Abstract
Recent years have seen a growing body of evidence that enzymatic remodeling of heparan sulfate proteoglycans profoundly affects a variety of physiological and pathological processes, including inflammation, neovasvularization and tumor development. Heparanase is the sole mammalian endoglycosidase that cleaves heparan sulfate. Extensively studied in cancer progression and aggressiveness, heparanase enzyme was recently implicated in several inflammatory disorders as well. Although the precise mode of heparanase action in inflammatory reactions is still not completely understood, the fact that heparanase activity is mechanistically important both in malignancy and in inflammation argues that this enzyme is a candidate molecule linking inflammation and tumorigenesis in inflammation-associated cancers. The elucidation of the specific effects of heparanase in cancer development, particularly when inflammation is a causal factor, will accelerate the development of novel therapeutic/chemopreventive interventions and help to better define target patient populations in which heparanase-targeting therapies could be particularly beneficial.
Keywords: Heparan sulfate, Heparanase, Extracellular matrix, Tumor microenvironment, Macrophages, Inflammation, Cancer
Introduction
Mammalian heparanase cleaves side chains of heparan sulfate (HS) proteoglycan, composed of repeating disaccharide units of hexuronic (either glucuronic or iduronic) acid linked to N-acetylglucosamine, with various degrees of O- and N-sulfation, deacetylation and epimerization. HS proteoglycans are ubiquitously found in the extracellular matrix (ECM) and plasma membrane of cells [1–4]. HS chains play key roles in numerous biological functions, including developmental processes, cytoskeleton organization, cell-cell and cell-ECM interactions [3,5,6]. The mechanisms underlying biologic action of HS include modulation of the activity of selected ligands, especially cytokines and growth factors, through either control of their cellular uptake, sequestration to the ECM and cell surface, or participation in receptor-ligand complex formation [1,3,6–9]. Additionally HS interacts with laminin, fibronectin, and collagens I and IV, through which HS contributes to the structural integrity of the ECM, preserves proper tissue organization and inhibits cellular invasion by promoting cell-cell and cell-ECM interactions [10,11]. In view of this functional diversity, it is not surprising that enzymatic degradation of HS may profoundly affect a variety of pathophysiological processes, including tumorigenesis, neovasvularization and inflammation [12–15]. While the precise mode of action of HS-degrading heparanase in inflammation until recently remained underinvestigated, the role of the enzyme in cancer was extensively studied. Cleavage of HS by heparanase promotes tumor progression via disassembly of extracellular barriers for cell invasion, release of HS–bound angiogenic and growth factors from the ECM depots, and generation of bioactive HS fragments which promote growth factor-receptor binding, dimerization and signaling [2,8,13,16,17]. Preferential expression of the enzyme was found in various cancer types [16], and patients bearing tumors with high levels of heparanase had a significantly shorter postoperative survival time as compared to patients with low heparanase levels in their tumors [18–24]. Furthermore, causative role for the enzyme in cancer progression was confirmed by both heparanase over-expression [25–27] and silencing/inhibition [27–31] studies. Notably, in recent years it has become apparent that along with the well documented extracellular catalytic aspects of heparanase action, heparanase protein can exert additional biological functions, such as regulation of gene transcription [32], facilitation of cell adhesion [33,34] and activation of several signaling pathways, either independent [35–37] or dependent [38] on its enzymatic activity. Additionally, the ongoing accumulation of data implicates heparanase in various non-cancerous pathologies (including inflammatory disorders [39–43], atherosclerosis [44,45], graft versus host disease [46], nephropathies [47–50], diabetes [51]). This review will primarily focus on heparanase’s role in inflammation and neoplastic progression, discussing the possible action of the enzyme in coupling inflammation and tumorigenesis in the setting of inflammation-triggered cancer.
Mammalian heparanase and its regulation
Heparanase (endo-β-glucuronidase) is the only known endoglycosidase enzyme capable of HS cleavage. It degrades HS side chains at sites of low sulfation, releasing saccharide products with appreciable size (4–7 kDa) that can still associate with protein ligands and facilitate their biological potency[17,52]. Its close homolog heparanase-2 (Hpa2), cloned based on sequence homology, exhibits no HS degrading enzymatic activity [53]. Intriguingly, Hpa2 was recently implicated in control of enzymatic action of heparanase [54]. Hpa2 protein appears to inhibit heparanase activity, most likely due to its high affinity to HS and ability to associate physically with heparanase [54]. Additional endogenous molecules controlling heparanase enzymatic activity include heparin [55] and eosinophilic major basic protein [56], released by activated mast cells and eosinophils. Inhibition of heparanase enzymatic activity represents only one type of regulatory mechanism. Since uncontrolled cleavage of HS could result in significant tissue damage, heparanase is kept tightly regulated at transcriptional and post-translational levels as well. With a few exceptions (placenta, activated immune cells, keratinocytes), in normal non-cancerous cells and tissues heparanase promoter is constitutively inhibited and the gene is not transcribed, largely due to promoter methylation [57–60] and action of wild type p53, which supresses transcription of the heparanase gene by direct binding to its promoter [61]. Thus, epigenetic changes and/or mutational inactivation of p53 during cancer development may provide molecular explanation for the induction of heparanase expression observed in many human tumors (reviewed in [16]). In addition, early growth response 1 (EGR1) transcription factor is an important regulator of heparanase promoter activity. EGR1 was shown to control heparanase expression acting as either an activator or repressor of heparanase transcription, depending on the cell and tissue type [62,63], while members of SP1 and Ets transcription factor families were associated with basal activity of heparanase promoter [64–66]. Heparanase expression is stimulated by high glucose, reactive oxygen species [44,67], estrogens [68,69] and inflammatory cytokines [41,70–72]
Post-translational processing represents an additional level of complexity in control of heparanase action: The heparanase mRNA encodes for a 61.2 kDa latent pro-enzyme with 543 amino acids which is cleaved into 8 and 50 kDa subunits that non-covalently associate to form an enzymatically active heterodimer [73–75]. Cellular processing of the latent 65 kDa pro-enzyme into its active 8+50 kDa heterodimer is inhibited by specific inhibitor of cathepsin L [76]. Moreover, multiple site-directed mutagenesis and cathepsin L gene silencing and knockout experiments strongly argue that cathepsin L is a predominant protease responsible for post-translational activation of pro-heparanase [77].
Heparanase in inflammation
Heparan sulphate glycosaminoglycans and their enzymatic remodelling are involved in shaping inflammatory responses through control of sequestration and release of cytokines/chemokines in extracellular space, modulation of leukocyte interactions with endothelial cells and ECM, and initiation of innate immune responses through recognition by toll-like receptor 4 (TLR4) [14,77–84]. Augmented expression/enzymatic activity of heparanase was reported in numerous inflammatory conditions, often associated with degradation of HS and remodelling of the ECM [15,39–43]. As can be expected from the diversity of HS functions in inflammation, its degradation by heparanase appears to affect several aspects of inflammatory reactions, such as leukocyte recruitment, extravasation and migration towards inflammation sites; release of cytokines and chemokines anchored within the ECM or cell surfaces, as well as activation of innate immune cells. In early studies heparanase activity originating in immunocytes (neutrophils, activated T-lymphocytes) was found to contribute primarily to their ability to penetrate blood vessel walls and accumulate in target organs [55,78–82]. However immunocytes are not the sole source of the enzyme in inflammation. Up-regulation of heparanase, locally expressed (i.e., by epithelial cells, vascular endothelium) at the site of inflammation, was demonstrated in multiple organ systems, including mouse models of delayed type hypersensitivity [71], vascular injury [83], chronic colitis [41], sepsis-associated lung injury [43], as well as in several auto-immune and auto-inflammatory human disorders, including rheumatoid arthritis [39], atherosclerosis [44,45], inflammatory lung disease [43], psoriasis (our unpublished results), ulcerative colitis and Crohn’s disease [40,41]. The role of heparanase as an important mediator of inflammation in multiple organ sites is further supported by anti-inflammatory effects of heparanase-inhibiting substances in mouse models of autoimmune encephalomyelitis, central nervous system inflammation, contact dermatitis, and colitis [71,84–86]. Yet the precise role of the enzyme in the inflammation process has proved difficult to pin down, as experimental evidence suggest that heparanase may act alternatively either augmenting or restraining inflammatory responses. It appears that tissue/organ specific contextual cues may dictate, at least in part, cellular interpretation and outcome of heparanase action in inflammation. In the following sections, the involvement of heparanase in modulation of immune responses will be discussed through a few illustrative examples of inflammatory disorders, including inflammatory bowel disease, inflammatory lung disease and neuroinflammation.
Ulcerative colitis and Crohn’s disease, represent two major forms of inflammatory bowel disease (IBD) - a chronic condition of the gastrointestinal tract resulting from inappropriate response of the mucosal immune system to commensal gut flora in genetically susceptible individuals [87]. In patients with IBD, an equilibrium between the immune response to microbial pathogens and tolerance to the normal flora becomes unbalanced, leading to the uncontrolled uptake of proinflammatory substances (i.e., bacteria, bacterial products) from the gut lumen and triggering immune activation, cytokine release, and further barrier dysfunction [87,88]. Given the important role of HS in maintaining the integrity of the gut wall [89,90], enzymatic degradation of HS is expected to significantly affect both the permeability of the colon and immune reactions. Indeed, analysis of glycosaminoglycan content in normal colonic tissue and colons of IBD patients revealed loss of HS from the subepithelial BM and from the vascular endothelium in submucosa [89,91–93]. In agreement, preferential expression of heparanase was reported in colonic epithelium of IBD patients during both acute and chronic phases of the disease [40,41]. Of note, immunocytes in the involved areas, showed little heparanase expression during all phases [40]. Similar temporo-spatial pattern of heparanase expression was observed in experimental dextran sodium sulfate(DSS) -induced chronic colitis in mice [41]. Although the evidence is still incomplete, it appears that in the setting of colitis (characterized by the presence of abundant luminal flora and activation of toll-like receptor [TLR] signaling pathways) heparanase enzyme of epithelial origin modulates inflammatory phenotype of innate immune cells (i.e., macrophages), preventing inflammation resolution and switching macrophage responses to chronic inflammation pattern [41]. In support of this notion, exacerbated chronic inflammatory phenotype and augmented recruitment and activation of macrophages were detected in colonic mucosa of heparanase-overexpressing transgenic (Hpa-tg) mice following induction of DSS colitis [41]. Of note, macrophage recruitment/activation in the presence of increased heparanase levels were also reported in the model of neointimal lesions following vascular injury [83] and implicated in atherosclerotic plaque progression toward vulnerability [94], further supporting the role of the enzyme in modulation of innate immune responses. Importantly, heparanase enzymatic activity strongly augmented activation of macrophages in vitro by lipopolysaccharide (LPS, a specific stimulator of TLR4 signaling), resulting in marked increase in production of TNFα and additional macrophage-derived proinflammatory cytokines [41]. In fact, it was previously shown that intact extracellular HS inhibits LPS-mediated TLR4 signaling and macrophage activation, and that its removal relieves this inhibition [95]. Thus, in the IBD setting, abnormal levels of heparanase may preserve inflammatory conditions by reprogramming macrophage responses from resolution of inflammation to unresolved chronic colitis. Activated macrophages, in turn, stimulate further production of heparanase by the epithelial cells of the inflamed colon via a TNFα-dependent mechanism, and proteolytic processing of the pro-enzyme via increased secretion of cathepsin L [41]. Thus, heparanase may participate in establishing a so called “multiplier” effect in IBD, in which even small elevation in initiating inflammatory stimuli gives rise to large increase in downstream cytokines.
An important addition to the emerging body of research exploring modulation of innate immune cell responses by heparanase is a recent report by Schmidt et al., focusing on enzymatic degradation of endothelial glycocalyx in pulmonary inflammatory disorders [43]. Glycocalyx, the thin gel-like endothelial layer that coats the luminal surface of blood vessels, is composed of HS and other glycosaminoglycans, proteoglycans and glycoproteins [96,97]. It serves as a barrier to circulating cells and controls availability of endothelial surface adhesion molecules to circulating leukocytes [96,97]. In the mouse model of sepsis-associated inflammatory lung disease, rapid induction of heparanase activity (through TNFα-dependent mechanism) was demonstrated in pulmonary microvascular endothelial cells [43]. Heparanase induction was also noted in biopsies of human inflammatory lung disease [43]. Consistent with the key role of HS in glycocalyx structure [96,97] and in control of immunocyte adhesion to endothelium [86], heparanase appears to contribute to acute inflammatory lung injury through degradation and loss of pulmonary endothelial glycocalyx, resulting in increased availability of endothelial surface adhesion molecules and fostering neutrophil recruitment to the endothelial surface [43]. Moreover, sepsis associated loss of pulmonary glycocalyx and endothelial hyperpermeability were attenuated in heparanase-null mice and in mice treated with enzymatic inhibitors of heparanase activity [43].
Perhaps paradoxically, constitutive overexpression of heparanase (i.e., in Hpa-tg mice) was shown to attenuate intraluminal crawling of neutrophils toward an extravascular chemokine source, reportedly due to reduction of endothelial surface HS chain length and altered ability of truncated HS to serve as a ligand for chemokines [98]. In addition, recent reports exploring acute inflammatory phenotypes of Hpa-tg mice in the models of inflammatory hyperalgesia [99] and neuroinflammation [42] demonstrated that neutrophil recruitment and activation were attenuated in the presence of constitutively increased levels of heparanase in Hpa-tg mice. It remains to be seen whether seemingly opposing effects of heparanase on acute inflammatory responses in different organ/tissue settings are a consequence of tissue-specific patterns reported for its enzymatic substrate, HS [100–102], or rather reflect different roles of the enzyme through distinctive phases of inflammatory cascade. Additionally, given anti-inflammatory effects of heparin [103], increased levels of highly sulfated, “heparin-like” HS fragments which are constantly present in Hpa-tg mice [104] may offer an alternative explanation for the observed effects of continuous heparanase overexpression on inflammatory hyperalgesia and neuroinflammation [42,99].
Heparanase in malignancy
The role of the enzyme in malignant tumor development and progression is, in contrast, better understood. For almost 3 decades heparanase enzymatic activity has been associated with the metastatic potential of tumor-derived cells [105,106]. More recently, demonstration of enhanced tumor growth [25–27,107,108] and increased metastatic ability [27,109] of myeloma, colon, breast, and prostate carcinoma cells following over-expression of the heparanase gene provided direct evidence for a causal role of heparanase in tumor progression. This notion was further supported by inhibition of the tumorigenic and metastatic abilities of cancer cells following heparanase gene silencing [27–29,31,110]. In addition, multiple lines of evidence mechanistically implicated heparanase in endothelial sprouting and establishment of a vascular network that accelerates primary tumor growth and provides a gateway for invading metastatic cells [13,111,112]. During tumor progression heparanase enzymatic action not only contributes to the breakdown of extracellular barriers for cell invasion, but also regulates the bioavailability and activity of growth factors. Various HS-binding growth factors (i.e., bFGF, VEGF, HGF), are sequestered by HS in the ECM protected from proteolytic degradation, and readily available to activate cells after being released by heparanase [1,6,9,111]. Heparanase enzymatic activity also generates HS fragments which potentiate growth factor - receptor binding, dimerization and signaling, and there is evidence that the fragments of HS generated by heparanase are more biologically active than the native HS chain from which they are derived [8,13].
In addition to the well-documented role of heparanase in creating a supportive microenvironment for tumor progression by enzymatic remodeling of HS glycosaminoglycan chains, some of the pro-cancerous effects of heparanase were recently attributed to its ability to enhance shedding of proteoglycan syndecan-1 from the tumor cell surface [113], promote cell adhesion and induce signal transduction, including activation of protein kinase B/Akt [35], Src [36], EGF [37] and HGF [114] and insulin receptor [38] signaling pathways. Preferential over-expression of the enzyme in human carcinomas of various origins, as well as association of augmented levels of heparanase with reduced patients’ survival post operation, and increased metastasis and higher microvessel density, further highlight the role of heparanase in sustaining the pathology of malignant tumors [12,16].
Heparanase as a candidate linking molecule between inflammation and cancer
Substantial evidence supports an association between inflammation and cancer [115,116]. Inflammatory conditions are present in the microenvironment of most tumors [117] and have been shown to contribute to cancer progression, among other mechanisms, through mobilization of tumor-supporting immunocyte populations (e.g., tumor associated macrophages, neutrophils) which supply bioactive molecules that foster survival, angiogenesis, invasion and metastasis [121–123]. Moreover, in several anatomic sites chronic inflammation is crucially implicated in tumor initiation, producing a mutagenic environment through release of reactive oxygen/nitrogen species from infiltrating immune cells (largely macrophages), generating cytokines, chemokines, growth factors, and anti-apoptotic proteins and activating tumor stimulating signaling pathways (e.g., NF-kB, STAT3) [115–118]. A key role of inflammation in tumor initiation is best exemplified by progression of Barrett’s oesophagus to adenocarcinoma [119]; chronic gastritis to intestinal-type gastric carcinoma, hepatitis to hepatocellular carcinoma [120]; pancreatitis to pancreas adenocarcinoma [121] and inflammatory bowel diseases to colorectal cancer [122].
Involvement of heparanase is well documented in the pathogenesis of above-mentioned tumor types, including Barrett’s oesophageal adenocarcinoma [123], gastric carcinoma [124], hepatocellular carcinoma [31,125,126], pancreatic carcinoma [21,127], colorectal cancer [23,24]. Strikingly, induction of heparanase was reported in essentially all inflammatory conditions implicated in initiation of these cancer types, namely in Barrett’s oesophagus [123,124], hepatitis C infection [125], chronic pancreatitis [21], Crohn disease and ulcerative colitis [40,41]. Induction ofheparanase in cancer-triggering inflammatory disorders prior to the appearance of malignancy, taken together with the role that the enzyme plays in tumor progression in tissues in which cancer-related inflammation typically occurs (i.e., gastrointestinal tract, pancreas, liver [21,23,24,31,123–127]), suggests that inflammation-stimulated heparanase expression may be mechanistically involved in coupling inflammation and cancer (Fig. 1). In fact, such a role for heparanase was recently demonstrated in a mouse model of at least one type of inflammation induced cancer (i.e., colitis-associated colon carcinoma). It appears that by sustaining continuous activation of macrophages that supply cancer-promoting cytokines (i.e., TNFα, IL-1, IL-6) heparanase (supplied by inflamed colonic epithelium) creates a tumorigenic microenvironment characterized by enhanced NF-kB and STAT3 signaling, augmented levels of cyclooxygenase 2 and increased vascularization [41]. Moreover, our ongoing experiments utilizing a mouse model of heparanase-overexpressing pancreatic carcinoma suggest that heparanase action in polarizing macrophages toward pro-tumorigenic phenotype is not limited solely to the phase of tumor initiation. The data is still fragmentary, but it appears that once the tumor is initiated and progresses toward malignancy, the enzyme may contribute to the switch of tumor-associated macrophages (TAMs) to a “trophic” phenotype [118], responsible for pro-tumorigenic action of TAMs at the later stages of tumor development.
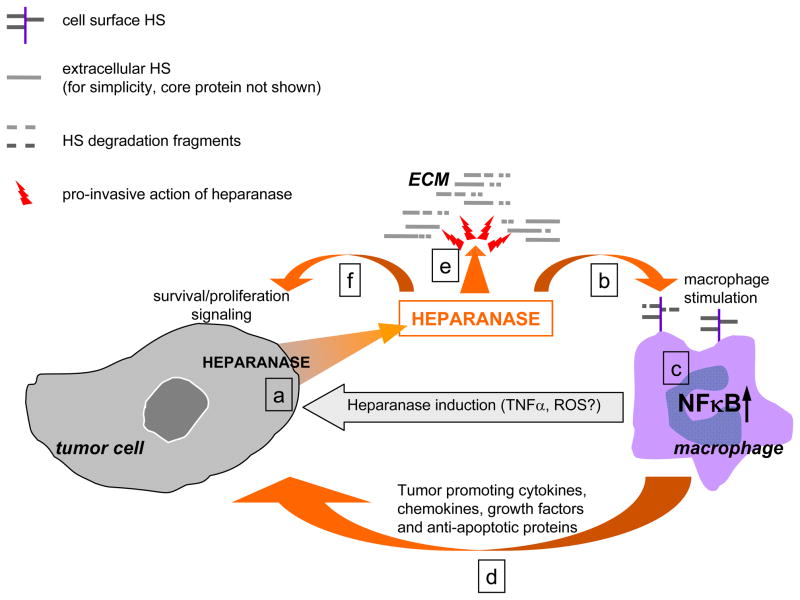
Figure 1. Heparanase may provide an important connection between inflammation and malignant development.
a. Macrophages infiltrating pre-neoplastic/neoplastic lesions induce heparanase expression in transformed cells (possibly via TNFα and/or ROS -dependent mechanisms [41,43,138]). b. In turn, heparanase degrades HS at the macrophage cell surface, relieving inhibitory action of intact HS on TLR signaling and generating HS degradation fragments which potentiate TLR activation [95,139], thus switching macrophage responses to the chronic inflammation pattern (c). d. As a result, macrophages continue to supply bioactive molecules that foster survival, angiogenesis, invasion and metastasis. e. Through degradation of HS in ECM heparanase facilitates breakdown of extracellular barriers for cell invasion and releases HS-bound growth factors (i.e., bFGF, VEGF, HGF) [9,16,17,111]. f. In addition, non--catalytic aspects of heparanase action may contribute to tumor progression through modulation of cell adhesion [33,34] and induction of signal transduction [35–37].
Conclusions and future prospects
Although the mechanisms of heparanase dependent polarization of innate immune cells toward a pro-inflammatory and/or pro-tumorigenic phenotype [41,43,83,94] are still obscure and just beginning to be elucidated, the emerging complexity of heparanase-driven responses in inflammatory and neoplastic disorders (Fig. 1) provides numerous avenues for therapeutic exploration. Therapeutic benefits are expected to be gained by strategies preventing enzymatic degradation of HS in ECM, inhibiting heparanase activated signaling pathways or reversing heparanase effects on immunocytes. During the last decade several inhibitory compounds are being developed and pre-clinically tested (Table 1), including small molecule inhibitors and polyanionic molecules such as laminaran sulfate, suramin, phosphomannopentaose sulfate (PI-88) synthetic fully sulfated HS mimetic PG545 and modified species of heparin [128–136]. Recently, a novel heparanase inhibitor SST0001 [134,137] (chemically modified non-anticoagulant heparin), has entered clinical testing in phase I trial for multiple myeloma. In depth recognition of the multilevel “fine tune” control that heparanase provides to heterotypic interactions among epithelial, endothelial, and immune cells in both inflammation and malignancy merits further systematic analysis and continuous searching for the effective heparanase- inhibiting compounds, toward future translation to the clinical setting.
Table 1.
Heparanase inhibitors in pre-clinical/clinical testing.
| Inhibitor | Experimental system | Effect | References/comments |
|---|---|---|---|
| Laminarin sulfate | Murine melanoma, mammary carcinoma. | Inhibition of experimental metastasis in vivo. | [123] |
| Suramin | Murine melanoma. | Inhibition of cell invasion through reconstituted basement membrane in vitro. | [133] |
| Phosphomanno pentaose sulfate (PI-88) | Post-operative hepatocellular carcinoma. | Preliminary efficacy in phase II trial. | [134] Currently in phase III trial for post-operative hepatocellular carcinoma |
| PG545 | Murine melanoma, carcinoma of breast, prostate, liver, lung, colon, head and neck cancer. | Inhibition of tumor growth, angiogenesis and metastasis in vivo. | [130] Phase I study in patients with advanced solid tumors has been terminated in 2012 due to unexpected injection site reactions. |
| SST0001 (a.k.a. G4000) | Myeloma, Ewing’s sarcoma, pancreatic carcinoma xenografts in immunodeficient mice. | Inhibition of cell invasion in vitro and tumor xenograft growth in vivo. | [30,127,129,132] Entered phase I trial for multiple myeloma. |
| M402 | Murine melanoma, mammary carcinoma. | Inhibition of experimental/spontaneous metastasis and prolonged survival. | [131] Phase I/II trial for metastatic pancreatic cancer (in combination with gemcitabine). |
Acknowledgments
This work was supported by the grants from the German-Israel Research Foundation (GIF), NIH (grant RO1-CA106456-09), Israel Science Foundation (grant 593/10) and by the EFSD/Novo Nordisk Award. We thank Prof. Israel Vlodavsky (Cancer and Vascular Biology Research Center, the Rappaport Faculty of Medicine, Technion, Haifa, Israel) for his continuous help and collaboration.
References
- 1.Bishop JR, Schuksz M, Esko JD. Nature. 2007;446:1030–7. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 2.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Annu Rev Biochem. 1999;68:729–77. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 3.Iozzo RV, Sanderson RD. J Cell Mol Med. 2011;15:1013–31. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Theocharis AD, Skandalis SS, Tzanakakis GN, Karamanos NK. FEBS J. 2010;277:3904–23. doi: 10.1111/j.1742-4658.2010.07800.x. [DOI] [PubMed] [Google Scholar]
- 5.Kramer KL, Yost HJ. Annu Rev Genet. 2003;37:461–84. doi: 10.1146/annurev.genet.37.061103.090226. [DOI] [PubMed] [Google Scholar]
- 6.Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Nat Rev Cancer. 2002;2:521–8. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 7.Belting M. Trends Biochem Sci. 2003;28:145–51. doi: 10.1016/S0968-0004(03)00031-8. [DOI] [PubMed] [Google Scholar]
- 8.Kato M, Wang H, Kainulainen V, Fitzgerald ML, Ledbetter S, Ornitz DM, Bernfield M. Nat Med. 1998;4:691–7. doi: 10.1038/nm0698-691. [DOI] [PubMed] [Google Scholar]
- 9.Vlodavsky I, Miao HQ, Medalion B, Danagher P, Ron D. Cancer Metastasis Rev. 1996;15:177–86. doi: 10.1007/BF00437470. [DOI] [PubMed] [Google Scholar]
- 10.Sanderson RD. Semin Cell Dev Biol. 2001;12:89–98. doi: 10.1006/scdb.2000.0241. [DOI] [PubMed] [Google Scholar]
- 11.Timar J, Lapis K, Dudas J, Sebestyen A, Kopper L, Kovalszky I. Semin Cancer Biol. 2002;12:173–86. doi: 10.1016/S1044-579X(02)00021-4. [DOI] [PubMed] [Google Scholar]
- 12.Vreys V, David G. J Cell Mol Med. 2007;11:427–52. doi: 10.1111/j.1582-4934.2007.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkin M, Ilan N, Ishai-Michaeli R, Friedmann Y, Papo O, Pecker I, Vlodavsky I. Faseb J. 2001;15:1661–3. doi: 10.1096/fj.00-0895fje. [DOI] [PubMed] [Google Scholar]
- 14.Gotte M. Faseb J. 2003;17:575–91. doi: 10.1096/fj.02-0739rev. [DOI] [PubMed] [Google Scholar]
- 15.Li JP, Vlodavsky I. Thromb Haemost. 2009;102:823–8. doi: 10.1160/TH09-02-0091. [DOI] [PubMed] [Google Scholar]
- 16.Ilan N, Elkin M, Vlodavsky I. Int J Biochem Cell Biol. 2006;38:2018–39. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Parish CR, Freeman C, Hulett MD. Biochim Biophys Acta. 2001;1471:M99–108. doi: 10.1016/s0304-419x(01)00017-8. [DOI] [PubMed] [Google Scholar]
- 18.Bar-Sela G, Kaplan-Cohen V, Ilan N, Vlodavsky I, Ben-Izhak O. Histopathology. 2006;49:188–93. doi: 10.1111/j.1365-2559.2006.02469.x. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Izhak O, Kaplan-Cohen V, Ilan N, Gan S, Vlodavsky I, Nagler R. Neoplasia. 2006;8:879–84. doi: 10.1593/neo.06382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doweck I, Kaplan-Cohen V, Naroditsky I, Sabo E, Ilan N, Vlodavsky I. Neoplasia. 2006;8:1055–61. doi: 10.1593/neo.06577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koliopanos A, et al. Cancer Res. 2001;61:4655–9. [PubMed] [Google Scholar]
- 22.Takaoka M, et al. Lab Invest. 2003;83:613–22. doi: 10.1097/01.lab.0000067482.84946.bd. [DOI] [PubMed] [Google Scholar]
- 23.Naomoto Y, Takaoka M, Okawa T, Nobuhisa T, Gunduz M, Tanaka N. Oncol Rep. 2005;14:3–8. [PubMed] [Google Scholar]
- 24.Nobuhisa T, et al. J Cancer Res Clin Oncol. 2005;131:229–37. doi: 10.1007/s00432-004-0644-x. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, et al. Blood. 2005;105:1303–9. doi: 10.1182/blood-2004-06-2141. [DOI] [PubMed] [Google Scholar]
- 26.Cohen I, et al. Int J Cancer. 2006;118:1609–17. doi: 10.1002/ijc.21552. [DOI] [PubMed] [Google Scholar]
- 27.Lerner I, Baraz L, Pikarsky E, Meirovitz A, Edovitsky E, Peretz T, Vlodavsky I, Elkin M. Clin Cancer Res. 2008;14:668–76. doi: 10.1158/1078-0432.CCR-07-1866. [DOI] [PubMed] [Google Scholar]
- 28.Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. J Natl Cancer Inst. 2004;96:1219–30. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]
- 29.Roy M, Reiland J, Murry BP, Chouljenko V, Kousoulas KG, Marchetti D. Neoplasia. 2005;7:253–62. doi: 10.1593/neo.04493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meirovitz A, Hermano E, Lerner I, Zcharia E, Pisano C, Peretz T, Elkin M. Cancer Res. 2011;71:2772–80. doi: 10.1158/0008-5472.CAN-10-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong Z, et al. Int J Oncol. 2012;40:1601–9. doi: 10.3892/ijo.2012.1338. [DOI] [PubMed] [Google Scholar]
- 32.He YQ, et al. Transcription. 2012;3:130–45. doi: 10.4161/trns.19998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldshmidt O, et al. Faseb J. 2003;17:1015–25. doi: 10.1096/fj.02-0773com. [DOI] [PubMed] [Google Scholar]
- 34.Zetser A, Bashenko Y, Miao HQ, Vlodavsky I, Ilan N. Cancer Res. 2003;63:7733–41. [PubMed] [Google Scholar]
- 35.Gingis-Velitski S, Zetser A, Flugelman MY, Vlodavsky I, Ilan N. J Biol Chem. 2004;279:23536–41. doi: 10.1074/jbc.M400554200. [DOI] [PubMed] [Google Scholar]
- 36.Zetser A, Bashenko Y, Edovitsky E, Levy-Adam F, Vlodavsky I, Ilan N. Cancer Res. 2006;66:1455–63. doi: 10.1158/0008-5472.CAN-05-1811. [DOI] [PubMed] [Google Scholar]
- 37.Cohen-Kaplan V, Doweck I, Naroditsky I, Vlodavsky I, Ilan N. Cancer Res. 2008;68:10077–85. doi: 10.1158/0008-5472.CAN-08-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purushothaman A, Babitz SK, Sanderson RD. J Biol Chem. 2012;287:41288–96. doi: 10.1074/jbc.M112.391417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li RW, Freeman C, Yu D, Hindmarsh EJ, Tymms KE, Parish CR, Smith PN. Arthritis Rheum. 2008;58:1590–600. doi: 10.1002/art.23489. [DOI] [PubMed] [Google Scholar]
- 40.Waterman M, Ben-Izhak O, Eliakim R, Groisman G, Vlodavsky I, Ilan N. Mod Pathol. 2007;20:8–14. doi: 10.1038/modpathol.3800710. [DOI] [PubMed] [Google Scholar]
- 41.Lerner I, et al. J Clin Invest. 2011;121:1709–21. doi: 10.1172/JCI43792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, et al. Acta Neuropathol. 2012;124:465–78. doi: 10.1007/s00401-012-0997-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt EP, et al. Nat Med. 2012;18:1217–23. doi: 10.1038/nm.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao G, et al. Diabetologia. 2011;54:1527–38. doi: 10.1007/s00125-011-2110-z. [DOI] [PubMed] [Google Scholar]
- 45.Osterholm C, Folkersen L, Lengquist M, Ponten F, Renne T, Li J, Hedin U. Atherosclerosis. 2013;226:67–73. doi: 10.1016/j.atherosclerosis.2012.09.030. [DOI] [PubMed] [Google Scholar]
- 46.Ostrovsky O, Shimoni A, Rand A, Vlodavsky I, Nagler A. Blood. 2010;115:2319–28. doi: 10.1182/blood-2009-08-236455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levidiotis V, et al. J Am Soc Nephrol. 2004;15:2882–92. doi: 10.1097/01.ASN.0000142426.55612.6D. [DOI] [PubMed] [Google Scholar]
- 48.Levidiotis V, Freeman C, Tikellis C, Cooper ME, Power DA. Nephrology (Carlton) 2005;10:167–73. doi: 10.1111/j.1440-1797.2005.00388.x. [DOI] [PubMed] [Google Scholar]
- 49.Gil N, et al. Diabetes. 2012;61:208–16. doi: 10.2337/db11-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shafat I, et al. PLoS One. 2012;7:e44076. doi: 10.1371/journal.pone.0044076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziolkowski AF, Popp SK, Freeman C, Parish CR, Simeonovic CJ. J Clin Invest. 2012;122:132–41. doi: 10.1172/JCI46177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zcharia E, Jia J, Zhang X, Baraz L, Lindahl U, Peretz T, Vlodavsky I, Li JP. PLoS ONE. 2009;4:e5181. doi: 10.1371/journal.pone.0005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McKenzie E, et al. Biochem Biophys Res Commun. 2000;276:1170–7. doi: 10.1006/bbrc.2000.3586. [DOI] [PubMed] [Google Scholar]
- 54.Levy-Adam F, et al. J Biol Chem. 2010;285:28010–9. doi: 10.1074/jbc.M110.116384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matzner Y, Bar-Ner M, Yahalom J, Ishai-Michaeli R, Fuks Z, Vlodavsky I. J Clin Invest. 1985;76:1306–13. doi: 10.1172/JCI112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Temkin V, Aingorn H, Puxeddu I, Goldshmidt O, Zcharia E, Gleich GJ, Vlodavsky I, Levi-Schaffer F. J Allergy Clin Immunol. 2004;113:703–9. doi: 10.1016/j.jaci.2003.11.038. [DOI] [PubMed] [Google Scholar]
- 57.Ateeq B, Unterberger A, Szyf M, Rabbani SA. Neoplasia. 2008;10:266–78. doi: 10.1593/neo.07947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shteper PJ, Zcharia E, Ashhab Y, Peretz T, Vlodavsky I, Ben-Yehuda D. Oncogene. 2003;22:7737–49. doi: 10.1038/sj.onc.1207056. [DOI] [PubMed] [Google Scholar]
- 59.Ogishima T, et al. Clin Cancer Res. 2005;11:1028–36. [PubMed] [Google Scholar]
- 60.Ogishima T, et al. Oncogene. 2005;24:6765–72. doi: 10.1038/sj.onc.1208811. [DOI] [PubMed] [Google Scholar]
- 61.Baraz L, Haupt Y, Elkin M, Peretz T, Vlodavsky I. Oncogene. 2006;25:3939–47. doi: 10.1038/sj.onc.1209425. [DOI] [PubMed] [Google Scholar]
- 62.de Mestre AM, Khachigian LM, Santiago FS, Staykova MA, Hulett MD. J Biol Chem. 2003;278:50377–85. doi: 10.1074/jbc.M310154200. [DOI] [PubMed] [Google Scholar]
- 63.de Mestre AM, Rao S, Hornby JR, Soe-Htwe T, Khachigian LM, Hulett MD. J Biol Chem. 2005;280:35136–47. doi: 10.1074/jbc.M503414200. [DOI] [PubMed] [Google Scholar]
- 64.Jiang P, Kumar A, Parrillo JE, Dempsey LA, Platt JL, Prinz RA, Xu X. J Biol Chem. 2002;277:8989–98. doi: 10.1074/jbc.M105682200. [DOI] [PubMed] [Google Scholar]
- 65.Lu WC, Liu YN, Kang BB, Chen JH. Oncogene. 2003;22:919–23. doi: 10.1038/sj.onc.1206201. [DOI] [PubMed] [Google Scholar]
- 66.Rao G, Liu D, Xing M, Tauler J, Prinz RA, Xu X. Neoplasia. 2010;12:946–56. doi: 10.1593/neo.10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maxhimer JB, Quiros RM, Stewart R, Dowlatshahi K, Gattuso P, Fan M, Prinz RA, Xu X. Surgery. 2002;132:326–33. doi: 10.1067/msy.2002.125719. [DOI] [PubMed] [Google Scholar]
- 68.Elkin M, Cohen I, Zcharia E, Orgel A, Guatta-Rangini Z, Peretz T, Vlodavsky I, Kleinman HK. Cancer Res. 2003;63:8821–6. [PubMed] [Google Scholar]
- 69.Xu X, Ding J, Rao G, Shen J, Prinz RA, Rana N, Dmowski WP. Hum Reprod. 2007;22:927–37. doi: 10.1093/humrep/del483. [DOI] [PubMed] [Google Scholar]
- 70.Chen G, Wang D, Vikramadithyan R, Yagyu H, Saxena U, Pillarisetti S, Goldberg IJ. Biochemistry. 2004;43:4971–7. doi: 10.1021/bi0356552. [DOI] [PubMed] [Google Scholar]
- 71.Edovitsky E, Lerner I, Zcharia E, Peretz T, Vlodavsky I, Elkin M. Blood. 2006;107:3609–16. doi: 10.1182/blood-2005-08-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hermano E, Lerner I, Elkin M. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-0930-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Levy-Adam F, Miao HQ, Heinrikson RL, Vlodavsky I, Ilan N. Biochem Biophys Res Commun. 2003;308:885–91. doi: 10.1016/s0006-291x(03)01478-5. [DOI] [PubMed] [Google Scholar]
- 74.McKenzie E, et al. Biochem J. 2003;373:423–35. doi: 10.1042/BJ20030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nardella C, Lahm A, Pallaoro M, Brunetti M, Vannini A, Steinkuhler C. Biochemistry. 2004;43:1862–73. doi: 10.1021/bi030203a. [DOI] [PubMed] [Google Scholar]
- 76.Abboud-Jarrous G, Rangini-Guetta Z, Aingorn H, Atzmon R, Elgavish S, Peretz T, Vlodavsky I. J Biol Chem. 2005;280:13568–75. doi: 10.1074/jbc.M413370200. [DOI] [PubMed] [Google Scholar]
- 77.Abboud-Jarrous G, Atzmon R, Peretz T, Palermo C, Gadea BB, Joyce JA, Vlodavsky I. J Biol Chem. 2008;283:18167–76. doi: 10.1074/jbc.M801327200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fridman R, Lider O, Naparstek Y, Fuks Z, Vlodavsky I, Cohen IR. J Cell Physiol. 1987;130:85–92. doi: 10.1002/jcp.1041300113. [DOI] [PubMed] [Google Scholar]
- 79.Vlodavsky I, et al. Invasion Metastasis. 1992;12:112–27. [PubMed] [Google Scholar]
- 80.Lider O, Baharav E, Mekori YA, Miller T, Naparstek Y, Vlodavsky I, Cohen IR. J Clin Invest. 1989;83:752–6. doi: 10.1172/JCI113953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Naparstek Y, Cohen IR, Fuks Z, Vlodavsky I. Nature. 1984;310:241–4. doi: 10.1038/310241a0. [DOI] [PubMed] [Google Scholar]
- 82.Lider O, Mekori YA, Miller T, Bar-Tana R, Vlodavsky I, Baharav E, Cohen IR, Naparstek Y. Eur J Immunol. 1990;20:493–9. doi: 10.1002/eji.1830200306. [DOI] [PubMed] [Google Scholar]
- 83.Baker AB, et al. Circ Res. 2009;104:380–7. doi: 10.1161/CIRCRESAHA.108.180695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parish CR, Hindmarsh EJ, Bartlett MR, Staykova MA, Cowden WB, Willenborg DO. Immunol Cell Biol. 1998;76:104–13. doi: 10.1046/j.1440-1711.1998.00722.x. [DOI] [PubMed] [Google Scholar]
- 85.Irony-Tur-Sinai M, Vlodavsky I, Ben-Sasson SA, Pinto F, Sicsic C, Brenner T. J Neurol Sci. 2003;206:49–57. doi: 10.1016/s0022-510x(02)00318-0. [DOI] [PubMed] [Google Scholar]
- 86.Floer M, et al. Am J Pathol. 2010;176:146–57. doi: 10.2353/ajpath.2010.080639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xavier RJ, Podolsky DK. Nature. 2007;448:427–34. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 88.Clayburgh DR, Shen L, Turner JR. Lab Invest. 2004;84:282–91. doi: 10.1038/labinvest.3700050. [DOI] [PubMed] [Google Scholar]
- 89.Belmiro CL, Souza HS, Elia CC, Castelo-Branco MT, Silva FR, Machado RL, Pavao MS. Int J Colorectal Dis. 2005 doi: 10.1007/s00384-004-0677-2. [DOI] [PubMed] [Google Scholar]
- 90.Bode L, Salvestrini C, Park PW, Li JP, Esko JD, Yamaguchi Y, Murch S, Freeze HH. J Clin Invest. 2008;118:229–38. doi: 10.1172/JCI32335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murch SH, MacDonald TT, Walker-Smith JA, Levin M, Lionetti P, Klein NJ. Lancet. 1993;341:711–4. doi: 10.1016/0140-6736(93)90485-y. [DOI] [PubMed] [Google Scholar]
- 92.Day R, Forbes A. Lancet. 1999;354:62–5. doi: 10.1016/S0140-6736(98)09267-8. [DOI] [PubMed] [Google Scholar]
- 93.Day R, Ilyas M, Daszak P, Talbot I, Forbes A. Dig Dis Sci. 1999;44:2508–15. doi: 10.1023/a:1026647308089. [DOI] [PubMed] [Google Scholar]
- 94.Blich M, et al. Arterioscler Thromb Vasc Biol. 2012;33:56–65. [Google Scholar]
- 95.Brunn GJ, Bungum MK, Johnson GB, Platt JL. Faseb J. 2005;19:872–4. doi: 10.1096/fj.04-3211fje. [DOI] [PubMed] [Google Scholar]
- 96.Weinbaum S, Tarbell JM, Damiano ER. Annu Rev Biomed Eng. 2007;9:121–67. doi: 10.1146/annurev.bioeng.9.060906.151959. [DOI] [PubMed] [Google Scholar]
- 97.Constantinescu AA, Vink H, Spaan JA. Arterioscler Thromb Vasc Biol. 2003;23:1541–7. doi: 10.1161/01.ATV.0000085630.24353.3D. [DOI] [PubMed] [Google Scholar]
- 98.Massena S, et al. Blood. 2010;116:1924–31. doi: 10.1182/blood-2010-01-266072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li L, et al. Neurosci Lett. 2012;511:4–7. doi: 10.1016/j.neulet.2011.12.038. [DOI] [PubMed] [Google Scholar]
- 100.Ledin J, Staatz W, Li JP, Gotte M, Selleck S, Kjellen L, Spillmann D. J Biol Chem. 2004;279:42732–41. doi: 10.1074/jbc.M405382200. [DOI] [PubMed] [Google Scholar]
- 101.Allen BL, Filla MS, Rapraeger AC. J Cell Biol. 2001;155:845–58. doi: 10.1083/jcb.200106075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guo CH, Koo CY, Bay BH, Tan PH, Yip GW. Int J Oncol. 2007;31:1415–23. [PubMed] [Google Scholar]
- 103.Young E. Thromb Res. 2008;122:743–52. doi: 10.1016/j.thromres.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 104.Escobar Galvis ML, et al. Nat Chem Biol. 2007;3:773–8. doi: 10.1038/nchembio.2007.41. [DOI] [PubMed] [Google Scholar]
- 105.Nakajima M, Irimura T, Di Ferrante N, Nicolson GL. J Biol Chem. 1984;259:2283–90. [PubMed] [Google Scholar]
- 106.Vlodavsky I, Fuks Z, Bar-Ner M, Ariav Y, Schirrmacher V. Cancer Res. 1983;43:2704–11. [PubMed] [Google Scholar]
- 107.Mahtouk K, et al. Blood. 2007;109:4914–23. doi: 10.1182/blood-2006-08-043232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Doviner V, Maly B, Kaplan V, Gingis-Velitski S, Ilan N, Vlodavsky I, Sherman Y. Mod Pathol. 2006;19:878–88. doi: 10.1038/modpathol.3800603. [DOI] [PubMed] [Google Scholar]
- 109.Vlodavsky I, et al. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 110.Jiang G, Zheng L, Pu J, Mei H, Zhao J, Huang K, Zeng F, Tong Q. PLoS One. 2012;7:e31379. doi: 10.1371/journal.pone.0031379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Joyce JA, Freeman C, Meyer-Morse N, Parish CR, Hanahan D. Oncogene. 2005;24:4037–51. doi: 10.1038/sj.onc.1208602. [DOI] [PubMed] [Google Scholar]
- 112.Purushothaman A, Uyama T, Kobayashi F, Yamada S, Sugahara K, Rapraeger AC, Sanderson RD. Blood. 2010;115:2449–57. doi: 10.1182/blood-2009-07-234757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yang Y, et al. J Biol Chem. 2007;282:13326–33. doi: 10.1074/jbc.M611259200. [DOI] [PubMed] [Google Scholar]
- 114.Ramani VC, Yang Y, Ren Y, Nan L, Sanderson RD. J Biol Chem. 2011;286:6490–9. doi: 10.1074/jbc.M110.183277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Grivennikov SI, Karin M. Curr Opin Genet Dev. 2010;20:65–71. doi: 10.1016/j.gde.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hanahan D, Weinberg RA. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 117.Mantovani A, Allavena P, Sica A, Balkwill F. Nature. 2008;454:436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 118.Qian BZ, Pollard JW. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Picardo SL, Maher SG, O’Sullivan JN, Reynolds JV. Dig Surg. 2012;29:251–60. doi: 10.1159/000341498. [DOI] [PubMed] [Google Scholar]
- 120.Chiba T, Marusawa H, Ushijima T. Gastroenterology. 2012;143:550–63. doi: 10.1053/j.gastro.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 121.Lowenfels AB, et al. N Engl J Med. 1993;328:1433–7. doi: 10.1056/NEJM199305203282001. [DOI] [PubMed] [Google Scholar]
- 122.Gupta RB, Harpaz N, Itzkowitz S, Hossain S, Matula S, Kornbluth A, Bodian C, Ullman T. Gastroenterology. 2007;133:1099–105. doi: 10.1053/j.gastro.2007.08.001. quiz 1340–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brun R, Naroditsky I, Waterman M, Ben-Izhak O, Groisman G, Ilan N, Vlodavsky I. Mod Pathol. 2009;22:1548–54. doi: 10.1038/modpathol.2009.115. [DOI] [PubMed] [Google Scholar]
- 124.Sonoda R, et al. Histopathology. 2010;57:90–100. doi: 10.1111/j.1365-2559.2010.03594.x. [DOI] [PubMed] [Google Scholar]
- 125.El-Assal ON, Yamanoi A, Ono T, Kohno H, Nagasue N. Clin Cancer Res. 2001;7:1299–305. [PubMed] [Google Scholar]
- 126.Zhang Y, Li L, Wang Y, Zhang J, Wei G, Sun Y, Shen F. Biochem Biophys Res Commun. 2007;358:124–9. doi: 10.1016/j.bbrc.2007.04.068. [DOI] [PubMed] [Google Scholar]
- 127.Hoffmann AC, et al. J Gastrointest Surg. 2008 Dec;:1674–81. doi: 10.1007/s11605-008-0628-2. discussion 1681–2. [DOI] [PubMed] [Google Scholar]
- 128.Miao HQ, Elkin M, Aingorn E, Ishai-Michaeli R, Stein CA, Vlodavsky I. Int J Cancer. 1999;83:424–31. doi: 10.1002/(sici)1097-0215(19991029)83:3<424::aid-ijc20>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 129.Ferro V, Hammond E, Fairweather JK. Mini Rev Med Chem. 2004;4:693–702. doi: 10.2174/1389557043403729. [DOI] [PubMed] [Google Scholar]
- 130.Pan W, et al. Bioorg Med Chem Lett. 2006;16:409–12. doi: 10.1016/j.bmcl.2005.09.069. [DOI] [PubMed] [Google Scholar]
- 131.Simizu S, Ishida K, Osada H. Cancer Sci. 2004;95:553–8. doi: 10.1111/j.1349-7006.2004.tb02485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Naggi A, et al. J Biol Chem. 2005;280:12103–13. doi: 10.1074/jbc.M414217200. [DOI] [PubMed] [Google Scholar]
- 133.McKenzie EA. Br J Pharmacol. 2007;151:1–14. doi: 10.1038/sj.bjp.0707182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ritchie JP, et al. Clin Cancer Res. 2011;17:1382–93. doi: 10.1158/1078-0432.CCR-10-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dredge K, et al. Br J Cancer. 2011;104:635–42. doi: 10.1038/bjc.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhou H, et al. PLoS One. 2011;6:e21106. doi: 10.1371/journal.pone.0021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shafat I, et al. J Cell Mol Med. 2011;15:1857–64. doi: 10.1111/j.1582-4934.2010.01190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.van den Hoven MJ, Waanders F, Rops AL, Kramer AB, van Goor H, Berden JH, Navis G, van der Vlag J. Nephrol Dial Transplant. 2009;24:2637–45. doi: 10.1093/ndt/gfp182. [DOI] [PubMed] [Google Scholar]
- 139.Akbarshahi H, Axelsson JB, Said K, Malmstrom A, Fischer H, Andersson R. J Transl Med. 2011;9:219. doi: 10.1186/1479-5876-9-219. [DOI] [PMC free article] [PubMed] [Google Scholar]