Abstract
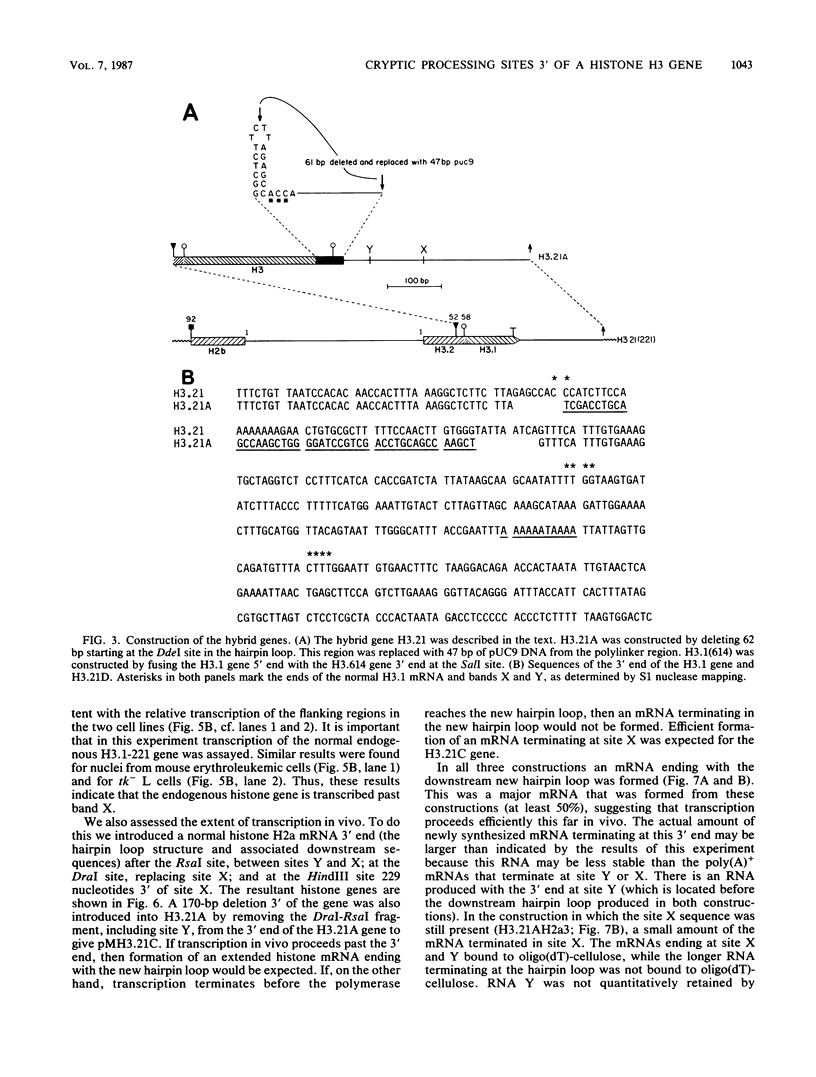
Introduction of the mouse histone H3.1 gene into tk- mouse L cells by cotransfection with the herpesvirus thymidine kinase gene resulted in the production of two mRNAs from the transfected gene, one with a normal 3' end and the other one with a longer 3'-untranslated region, ending at site X, which was poly(A)+. In contrast, the endogenous histone H3.1 gene only produced a single mRNA. The cryptic poly(A)+ site was only used when the histone H3.1 gene was transfected. To localize possible downstream cryptic processing sites, the hairpin loop at the end of the histone gene was deleted and the resulting deletions were introduced into L cells. Two major mRNAs were produced from this gene, one ending at site X and the major one ending at site Y, which was located 150 nucleotides before site X. Transcription extended downstream of site X efficiently in the endogenous gene, as judged by the extent of transcription of downstream sequences in isolated nuclei. Transcription extended downstream of site X in the transfected gene because the placement of a normal histone 3' end downstream of site X resulted in transcripts that ended at site X and longer transcripts that ended with the new histone 3' end. These results indicate that transcription may normally proceed a substantial distance past the hairpin loop (greater than 500 bases). The formation of the different 3' ends in these transfected genes was due to competition between different processing mechanisms.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alterman R. B., Ganguly S., Schulze D. H., Marzluff W. F., Schildkraut C. L., Skoultchi A. I. Cell cycle regulation of mouse H3 histone mRNA metabolism. Mol Cell Biol. 1984 Jan;4(1):123–132. doi: 10.1128/mcb.4.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alterman R. B., Sprecher C., Graves R., Marzluff W. F., Skoultchi A. I. Regulated expression of a chimeric histone gene introduced into mouse fibroblasts. Mol Cell Biol. 1985 Sep;5(9):2316–2324. doi: 10.1128/mcb.5.9.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantine J. E., Woodland H. R. Polyadenylation of histone mRNA in Xenopus oocytes and embryos. FEBS Lett. 1985 Jan 28;180(2):224–228. doi: 10.1016/0014-5793(85)81075-9. [DOI] [PubMed] [Google Scholar]
- Bannon G. A., Calzone F. J., Bowen J. K., Allis C. D., Gorovsky M. A. Multiple, independently regulated, polyadenylated messages for histone H3 and H4 in Tetrahymena. Nucleic Acids Res. 1983 Jun 25;11(12):3903–3917. doi: 10.1093/nar/11.12.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier C., Folk W., Birnstiel M. L. The terminal RNA stem-loop structure and 80 bp of spacer DNA are required for the formation of 3' termini of sea urchin H2A mRNA. Cell. 1983 Dec;35(2 Pt 1):433–440. doi: 10.1016/0092-8674(83)90176-9. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Schümperli D., Sconzo G., Birnstiel M. L. 3' editing of mRNAs: sequence requirements and involvement of a 60-nucleotide RNA in maturation of histone mRNA precursors. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1057–1061. doi: 10.1073/pnas.81.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Breindl M., Gallwitz D. On the translational control of histone synthesis. Quantitation of biologically active histone mRNA from synchronized HeLa cells and its translation in different cell-free systems. Eur J Biochem. 1974 Jun 1;45(1):91–97. doi: 10.1111/j.1432-1033.1974.tb03533.x. [DOI] [PubMed] [Google Scholar]
- Brush D., Dodgson J. B., Choi O. R., Stevens P. W., Engel J. D. Replacement variant histone genes contain intervening sequences. Mol Cell Biol. 1985 Jun;5(6):1307–1317. doi: 10.1128/mcb.5.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler W. B., Mueller G. C. Control of histone synthesis in HeLa cells. Biochim Biophys Acta. 1973 Feb 4;294(1):481–496. doi: 10.1016/0005-2787(73)90104-4. [DOI] [PubMed] [Google Scholar]
- DeLisle A. J., Graves R. A., Marzluff W. F., Johnson L. F. Regulation of histone mRNA production and stability in serum-stimulated mouse 3T6 fibroblasts. Mol Cell Biol. 1983 Nov;3(11):1920–1929. doi: 10.1128/mcb.3.11.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J. D., Sugarman B. J., Dodgson J. B. A chicken histone H3 gene contains intervening sequences. Nature. 1982 Jun 3;297(5865):434–436. doi: 10.1038/297434a0. [DOI] [PubMed] [Google Scholar]
- Fahrner K., Yarger J., Hereford L. Yeast histone mRNA is polyadenylated. Nucleic Acids Res. 1980 Dec 11;8(23):5725–5737. doi: 10.1093/nar/8.23.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gick O., Krämer A., Keller W., Birnstiel M. L. Generation of histone mRNA 3' ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986 Jun;5(6):1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves R. A., Marzluff W. F. Rapid reversible changes in the rate of histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol Cell Biol. 1984 Feb;4(2):351–357. doi: 10.1128/mcb.4.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves R. A., Wellman S. E., Chiu I. M., Marzluff W. F. Differential expression of two clusters of mouse histone genes. J Mol Biol. 1985 May 25;183(2):179–194. doi: 10.1016/0022-2836(85)90211-6. [DOI] [PubMed] [Google Scholar]
- Heintz N., Sive H. L., Roeder R. G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983 Apr;3(4):539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Robins A. J., Colman A., Wells J. R. Chicken histone H5 mRNA: the polyadenylated RNA lacks the conserved histone 3' terminator sequence. Nucleic Acids Res. 1982 Nov 11;10(21):6777–6785. doi: 10.1093/nar/10.21.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Stauber C., Schindler R., Schümperli D. Faithful cell-cycle regulation of a recombinant mouse histone H4 gene is controlled by sequences in the 3'-terminal part of the gene. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4389–4393. doi: 10.1073/pnas.82.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molgaard H. V., Perucho M., Ruiz-Carrillo A. Histone H5 messenger RNA is polyadenylated. Nature. 1980 Jan 31;283(5746):502–504. doi: 10.1038/283502a0. [DOI] [PubMed] [Google Scholar]
- Price D. H., Parker C. S. The 3' end of drosophila histone H3 mRNA is produced by a processing activity in vitro. Cell. 1984 Sep;38(2):423–429. doi: 10.1016/0092-8674(84)90497-5. [DOI] [PubMed] [Google Scholar]
- Ruderman J. V., Pardue M. L. A portion of all major classes of histone messenger RNA in amphibian oocytes is polyadenylated. J Biol Chem. 1978 Mar 25;253(6):2018–2025. [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Structure of a cluster of mouse histone genes. Nucleic Acids Res. 1983 Oct 11;11(19):6679–6697. doi: 10.1093/nar/11.19.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K., Galli G., Busslinger M., Birnstiel M. L. The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J. 1984 Dec 1;3(12):2801–2807. doi: 10.1002/j.1460-2075.1984.tb02212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. D., Wellman S. E., Marzluff W. F. Sequences of four mouse histone H3 genes: implications for evolution of mouse histone genes. J Mol Evol. 1986;23(3):242–249. doi: 10.1007/BF02115580. [DOI] [PubMed] [Google Scholar]
- Wells D., Kedes L. Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc Natl Acad Sci U S A. 1985 May;82(9):2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudt L. P., Pastink A., Kempers-Veenstra A. E., Jansen A. E., Mager W. H., Planta R. J. The genes coding for histone H3 and H4 in Neurospora crassa are unique and contain intervening sequences. Nucleic Acids Res. 1983 Aug 25;11(16):5347–5360. doi: 10.1093/nar/11.16.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]