G/C, −141G/C, and −241T/C genotypes of survivin were determined by polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP). The PCR primers were designed by Primer design software primer3 and provided by Shanghai Sangon Biological Engineering Company. The primers for PCR, the restriction enzymes (MBI Fermentas), and the fragments length after digestion are shown in Table 2. Amplification was carried out on Applied Bio-systems AB (USA). The annealing temperature was 53°C, 55°C, and 56°C for −31G/C, −141G/C, and −241T/C, respectively. After digested by restriction enzymes, the digested products were analyzed on 8% nondenaturing polyacrylamide gel electrophoresis followed by silver staining. The genotyping method was confirmed by the DNA sequencing analysis.
Table 2.
Polymerase Chain Reaction–Restriction Fragment Length Polymorphism-Based Assay of Survivin SNPs
| SNP | Position | Sequence of primer | Enzyme | Interpretation (bp) |
|---|---|---|---|---|
| rs9904341 | −31 | 5-GACTACAACTCCCGGCACAC-3 5-TGTAGAGATGCGGTGGTCCT-3 |
EcoO109I | CC: 226 GC: 226, 134, 92 GG:134, 92 |
| rs17882312 | −141 | 5-GATTACAGGCGTGAGCCACT-3 5-TCAAATCTGGCGGTTAATGG-3 |
BanI | GG:162, 104 GC: 266, 162, 104 CC: 266 |
| rs17878467 | −241 | 5-CTCAGCCTCCCGAGTAGTTG-3 5-TCAAATCTGGCGGTTAATGG-3 |
HaeI | CC: 220, 202 TC: 422, 220, 202 TT: 422 |
SNP, single-nucleotide polymorphism.
Immunohistochemistry
Paraffin sections (3 μm) were dried, deparaffinized, and rehydrated in graded alcohol to water. Heat-mediated antigen retrieval was performed using pressure cooker treatment for 5 min in the EDTA buffer (pH 8.0). Endogenous peroxide activity was quenched by applying 3% hydrogen peroxide for 10 min. The slides were incubated for 40 min at 37°C with a primary antibody for Survivin (rabbit polyclonal Survivin antibody, 1:100; Bio-World) and Ki-67 (clone: MIB-1, 1:200; Dako). After washing, the tissue sections were incubated in the second antibodies (Envision™ and HRP rabbit/mouse; Dako) for 20 min at 37°C. In the negative controls, the primary antibody was omitted. The reaction products were visualized by DAB.
Expression of Survivin was determined in the nucleus and in the cytoplasm by assessing semiquantitatively the percentage of marked tumor cells and the staining intensity. Score for percentage of positive cells is as follows: 1, 1%–10% positive cells; 2, 11%–50%; 3, 51%–80%; and 4, >80% positive cells. Staining intensity was scored as 1, weak; 2, moderate; and 3, intensive. Scores for the percentage of positive cells and scores for expression intensities were multiplied to calculate an immunoreactivity score. The score ≤3 considered negative (−) and 4–12 considered positive (+) (Taubert et al., 2010). Ki-67 was evaluated by counting the number of positive nuclei in 500 tumor cells, and the percentage was calculated as the Ki-67-labeling index. Ki-67 negativity was defined as ≤10% (−), and positivity was defined as >10% (+) (Al Gizawy et al., 2011).
Statistical analysis
Genotypes were tested for the Hardy–Weinberg equilibrium test. The numeration data groups were compared using the chi-square test; relationship between Survivin and Ki-67 was assessed using the Spearman's rank correlation analysis. The odds ratios and 95% confidence intervals were calculated by multivariate logistic regression model test, adjusted for sex, age, smoking, and alcohol consumption status. For all hypothesis tests, a two-tailed significance level of p<0.05 was considered statistically significant. All the data were analyzed using the SPSS 11.5 statistical package.
Results
Survivin gene polymorphisms of the subjects
The genotype distributions of −31G/C and −241T/C polymorphisms among the controls and the cases were in the Hardy–Weinberg equilibrium (−31G/C: χ2=0.128, p=0.720; −241T/C: χ2=1.989, p=0.158). Genotype and allele frequencies of −31G/C and −241T/C among the cases and controls are summarized in Table 3. The frequencies of the CC, GC and GG genotypes for the −31G/C polymorphism were 40.0%, 44.7%, and 15.3% among cases and 28.5%, 51.1%, and 20.4% among controls, respectively. It showed significant differences in the −31G/C genotypic distributions (χ2=8.383, p=0.015). The −31C allele was over-represented among the CRC cases (χ2=7.699, p=0.006). Adjusted for sex, age, smoking, and alcohol consumption status, the risks of CRC for individuals carrying the GC and GG genotypes were decreased, and they were 0.618-fold and 0.519-fold compared with the individuals carrying the CC genotype. The −31CC genotype may increase the risk of CRC.
Table 3.
Frequency Distributions of Survivin Polymorphisms Among Cases and Controls
| Polymorphisms | Cases, n=275(%) | Controls, n=270(%) | OR (95% CI)a | p |
|---|---|---|---|---|
| −31G/C Genotype | ||||
| CC | 110 (40.0) | 77 (28.5) | 1.000 | 0.015 |
| GC | 123 (44.7) | 138 (51.1) | 0.618 (0.420–0.907) | |
| GG | 42 (15.3) | 55 (20.4) | 0.519 (0.312–0.865) | |
| Allele frequency | ||||
| C | 343 (62.4) | 292 (54.1) | 1.000 | 0.006 |
| G | 207 (37.6) | 248 (45.9) | 0.711 (0.558–0.905) | |
| −241T/C genotype | ||||
| CC | 166 (60.4) | 165 (61.1) | 1.000 | 0.919 |
| TC | 92 (33.4) | 88 (32.6) | 1.083 (0.732–1.602) | |
| TT | 17 (6.2) | 17 (6.3) | 1.084 (0.509–2.308) | |
| Allele frequency | ||||
| C | 424 (77.1) | 418 (77.2) | 1.000 | 0.901 |
| T | 126 (22.9) | 122 (22.8) | 1.018 (0.767–1.352) | |
Adjusted for sex, age, smoking, and alcohol consumption status.
OR, odds ratios; CI, confidence intervals.
Comparisons between the case and control groups showed no significant differences in the −241T/C genotypic distributions. In this study, no polymorphism of −141G/C was found, and the genotype was CC.
The associations of clinicopathologic parameters of CRC and −31G/C polymorphisms
The association of the genotypic distribution of the −31G/C polymorphism and clinicopathologic features of CRC was analyzed. Results showed that the location of tumor, gross type, tumor size, histologic type, depth of invasion, differentiation degree, lymph node metastasis, and TNM stage were not associated with the −31G/C polymorphism. The data of the −31G/C genotype and clinicopathologic parameters are summarized in Table 4.
Table 4.
The Associations of the Clinicopathologic Parameters of Colorectal Cancer and the Survivin 31G/C Single-Nucleotide Polymorphisms
| |
−31G/C genotype |
|
|
||
|---|---|---|---|---|---|
| Clinicopathological parameters | CC n=110 (%) | GC n=123 (%) | GG n=42 (%) | χ2 | p |
| Location | |||||
| Rectum | 70 (63.6) | 65 (52.8) | 22 (52.4) | 3.209 | 0.201 |
| Colon | 40 (36.4) | 58 (47.2) | 20 (47.6) | ||
| Gross type | |||||
| Ulcerated | 61 (55.5) | 72 (58.5) | 28 (66.7) | 1.574 | 0.455 |
| Elevated type | 49 (44.5) | 51 (41.5) | 14 (33.3) | ||
| Tumor size (cm) | |||||
| ≥3 | 72 (65.5) | 89 (72.4) | 29 (69.0) | 1.296 | 0.523 |
| <3 | 38 (34.5) | 34 (27.6) | 13 (31.0) | ||
| Histologic type | |||||
| Adenocarcinoma | 101 (91.8) | 108 (87.8) | 34 (81.0) | 3.558 | 0.169 |
| Mucinous adenocarcinoma | 9 (8.2) | 15 (12.2) | 8 (19.0) | ||
| Depth of invasion | |||||
| Within serosa | 66 (60.0) | 78 (63.4) | 29 (69.0) | 1.091 | 0.580 |
| Serosa and beyond | 44 (40.0) | 45 (36.6) | 13 (31.0) | ||
| Lymph node metastasis | |||||
| Yes | 49 (44.5) | 51 (41.5) | 18 (42.9) | 0.225 | 0.893 |
| No | 61 (55.5) | 72 (58.5) | 24 (57.1) | ||
| TNM stage | |||||
| I+II | 63 (57.3) | 81 (65.9) | 26 (61.9) | 1.812 | 0.404 |
| III+IV | 47 (42.7) | 42 (34.1) | 16 (38.1) | ||
| Differentiation (adenocarcinoma) | |||||
| Well | 49 (44.5) | 69 (56.1) | 23 (54.8) | 3.343 | 0.188 |
| Poor | 61 (55.5) | 54 (43.9) | 19 (45.2) | ||
The expression of Survivin and Ki-67 in CRC with various −31G/C genotypes
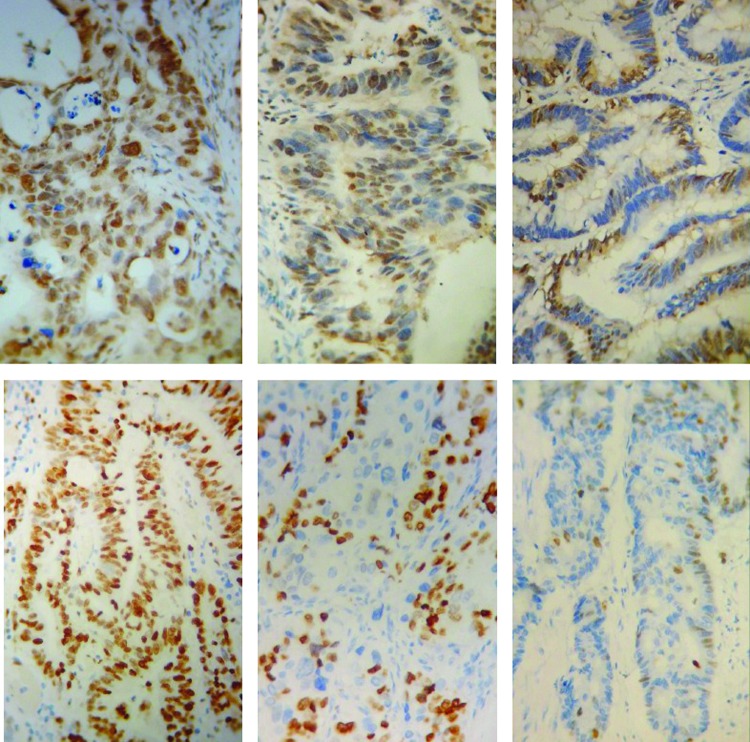
Expression of Survivin was determined most in the nucleus, small part in the cytoplasm. The Ki-67 immunostaining was confined to the nucleus. The positive cells of Survivin and Ki-67 decreased gradually from CC to GC, GG (Fig. 1). The expression of Survivin and Ki-67 was associated with the −31G/C polymorphism (Table 5, p<0.05). Survivin and Ki-67 expression of patients with the CC genotype was significantly higher than that in patients with the GC and GG genotypes (p<0.05). There were no significant differences of Survivin and Ki-67 expression between patients with the GC and GG genotype.
FIG. 1.
Immunohistochemical staining for Survivin (above row) and Ki-67 (following row) in colorectal cancer with the CC genotype (left column), GC genotype (center column), and GG genotype (right column) (×400). Color images available online at www.liebertpub.com/dna
Table 5.
The Relationship of the Survivin −31G/C Polymorphism and Survivin, Ki-67 in Colorectal Cancer Patients
| |
Survivin, n (%) |
|
|
Ki-67, n (%) |
|
|
||
|---|---|---|---|---|---|---|---|---|
| Polymorphism | + | − | χ2 | p | + | − | χ2 | p |
| CC | 54 (90.0) | 6 (10.0) | 5.356a | 0.021a | 59 (98.3) | 1 (1.7) | 5.758a | 0.016a |
| GC | 45 (73.8) | 16 (26.2) | 0.210b | 0.646b | 53 (86.9) | 8 (13.1) | 0.154b | 0.695b |
| GG | 22 (71.0) | 9 (29.0) | 5.378c | 0.020c | 26 (83.9) | 5 (16.1) | 6.942c | 0.008c |
| 6.697d | 0.035d | 6.970d | 0.031d | |||||
+, Survivin positivity was defined as score 4–12; Ki-67 positivity was defined as >10%.
−; Survivin negativity was defined as score ≤3; Ki-67 negativity was defined as ≤10%.
Compare with CC and GC groups.
Compare with GC and GG groups.
Compare with GG and CC groups.
Compare with all of three groups.
After Spearman's rank correlation analysis (Table 6), the expression of Survivin in CRC was positively correlated with the expression of Ki-67 (r=0.178, p=0.029).
Table 6.
The Relationship Between Survivin and Ki-67 in Colorectal Cancer Patients
| |
Ki-67 |
|
|
|
|---|---|---|---|---|
| Survivin | + | − | r | p |
| + | 113 | 8 | ||
| − | 25 | 6 | 0.178 | 0.029 |
+, Survivin positivity was defined as score 4–12. Ki-67 positivity was defined as >10%.
−; Survivin negativity was defined as score ≤3. Ki-67 negativity was defined as ≤10%.
Discussion
CRC is characterized by the partial suppression of apoptosis, which in turn gives tumor a selective advantage for survival and can cause current chemotherapy approaches to be ineffective (Li and Brattain, 2006). Survivin promotes tumor development and progression by inhibiting apoptosis and increasing cell proliferation (Altieri, 2008). In the above case–control study, we evaluated the relationship between the polymorphisms of −31G/C, −141G/C, and −241T/C and CRC risk in a Chinese Han population. In addition, Survivin and Ki-67 expression had been analyzed.
Results showed that the −241T/C polymorphism was not associated with the susceptibility of CRC, and there was no −141G/C polymorphism of survivin in our study. However, the −31CC genotype could increase the risk of CRC. We also showed that the −31G/C SNP was not associated with any clinicopathologic features of CRC, such as the location of tumor, depth of invasion, tumor size, histologic type, lymph node metastasis, and TNM stage. For the clinicopathologic features of CRC, inconsistent with our results, Gazouli et al. (2009) showed the −31CC genotype and the −31C allele were significantly more frequent in stage grouping III and IV than in stage grouping I and II. Han et al. (2009) showed 31G/C was significantly associated with the age of disease onset of ovarian cancer. It is hard to decipher the reason for the discrepancy. It may be due to the different regions and malignancy, and the inadequate study design such as a limited sample size or nonrandom sampling should be considered.
In a southern Chinese population, Huang et al. (2009) found that the survivin gene −31G/C polymorphism is associated with sporadic CRC risk, and the G-variant genotype is the independent protective factor against sporadic CRC. In recent years, some research showed that the −31CC genotype of survivin was associated with gastric cancer, ESCC, and oral cancer and may be a risk factor of them in the Chinese population (Cheng et al., 2008; Yang et al., 2009; Weng et al., 2012), as Srivastava et al. (2012) suggested an important role of the survivin −31G/C polymorphism with cancer risk, especially in the Asian population. However, the mechanism is still unknown. Survivin is involved in the regulation of cell division and survival, two key processes in cancer (Antonacopoulou et al., 2011). In the progress of the CRC, it is not clear whether closer relationship between the survivin gene and the cell proliferation.
In the present study, we demonstrated that the expression of Survivin in the cancer tissue of CRC patients with the −31CC genotype was significantly higher than that with the GC and GG genotype, but there were no differences between the GC and GG genotype. Xu et al. (2004) reported that the presence of the −31CC genotype was more frequent in cancer cell lines, and mRNA and protein levels of survivin increased in these cell lines. In a M Gazouli et al. (2009) study, they observed that the survivin mRNA level of CRC cases with the −31CC homozygous genotype exhibited ∼1.6-fold higher than the carriers of the −31GG and −31GC genotypes. However, Cheng et al. (2008) found that survivin mRNA was overexpressed in gastric cancer cases, but with no significant difference in various −31G/C polymorphism genotypes. Our results showed that the Survivin protein expression level of CRC cases with the −31CC genotype was significantly higher than that with the −31GG and −31GC genotypes. This finding confirmed previous studies (Xu et al., 2004; Jang et al., 2008; Yang et al., 2009). The −31G/C polymorphism located at the cell cycle-dependent element/cell cycle homology (CDE/CHR) region repressor elements and conferred aberrant cell cycle-dependent transcription, mediated through the functional disruption of binding at the CDE/CHR repressor motifs (Xu et al., 2004). We speculate that the −31G/C polymorphism may influence survivin transcription and translation by modifying the banding motif of the CDE/CHR repressor. Further studies should be needed to confirm this surmise.
Nucleocytoplasmic localization of Survivin in tumor cells, determined by immunohistochemistry, has been reported to provide prognostic information in several types of cancer (Fortugno et al., 2002; Mohamed et al., 2009). Regarding the Survivin immunostaining, Aune et al. (2011) reported that the nuclear staining was recorded in ovarian carcinomas, and this pattern is obviously related to proliferation, whereas the role of the cytoplasmatic immunoreactivity is not fully clarified. Further, the Survivin indices paralleled those of Ki-67 and PHH3, with a considerable overlap between the malignancy groups (Li, 2005; Li et al., 2005). In our research, the expression of Survivin was determined most in the nucleus, small part in the cytoplasm. Moreover, we found that the expression of Survivin was positively correlated with the expression of Ki-67 in CRC tissues with various −31G/C genotypes. In consistent with the expression of Survivin, the Ki-67 expression of CRC patients with the −31CC genotype was significantly higher than those with the GC and GG genotype. Survivin activates aurora-B kinase (ABK) catalyzing mitosis, and overexpression of Survivin provides activation of ABK, which results in increased mitosis and proliferation (Boman et al., 2007). This effect may be synergistic with Survivin's ability to prevent apoptosis, so both effects would promote tumor growth. Ultimately, both these mechanisms contribute to the exponential increase of proliferative cell populations (Zhang et al., 2010).
Conclusion
The survivin 31G/C polymorphism may adjust the Survivin protein expression, and it might contribute to a risk of developing CRC. The survivin 31G/C polymorphism may be a genetic modifier for CRC prognosis in the Chinese population. Further rigorous studies with larger sample sizes and studies of biological functions of these SNPs are needed to validate the role of the survivin promoter SNPs in the development of CRC.
Acknowledgments
This work was financially supported by the State Natural Sciences Foundation Projects of China (No. 81101679) and the Programs of Health Bureau of Sichuan Province (No. 100240 and 120365).
Disclosure Statement
No competing financial interests exist.
References
- Al Gizawy S.M. Essa H.H. Refaiy A.M., et al. Prognostic value of expression of survivin And Ki-67 in head and neck squamous cell carcinoma treated by chemoradiotherapy. Life Sci J. 2011;8:724–733. [Google Scholar]
- Altieri D.C. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22:8581–8589. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- Altieri D.C. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- Altieri D.C. Survivin, cancer networks and pathwaydirected drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- Ambrosini G. Adida C. Sirugo G., et al. Induction of apoptosis and inhibition of cell proliferation bysurvivin gene targeting. J Biol Chem. 1998;273:11177–11182. doi: 10.1074/jbc.273.18.11177. [DOI] [PubMed] [Google Scholar]
- Antonacopoulou A.G. Floratou K. Bravou V., et al. The survivin-31 SNP in human colorectal cancer correlates with survivin splice variant expression and improved overall survival. Cell Oncol (Dordr) 2011;34:381–391. doi: 10.1007/s13402-011-0038-4. [DOI] [PubMed] [Google Scholar]
- Aune G. Stunes A.K. Tingulstad S., et al. The proliferation markers Ki-67/MIB-1, phosphohistone H3, and survivin may contribute in the identification of aggressive ovarian carcinomas. Int J Clin Exp Pathol. 2011;4:444–453. [PMC free article] [PubMed] [Google Scholar]
- Boman B.M. Wicha M.S. Fields J.Z. Runquist O.A. Symmetric division of cancer stem cells a key mechanism in tumor growth that should be targeted in future therapeutic approaches. Clin Pharmacol Ther. 2007;81:893–898. doi: 10.1038/sj.clpt.6100202. [DOI] [PubMed] [Google Scholar]
- Cheng Z.J. Hu L.H. Huang S.J., et al. Correlation of −31G/C polymorphisms of survivin promoter to tumorigenesis of gastric carcinoma. Ai Zheng. 2008;27:258–263. [PubMed] [Google Scholar]
- Fortugno P. Wall N.R. Giodini A., et al. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J Cell Sci. 2002;115:575–585. doi: 10.1242/jcs.115.3.575. [DOI] [PubMed] [Google Scholar]
- Gazouli M. Tzanakis N. Rallis G., et al. Survivin −31G/C promoter polymorphism and sporadic colorectal cancer. Int J Colorectal Dis. 2009;24:145–150. doi: 10.1007/s00384-008-0601-2. [DOI] [PubMed] [Google Scholar]
- Gzy Zew Ska J. Guzinska-Ustymow Icz K. Lebelt A., et al. Evaluation of proliferating markers K-i 67, PCNA in gastric cancers. Rocz Akad Med Bialymst. 2004;49:64–66. [PubMed] [Google Scholar]
- Han C.H. Wei Q. Lu K.K., et al. Polymorphisms in the survivin promoter are associated with age of onset of ovarian cancer. Int J Clin Exp Med. 2009;2:289–299. [PMC free article] [PubMed] [Google Scholar]
- Huang J. Wang J.-P. Wang L., et al. Association between survivin promoter −31G/C polymorphism and genetic susceptibility to sporadic colorectal cancer. Chin J Pathophysiol. 2009;25:2344–2348. [Google Scholar]
- Jang J.S. Kim K.M. Kang K.H., et al. Polymorphisms in the survivin gene and the risk of lung cancer. Lung Cancer. 2008;60:31–39. doi: 10.1016/j.lungcan.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Li D.X. Zeng B. Xie L.Q. Expression and the clinical significance of survivin protein and β-catenin protein in primary hepatocellular carcinoma (HCC) J Mod Oncol. 2008;16:62–66. [Google Scholar]
- Li F. Role of survivin and its splice variants in tumorigenesis. Br J Cancer. 2005;92:212–216. doi: 10.1038/sj.bjc.6602340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Brattain M.G. Role of the survivin gene in pathophysiology. Am J Pathol. 2006;169:1–11. doi: 10.2353/ajpath.2006.060121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Yang J. Ramnath N., et al. Nuclear or cytoplasmic expression of survivin:what is the significance? Int J Cancer. 2005;114:509–512. doi: 10.1002/ijc.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed S. Yasufuku K. Nakajima T. Nuclear survivin in pN2 nonsmall cell lung cancer: prognostic and clinical implications. Eur Respir J. 2009;33:127–133. doi: 10.1183/09031936.00068708. [DOI] [PubMed] [Google Scholar]
- Montorsi M. Maggioni M. Falleni M., et al. Survivin gene expression in chronic liver disease and hepatocellular carcinoma. Hepatogastroenterology. 2007;54:2040–2044. [PubMed] [Google Scholar]
- Pennati M. Folini M. Zaffaroni N. Targeting survivin in cancer therapy. Expert Opin Ther Targets. 2008;12:463–476. doi: 10.1517/14728222.12.4.463. [DOI] [PubMed] [Google Scholar]
- Raff M. Cell suicide for beginners. Nature. 1998;396:119–122. doi: 10.1038/24055. [DOI] [PubMed] [Google Scholar]
- Srivastava K. Srivastava A. Mittal B. Survivin promoter −31G/C polymorphism and cancer susceptibility: a meta-analysis. Mol Biol Rep. 2012;39:1509–1516. doi: 10.1007/s11033-011-0889-9. [DOI] [PubMed] [Google Scholar]
- Taubert H. Heidenreich C. Holzhausen H.J., et al. Expression of survivin detected by immunohistochemistry in the cytoplasm and in the nucleus is associated with prognosis of leiomyosarcoma and synovial sarcoma patients. BMC Cancer. 2010;24:65. doi: 10.1186/1471-2407-10-65. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C.B. Apoptosis is the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- Weng C.J. Hsieh Y.H. Chen M.K., et al. Survivin SNP-carcinogen interactions in oral cancer. J Dental Res. 2012;91:358–363. doi: 10.1177/0022034512438402. [DOI] [PubMed] [Google Scholar]
- Xu Y. Fang F. Ludewig G., et al. A mutation found in the promoter region of the human survivin gene is correlated to overexpression of survivin in cancer cells. DNA Cell Biol. 2004;23:527–537. doi: 10.1089/dna.2004.23.527. [DOI] [PubMed] [Google Scholar]
- Yang X. Xiong G. Chen X., et al. Polymorphisms of survivin promoter are associated with risk of esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2009;135:1341–1349. doi: 10.1007/s00432-009-0575-7. [DOI] [PubMed] [Google Scholar]
- Zhang T. Fields J.Z. Opdenaker L., et al. Survivin-induced aurora-B kinase activation: a mechanism by which APC mutations contribute to increased mitoses during colon cancer development. Am J Pathol. 2010;177:2816–2826. doi: 10.2353/ajpath.2010.100047. [DOI] [PMC free article] [PubMed] [Google Scholar]