Abstract
Pyrrole-imidazole (Py-Im) polyamides are a class of programmable DNA minor groove binders capable of modulating the activity of DNA-binding proteins and affecting changes in gene expression. Estrogen Receptor Alpha (ERα) is a ligand-activated hormone receptor that binds as a homodimer to estrogen response elements (EREs) and is a driving oncogene in a majority of breast cancers. We tested a selection of structurally similar Py-Im polyamides with differing DNA sequence specificity for activity against 17β-estadiol (E2) induced transcription and cytotoxicity in ERα positive, E2 stimulated T47D-KBLUC cells, which express luciferase under ERα control. The most active polyamide targeted the sequence: 5’-WGGWCW-3’ (W = A or T), which is the canonical ERE-half site. Whole transcriptome analysis using RNA-Seq revealed that treatment of E2-stimulated breast cancer cells with this polyamide reduced the effects of E2 on the majority of those most strongly affected by E2, but had much less effect on the majority of E2 induced transcripts. In vivo, this polyamide circulated at detectable levels following subcutaneous injection and reduced levels of ER-driven luciferase expression in xenografted tumors in mice after subcutaneous compound administration without significant host toxicity.
Keywords: polyamide, minor groove binder, estrogen receptor, breast cancer, gene expression
Introduction
Estrogen receptor alpha (ERα) is a member of the nuclear hormone receptor family of transcription factors and is active in a majority of breast adenocarcinomas (1, 2). Breast tumors that express ERα and are sensitive to circulating estrogens respond to therapeutics that modulate ERα activity (3). Such therapeutics include tamoxifen, a selective estrogen receptor modulator that acts as a weak agonist/antagonist by binding to the ERα ligand binding pocket, and the aromatase inhibitors that inhibit synthesis of E2 (3). A different strategy for modulation of ERα activity is inhibition of the ERα-ERE interface by a DNA-binding molecule.
Py-Im polyamides are a class of synthetic, minor groove binding ligands inspired by the natural product distamycin A (4, 5). Py-Im polyamides are oligomers of aromatic amino acids linked in series to fold in an anti-parallel fashion when bound in the minor groove of DNA (4, 5). Sequence specificity is programmed through side-by side pairings of the Py and Im subunits that recognize differences in the shape and hydrogen bonding pattern presented by the edges of the Watson Crick base pairs in the floor of the minor groove (6, 7). Binding specificity has been extensively characterized by DNAse I footprinting titrations and other methods. An Im:Py pair preferentially recognizes G:C; Py:Im prefers C:G, and Py:Py is degenerate for A:T and T:A (6, 7). Py-Im polyamide binding in the minor groove also induces allosteric changes to the major groove (8, 9) and binding affinity is sufficient to modulate the binding of DNA-binding proteins (8–11).
In cell culture, selected polyamides have been used to modulate expression of genes induced by testosterone (11), TNF-alpha (12), hypoxia (13, 14), and dexamethasone (10). The mechanisms by which polyamides affect gene expression changes in cell culture is still not well understood and may involve direct effects on multiple DNA-dependent processes including transcription factor occupancy, chromatin structure, RNA polymerases, and DNA replication (15). The pharmacokinetics and toxicity of a number of polyamides after intravenous and subcutaneous injection in mice and rats have been described (16–18). In mice, a selected polyamide was reported to induce changes in TGF-beta expression in kidney glomeruli, and a fluorescent analog of this polyamide was observed in kidney glomeruli after tail vein injection in rats (19). Gene expression changes have also been observed in tumor xenografts in immune compromised mice treated with a hairpin-polyamide (20).
In this study, our goal was to identify a Py-Im polyamide capable of affecting E2 stimulated gene expression in breast cancer cells and characterize its activity in cell culture and in tumor xenografts. To do this, we drew from an earlier study that reported a polyamide targeted to the ERE consensus half site 5’-WGGWCW-3’ inhibited ERα binding to DNA in cell-free systems (21). The DNA-binding affinity and specificity of the ERE-targeted polyamide was characterized in this and other studies (21, 22). Since those publications, we have improved the nuclear uptake of polyamides via modification of the C-terminus (23). We have also demonstrated that polyamides are bioavailable after intravenous and subcutaneous injection in mice (20, 24). We then decided to re-examine the activity of polyamides capable of disrupting ERE-driven gene expression for use in vivo. We have screened a focused library of polyamides for cytotoxicity and inhibition of luciferase activity using the breast cancer cell line T47D-KBLUC that expresses luciferase under the control of three tandem, canonical EREs (25). The most active polyamide identified, which the consensus ERE, was further evaluated and showed a partial suppression of E2 stimulated gene expression in cell culture. This polyamide circulated in mouse serum after subcutaneous injection and demonstrated activity against E2-induced luciferase expression in T47D-KBLUC tumor xenografts in mice with minimal host toxicity. A fluorescent analog of this polyamide distributed widely in both tumor and mouse tissue after subcutaneous injection.
Materials and Methods
Polyamide Synthesis and Characterization
The polyamides 1–5 were synthesized following previously published solid phase synthesis protocols (26). Compound purities were confirmed by analytical HPLC and MALDI-TOF MS. Melting temperature analysis was performed on a Varian Cary 100 spectrophotometer with temperature control. Oligonucleotides (IDTDNA) were dissolved in 10 mM sodium cacodylate, 10 mM KCl, 10 mM MgCl2 and 5 mM CaCl2 at pH 7.0 at a concentration of 2 µM. Polyamides were added to oligo solution to a final concentration of 4 µM in 0.1% DMSO. Oligonucleotides were annealed from 25–90°C and then back to 25°C at 5° C/min. Subsequently, the temperature was elevated at a rate 0.5° C/min between 25–90° C. Melting temperatures are defined as a maximum of the first derivative of absorbance at 260 nm over the range of temperatures.
Cell culture and imaging
Cell lines used were purchased directly from ATCC and used within 6 months. No subsequent authentications were done by the authors. All experiments were conducted with T47D-KBLuc cells (ATCC), unless specifically mentioned otherwise. Cells were grown in RPMI-1640 held at 37° C in 5% CO2. Media was supplemented with 10 % FBS and 1 % Penicillin / Streptamycin. Before imaging, cells were plated in 35mm optical dishes (MatTek) at 5×104 cells per dish in the presence of 10 nM E2. Cells were dosed with polyamide for 24 hours. Cells were then washed twice with PBS and imaged on a confocal microscope (Exciter, Zeiss) using a 63× oil immersion lens in a method previously described. Confocal imaging was performed following our previously published protocols (27, 28).
Tissue processing for fluorescence imaging
The tissue sections for fluorescent imaging were obtained by fixing the tumors in 10% formaldehyde solution for 24 hours, and subsequent cryoprotection in 15 % sucrose (24h) and 30 % sucrose (24h). The tumors were frozen in Tissue-Tek O.C.T. (Sakura Finetek) and 50 µm (for T47D-KBLuc xenograft) or 10 µm (for other tissues) sections were obtained using a Leica CM 1800 cryotome. Imaging was performed as described above.
Cell toxicity and luciferase assays
T47D-KBLUC cells were plated at 3×103 cells/well in 96-well plates, incubated in standard growth media containing 10 nM E2 for 48 hours and then dosed with medium containing 10 nM E2 and between 2 nM and 50 µM polyamides. The cells were then incubated for 96 hours and analyzed using either WST-1 assay (Roche) or luciferase assay system (Promega) according to the manufacturers’ instructions.
Gene expression analysis by qRT-PCR
Cells were plated in 12 well-plates at 1.1×105 cells/well and incubated in the growth medium supplemented with 10 nM E2 for 24 hours. Afterwards, medium was exchanged with the growth medium supplemented with polyamides and 10 nM E2. RT-qPCR has been performed according to previously established protocols (3–6). Confirmation of inhibition of TFF1 expression by polyamides 1 – 4 was performed qRT-PCR was done following the same timeline as cell toxicity and luciferase assays. Gene expression was normalized against GUSB as housekeeping gene. All primers yielded single amplicons as determined by both melting denaturation analysis and agarose gel electrophoresis. The following primer pairs were used. GUSB: fwd. 5'-CTC ATT TGG AAT TTT GCC GAT T-3' rev. 5'-CCC AGT GAA GAT CCC CTT TTT A-3'. DOK7: fwd. 5’-GAC AAG TCG GAG CGT ATC AAG-3’ rev. 5’-ATG TCC TCT AGC GTC AGG CT-3’. WT1: fwd. 5’-CAC AGC ACA GGG TAC GAG AG-3’ rev. 5’-CAA GAG TCG GGG CTA CTC CA-3. TGFB2: fwd. 5’-CAG CAC ACT CGA TAT GGA CCA-3’ rev. 5’-CCT CGG GCT CAG GAT AGT CT-3’.
Chromatin immunoprecipitation experiments
T47D-KBLUC cells were plated into 500 square centimeter plates and grown in RPMI-1640 with 10% FBS until 75% confluence was reached. Plates were washed with RPMI-1640 with charcoal treated 10% FBS and then the media replaced with RPMI-1640 with charcoal treated 10% FBS with 2 µM polyamide 1 and incubated for 48 hours. Plates were then treated with 10 nM E2 or vehicle for 45 minutes. Crosslinked chromatin was obtained using the 2-step crosslinking methods previously described (29). Chromatin was isolated and sheared. Antibodies to ERα (AC-066-100, Diagenode) were used to immunoprecipitate ERα-bound DNA fragments. Crosslinks were reversed, and PCRs using primers targeted to the regions of interest were used to assess enrichment of bound fragments as compared to negative controls. TFF1 promoter: fwd. 5’-TCA GAT CCC TCA GCC AAG AT-3’ rev. 5’-TGG TCA AGC TAC ATG GAA GG-3’ Negative loci control. fwd. 5’-AAA GAC AAC AGT CCT GGA AAC A-3’ rev. 5’-AAA AAT TGC TCA TTG GAG ACC-3’.
Circulation and toxicity in vivo
All animal experiments were conducted according to approved IACUC protocols at the California Institute of Technology. Circulation studies were done as previously described (30). Briefly, 120 nmol of polyamide 1 was injected subcutaneously into the right flank of four female C57BL/6 mice in a total of 200 uL of a 20% DMSO/PBS vehicle. Blood was collected retroorbitally at serial time points. Serum was treated with methanol, analyzed via HPLC, and quantified against a standard curve of concentration versus peak area, all as previously described to determine approximate serum concentrations (24). For toxicity studies, five female C57BL/6 mice were injected with 20 nmol of polyamide 1 in a total of 200 uL of a 20% DMSO/PBS vehicle on days 1, 3, 5, 8, 10, 12, and then with 30 nmol on days 15, 17, 19, 22, 24, 26 and were weighed prior to each treatment day. Mice were euthanized if weight loss was >15% of initial body weight, if dehydration was over 10 %, or moribund behavior was observed. None were observed in this experiment.
Xenografts
Engraftment of T47D-KBluc. Experiments were performed in appropriately shaved female NSG mice (JAX) between 8 and 12 weeks of age. Cells were injected into the left flank area of the animals as suspensions of 5.0×106 mL−1 in 50% RPMI 1640 growth medium and 50% matrigel, 200 µL per injection. Mice also received a subcutaneous E2 pellet (0.72 mg, 60 day slow release, Innovative Research of America, Sarasota, Florida) implanted into the right flank on the day of engraftment. Treatment and tumor monitoring. Mice were treated with either 25 nmoles of 1 or 50 nmoles of 5. For the short term and fluorescent imaging the studies they were treated 8 days after engraftment, every second day for a total of 4 injections. For long term treatment, injections started 16 days after engraftment and were continued twice a week for the following 4 weeks. Imaging was accomplished using the IVIS imaging system (Caliper). The animals were anesthetized with 2–3 % isoflurane and injected intraperitoneally with 150 µL of RediJect D-Luciferin (Caliper) and subsequently transferred to the imaging chamber while isoflurane levels were reduced to 1–2.5 %. The floor of the imager was heated to +37 °C to avoid animal hypothermia. Breathing frequency was monitored and not allowed to drop below one per second, adjusting the isoflurane levels accordingly at all times. Endpoint criteria and euthanasia. Animal endpoint criteria encompassed weight loss of over 15 %, restriction of motoric function by the engrafted tumor, dehydration of over 10 % and moribund behavior. Where appropriate, the animals were euthanized by asphyxiation in a CO2 chamber. Tumor tissue harvest. Animals were resected and tumors excised using standard forceps, scissors and surgical blades. The tumors were weighed immediately afterwards. For studies with FITC-conjugate 5, resected tumor tissue was homogenized via blunt force and then pushed through a microfilter to achieve single cell suspensions which were plated on glass microscopy slides for 6 hours prior to imaging using a Zeiss Exciter fluorescence confocal microscope.
RNA-Seq sample preparation and data processing
Cells for gene expression analysis were plated in 10 cm diameter dishes at 1.1×106 cells/dish and incubated in the growth medium supplemented with 10 nM E2 for 24 hours. Afterwards, medium was exchanged with the growth medium supplemented with polyamides and 10 nM E2 and incubated for 48 hours in 5% CO2 and 37°C. The RNA was then harvested using an RNEasy kit (Qiagen). Subsequently, a Riboguard RNAse inhibitor was added and samples were treated with TurboDNA Free DNAse (Ambion), according to manufacturers’ instructions. RNA-Seq libraries were prepared using standard Illumina reagents and protocols. Single read sequencing with the read length of 50 nucleotides were performed on the Illumina HiSeq2000 sequencer, following manufacturers’ instructions, producing 35–50 million reads per library. Sequencing data were mapped against the combined human (hg19) transcriptome, using the Bowtie program package 0.12.7 (31) and the refseq annotation. The open access processing package Cuffdiff was employed to calculated differential gene expression. Inter-replicate statistical significance was calculated with the DEseq module (32).
Results
Design of polyamides
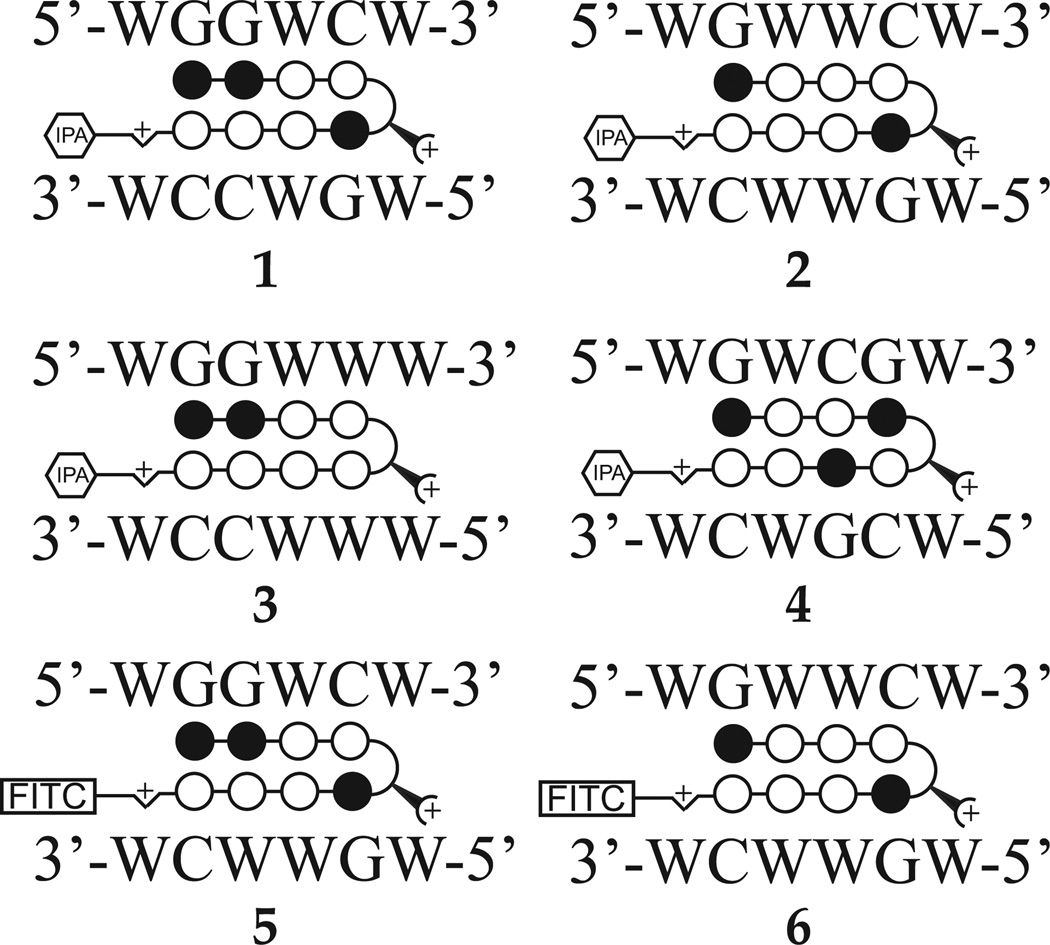
We synthesized four eight-ring hairpin Py-Im polyamides to screen for activity against E2 stimulated gene expression (Figure 1, Supplementary Figure S1). Polyamide 1 targets 5’-WGGWCW-3’, which is the half-site ERE consensus. Polyamide 2 was previously reported to inhibit a subset of DHT-induced gene expression in cultured prostate cancer cells (11). Polyamide 3 was recently characterized in cultured lung cancer cells and used to partially abrogate TNF-stimulated transcription (12). Polyamide 4 targets the sequence 5’-WGWCGW-3’. Polyamide 5 and 6 are FITC-conjugated analogs of 1 and 2respectively, used to visualize cellular uptake and distribution in this study.
Figure 1.
Ball-and-stick models of polyamides 1–6 with DNA target sequences as follows: 1, 5’-WGGWCW-3’; 2, 5’-WGWWCW-3’; 3, 5’-WGGWWW-3’; 4, 5’-WGWCGW-3’. 5 and 6 are FITC-analogs of 1 and 2, respectively, used for fluorescence microscopy experiments. Chemical structures of 1–6 are in Supplementary Figure S1.
Evaluation of binding of polyamides to an ERE half site by DNA thermal denaturation assays
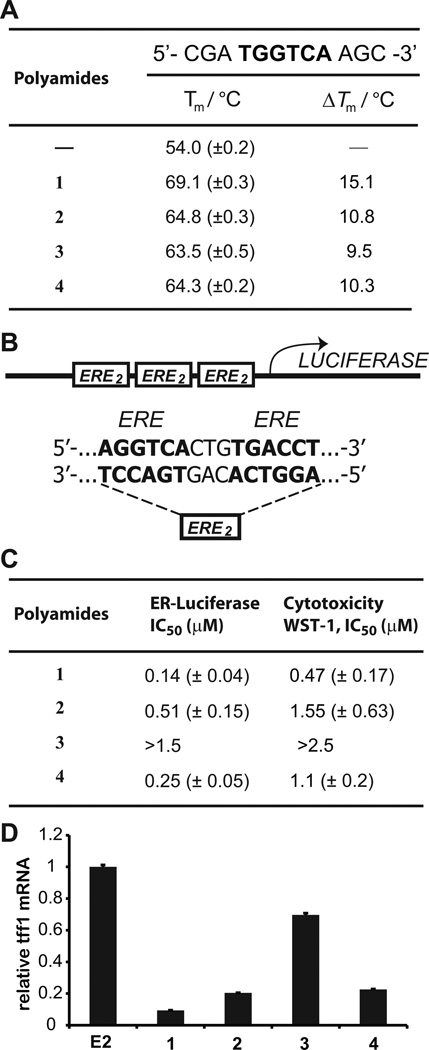
Polyamides 1–4 were incubated with duplex DNA 5’-CGATGGTCAAGC-3’, which contains an ERE half-site consensus, and melting temperatures measured (Figure 2A). Duplex stabilization was greatest for 1a polyamide that was predicted to bind this sequence based on established Py-Im polyamide pairing rules (6, 7). The other polyamides demonstrated less stabilization of this duplex.
Figure 2.
A, Thermal denaturation assays of a duplex DNA oligonucleotide containing a half site ERE. Polyamide 1 demonstrates the most stabilization. B, Sequence of ERE-driven luciferase in T47D-KBLUC cells. C, Polyamides 1–4 were screened for cytotoxicity and suppression of ER-driven luciferase. Polyamide 1 is most potent by both measures. Representative isotherms are displayed in Supplementary Figure S2. D, Polyamides dosed at 0.3 µM were screened for activity against TFF1 expression, a known ERE driven gene. The relative activities of 1–4 approximately mirror what is seen in the luciferase assay at this concentration. At higher concentrations (~1 µM), all 4 polyamides demonstrate activity.
Luciferase activity and cytotoxicity in T47D-KBLUC cells
The ERα positive cell line T47D-KBLUC expresses luciferase under the control of three tandem repeats of the sequence 5’-AGGTCACTTGACCT-3’ (25), which is the consensus sequence for the ERα-DNA homodimer (Figure 2B). T47D-KBLUC cells were grown in 10% FBS/RPMI-1640 media with 10 nM E2 for 48 hours. Then, media was replenished with varying concentrations of polyamides 1–4 for 96 hours. An extended incubation time with E2 was used to approximate the in vivo condition of continued E2 circulation. Cell proliferation and viability was assayed using WST-1 (Roche), and luciferase output was measured (Figure 2C). Both luciferase output and proliferation were affected most by treatment with 1 (IC50 0.47 µM for viability, 0.14 µM for luciferase suppression), and least by 3 (IC50 > 2.5 and 1.5 µM, respectively). The representative data for luciferase and WST-1 assay shown in supplementary figure S2. We identified TFF1 as one of the most highly induced transcripts by E2 based on published reports (33).The effects of 1–4 no E2 stimulated TFF1 expression were measured to validate the luciferase screen. Polyamide 1 was again found most potent, although 2 and 4 demonstrated significant inhibition of TFF1 as well (Figure 2D). Inhibition of TFF1 mRNA by 1 is dose responsive (Supplementary Figure S3). In addition, 1 demonstrates significantly less toxicity to LNCaP, U251, and A549 cell lines (Supplementary Figure S4), which have low expression of ER-α (34–37). Chromatin immunoprecipitation of ERα at the TFF1 promoter after E2 stimulation of cells pre-treated with 1 showed reduced occupancy as compared to vehicle treated cells (Supplementary Figure S5).
Genome-wide polyamide effects on E2 induced gene expression
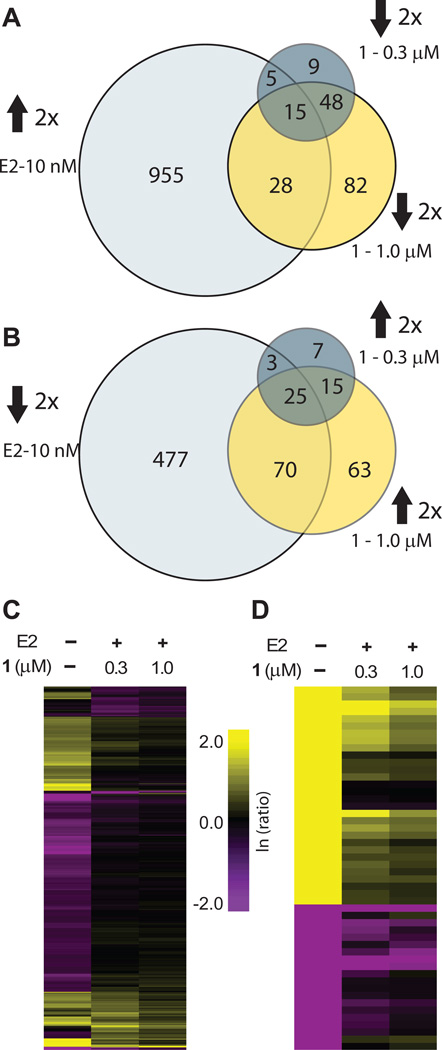
Effects of hairpin polyamide 1 at 0.3 and 1 µM on the transcriptome of E2 induced cells were measured using RNA-Seq. Reads were mapped using Hg19 reference human genome and data was analyzed using the Bowtie and CuffDiff packages (38). Only the genes with fragments per kilobase of exon per million fragments mapped (FPKM) ≥ 20 and at least two-fold change in gene expression upon treatment with either 1 or E2 were used in the analysis (Supplementary Table S1). Among those genes, at 1.0 µM, 1 affected expression of 346 genes (0.7% of total) at least two-fold as compared to E2 treated control. Of these genes, an equal number of genes were up- and down-regulated (173 in each case). At the lower concentration of 0.3 µM, expression of 127 genes (0.3% of total) was affected at least two-fold, and a majority of these genes (77 vs 50) were downregulated. At the same threshold, E2 upregulated 1003 genes (2.0%) (Figure 3A) and downregulated 575 genes (1.2%) (Figure 3B). A fraction of expression changes induced by E2 were reversed by 1 (Supplementary Table S2), and this fraction was greater for E2 repressed genes. Among E2 upregulated genes 43 (4.3%) were repressed by 1 at least two fold at 1.0 µM. Among those 575 genes that were downregulated by E2, 95 (16.5%) were de-repressed by 1 at 1.0 µM at least two fold (Figure 3A–B). Overall, of the 346 genes affected by 1 at 1.0 µM, 138 (39.9%) represent genes whose up- or down-regulation by E2 was abrogated by polyamide treatment. Genes whose expression was affected by 1 at a lower concentration (0.3 µM) were largely a subset of the genes affected at 1.0 µM, 103 of which (81.1%) were affected by 1 at both concentrations.
Figure 3.
RNA-seq global transcriptome analysis. All ratios are normalized to the induced control (10 nM E2). A, Venn diagrams show the overlap between genes upregulated by E2 at least two-fold and genes downregulated by 1 at 0.3 µM or 1.0 µM. B, Venn diagrams for the overlap of genes downregulated by E2 at least two-fold and de-repressed by 1 at 0.3 µM or 1.0 µM. C, Hierarchical clustering (Euclidian distance, complete linkage) of genes changed at least two-fold as compared to the induced state. D, 50 genes that were most changed by E2 induction were clustered (Eucludian distance, complete linkage). Of those genes 30 were upregulated and 20 were downregulated by E2. Fold-changes are relative to E2-induced control.
Further analysis was performed using Euclidian distance clustering with complete linkage (Figure 3C). Interestingly, while the majority of E2 affected genes are not affected by 1out of the top 50 genes most strongly affected by E2, 28 (56%) are inhibited at least two-fold, and 38 out of 50 (78%) genes are inhibited at least 1.5-fold by 1 (Figure 3D).
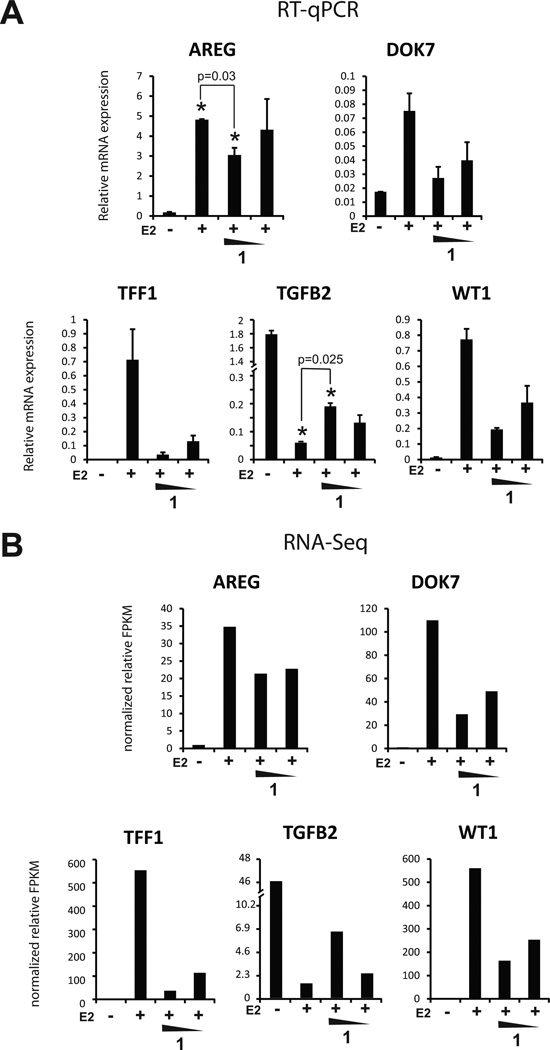
Five transcripts were selected for verification by qRT-PCR and all five showed good reproducibility of the expression changes seen by RNA-Seq (Figure 4). Four were upregulated by E2 (AREG, DOK7, TFF1, WT1) and one downregulated by E2 (TGFB2).
Figure 4.
Confirmation of genome-wide polyamide effects observed by RNA-seq. Five genes (four induced and one repressed by estrogen) were interrogated. A, Relative mRNA levels of selected genes measured by qRT-PCR. B, Relative mRNA expression values as measured by fragments per kilobase of exon million (FPKM) from RNA-seq. Concentrations of 1 are 1.0 µM and 0.3 µM.
Circulation and toxicity of polyamide 1 in mice
To assess serum concentrations of 1 after subcutaneous injection, four female C57BL/6 mice were injected subcutaneously into the left flank with 120 nmol of 1 in a 200 µL 20% DMSO/PBS vehicle. Serial serum samples were taken via retroorbital draw and processed by methods previously described (30). Polyamide 1 was detectable in serum for up to 24 hours after injections, and reached a maximum concentration of 3 µM at 6 hours after injection (Supplementary Figure S6A). Toxicity after repeated injections of 1 was assessed by daily weights and visual inspection of treated mice. Five female C57BL/6 mice were injected with 20 nmol of 1 subcutaneously to the left flank three times a week for two weeks without measurable weight loss. The regimen was then increased to 30 nmol for two weeks, again without measurable weight loss or changes in animal behavior (Supplementary Figure S6B).
Effects on ERα mediated transcription in T47D-KBLUC tumor bearing mice after short-term treatment
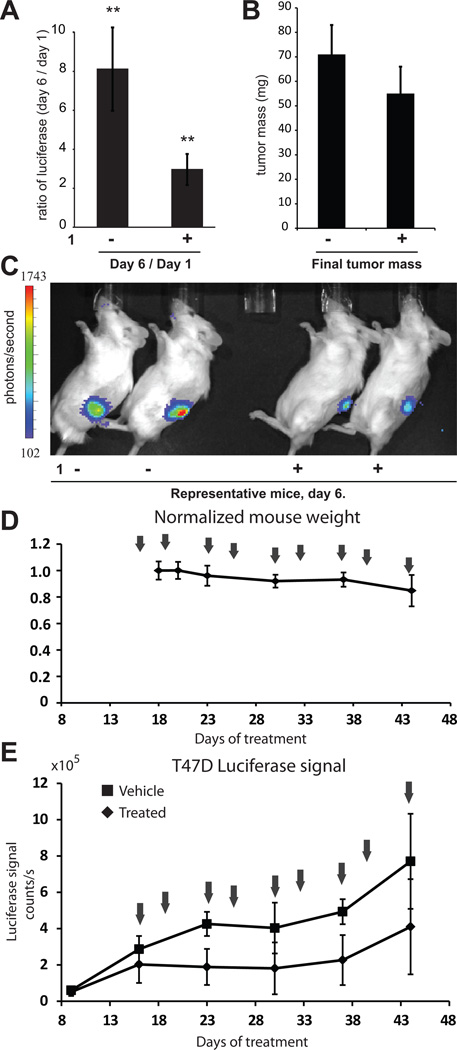
To measure the efficacy of 1 in vivo against E2-induced transcription, T47D-KBLUC cells were engrafted into female nod-scid-gamma (NSG) immunocompromised mice supplemented with a slow-release subcutaneous E2 pellet in the right flank to facilitate E2-induced growth. After one week of growth, mice were imaged utilizing the IVIS imaging system (Caliper) and stratified into groups of 12 mice each for vehicle and polyamide treatment. Polyamide 1 (25 nmol) in 200 µL 20% DMSO/PBS was injected subcutaneously into the left shoulder every other day for a total of four injections. Vehicle treated mice received 20% DMSO/PBS alone. After three injections, mice were re-imaged. Luciferase output increased an average of eight-fold for the vehicle treated mice and three-fold for the mice treated with 1 (Figure 5A). Mice were euthanized the day following the fourth injection for tumor resection. Tumors from vehicle treated mice were 71±12 mg and tumors from polyamide treated mice 55 ±11 mg (Figure 5B), which does not explain the differences seen in luciferase expression. Representative images of mice treated with 1 or vehicle at day 6 are shown (Figure 5C).
Figure 5.
Xenograft studies. A, Treatment of T47D-KBLUC bearing mice with 1 results in suppression of ER-driven luciferase. **p<0.01. n = 12 mice per group. Errors are 95% CI. B, Tumors masses at experimental endpoint were vehicle: 71±12 mg (95% CI); 1: 55 ±11 mg (95% CI). C, Representative luciferase output of vehicle and treated mice on day 6. D, Treatment schedule for extended time-course experiments with normalized mouse weights over time. Arrows indicate treatment days. E, Luciferase signal for polyamide 1 and vehicle treated groups. Error bars are standard deviations. Mean tumor masses at the endpoint for animals treated with vehicle is 165±27mg (95% CI) and for animals treated with 1 is 128±54mg (95% CI).
Effects on ERα mediated transcription in T47D-KBLUC tumor bearing mice after long-term treatment
To investigate the effects of 1 in tumor bearing mice after extended treatment, T47D-KBLUC cells were again engrafted into female NSG mice supplemented with a subcutaneous E2 pellet in the right flank. Tumors were grown for nine days prior to stratification of five mice each into polyamide 1 and vehicle treatment groups. Mice were treated with vehicle or 25 nmol of 1 in 20% DMSO/PBS, subcutaneously into the left shoulder twice a week for a course of 9 injections (25 days), beginning on day 16 after engraftment (Figure 5D). Treated mice maintained their weights at >90% until the final days of treatment when their weights decreased to >85% prior to euthanasia. Luciferase was monitored weekly utilizing the IVIS imaging system. Luciferase output in the polyamide treated mice was consistently lower than vehicle treated mice (Figure 6E). At the experimental endpoint, tumors from vehicle treated mice were 165 ± 27 mg and tumors from polyamide treated mice 128 ± 54 mg.
Figure 6.
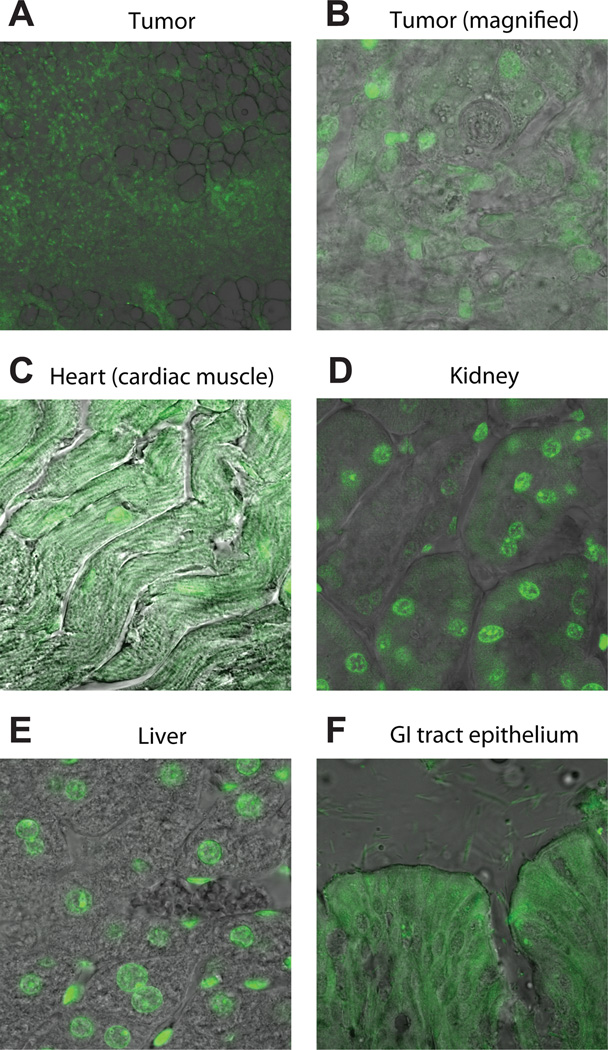
Tissue frozen sections of tissue extracted from xenograft bearing mouse treated with 5. A, FITC-labeled Py-Im polyamide 5 distributes widely in sections of the T47D-KBLUC tumor xenograft. B, High magnification shows nuclear localization of 5 in tumor cells. C, Cardiac muscle sections show a fibrous pattern of fluorescence in the cytoplasm as well as nuclear staining. D, Kidneys demonstrate nuclear localization of 5. E, Liver sections show nuclear localization of 5. F, Bowel epithelia show cytoplasmic fluorescence.
Tissue distribution of FITC-conjugated polyamide 5 in mice bearing T47D-KBLUC xenografts
Py-Im polyamide 5 is a FITC labeled conjugate of hairpin 1 that was synthesized to evaluate tissue and sub-cellular localization via fluorescence microscopy. T47D-KBLUC cells cultured in vitro and then treated with 5 showed nuclear fluorescence similar to what has been reported in other cell lines (27, 28) treated with FITC-conjugated polyamides in cell culture (Supplementary Figure S7). An NSG mouse engrafted with T47D-KBLUC cells as described in the previous section was treated with polyamide 5 in a manner identical to that of 1except at a dose of 50 nmol per injection. After three injections, the mouse was euthanized, the tumor resected, and internal organs dissected. Tissue was fixed, cryoprotected, sectioned and imaged immediately. Fluorescence signal was evenly distributed throughout multiple sections of the tumor xenograft. A representative section is shown (Figure 6A). High magnification reveals nuclear localization in tumor tissue (Figure 6B). Sections of cardiac muscle show significant cytoplasmic fluorescence in a fibrous pattern (Figure 6C). Sections of kidney and liver both show nuclear fluorescence localization, with minimal cytoplasmic fluorescence (Figure 6D–E) Small bowel epithelia show diffuse cellular fluorescence (Figure 6F).
To ensure that nuclear fluorescence in the xenografts was not an artifact of the fixation process, we extracted live cells from T47D-KBLUC xenografted tumors from mice treated with 5 as above. In this experiment, cells were isolated via filtration and plated on microscope slides, and incubated for six hours before imaging. Cells derived from the tumor demonstrated nuclear staining in a pattern similar to that seen in the fixed tumor sections as well as cultured cells treated with 5 in vitro (Supplementary Figure S8).
Discussion
In order for DNA-binding Py-Im polyamides to be considered for therapeutic application, these molecules must possess favorable pharmacokinetic and pharmacodynamic properties and exert a desired effect in target tissues. In this study, ERα-induced transcription in xenografted breast cancer tumors was the target. A polyamide targeted to the ERE half site 5’-WGGWCW-3’ was identified from a focused screen for activity against ER mediated transcription and cytotoxicity against ERα positive breast cancer cells. This polyamide was further tested for its global effects on the transcriptome of E2 induced T47D-KBLUC cells. Hairpin polyamide 1 demonstrated limited toxicity and circulated at therapeutic levels in serum after subcutaneous injection. It also showed activity against ER driven luciferase expression in xenografted tumors in immunocompromised mice. FITC-polyamide conjugate 5 shows widespread localization in body tissues including sections through the xenografted tumor, which reveal nuclear fluorescence.
Suppression of ER-induced gene expression
We screened for both suppression of E2 induced luciferase expression and for anti-proliferation by WST-1 assay using T47D-KBLUC cells. Polyamide 1 was the most active by both measures while 3 was inactive by either measure. These molecules differ only at a single atom, which represents the difference between a Py and Im heterocycle. Although polyamides have been shown to have differing uptake properties depending on Py-Im content and sequence (27, 28), the differences in activity in this series is likely not explained by differing uptake efficiency since confocal microscopy of FITC-polyamide conjugates 5 and 6 are similar (Supplementary Figure 7).
Global effects on the E2 stimulated transcriptome
E2 exerts its effects through direct DNA binding and less frequently extranuclear pathways that do not involve ERα-DNA binding (39). Our genome-wide transcriptome analysis (Figure 4) revealed that a small fraction of gene expression changes induced by E2 treatment is suppressed by polyamide 1. From a total of 1003 E2 up-regulated genes, only 43 (0.43%) are repressed by 1 at 1.0 µM, and from a total of 575 E2 downregulated genes, 95 (16.5%) were de-repressed. However, among the 50 genes most strongly either induced or repressed by E2, a majority were significantly affected by 1. Among these top 50, 28 (56%) were downregulated by 1 (1.0 µM) at least two-fold. At a lower cut-off of 1.5-fold, 38 (76%) of E2 induced gene expression changes were abrogated by the action of 1 at 1.0 µM. Many of these strongly E2-responsive genes play important roles in the development of tumors and are therapeutically relevant. Among them is Wilms Tumor 1 (WT1), a gene originally identified as a tumor suppressor (40), however more recently it has become apparent that can also act as an oncogene (41). WT1 expression is detectable in 90% of breast cancers (42) and high levels of WT1 expression are correlated with poor patient survival (43). TFF1 is a predictor for breast cancer patient survival (36). Transforming growth factor-β2 (TGF-β2) was observed among the most strongly E2-repressed genes and was also over 3-fold de-repressed by 1 at 1.0 µM. TGF-β2 is involved in cancer development that is also de-repressed by traditional anti-estrogens (44). We conclude that 1 acts in an anti-estrogenic fashion among genes that are most potently affected by E2, but is less active for the majority of E2 responsive genes. If the mechanism by which 1 interferes with estrogen driven gene expression is through direct interference with ER-DNA interfaces, we would not expect to affect ER-driven transcription at loci where ER signals through a tethering complex (45), such as with Ap1 and Sp1. Indirect interactions between estrogen receptor and DNA through tethering with other proteins offer a partial explanation for the limited number of ER driven transcripts affected by 1.
Most transcripts affected by 1 are not explained on the basis of E2 antagonism; 295 genes that are either up- or down- regulated by 1 at least 2-fold that are not explained by effects on ER activity. Of these, 164 are up-regulated and 131 down-regulated by 1. To further characterize these effects, we utilized the DAVID functional annotation tool (46, 47). For the up-regulated transcripts, enriched biological processes include those involved with the regulation of apoptosis and cell-death, as well as responses to endogenous and hormone stimuli whereas downregulated genes suggested that 1 is involved in regulation of GTPase mediated signal transduction and protein transport and biosynthesis (Supplementary Table S3). The mechanisms of cell-death may include the inhibition of transcription (10, 11, 13, 15), but other DNA dependent processes may contribute, and are an area of current investigation. Whether or not the effects of 1 on these biological processes are specific to polyamide 1 or may represent a class-effect is unknown but is also under study.
Polyamide treatment suppresses E2 simulated luciferase expression in vivo
T47D-KBLUC cells were chosen as the cell line for our study based on previous work using T47D cells as a model for ERα positive breast cancer. Both vehicle and polyamide treated groups showed an increase in total luciferase expression from the baseline measurement immediately prior to treatment on day 1. However, on day 6, after three sequential injections, this increase was significantly blunted in the polyamide treated group as compared to vehicle (from ~8-fold to ~3-fold), suggesting that 1 was able to reach sufficient concentrations in tumor tissue to affect luciferase expression. The approximately 2.5-fold difference in luciferase between polyamide treated and vehicle treated groups, if interpolated to the in vitro data, suggest an approximate concentration of 0.3 µM within the xenograft tissue. Tumor masses did not differ significantly between polyamide treated and vehicle treated mice in this experiment. However, this six-day experiment may be too brief to adequately assess effects on tumor growth.
Effects on tumor size
Although there are no published reports on the growth of T47D-KBLUC xenografts in mice, data from parental T47D xenografts show a slow, linear growth pattern rather than exponential (48). To better assay for antitumor activity of 1we conducted similar experiments over a longer period of time. T47D-KBLUC xenografted tumors were grown for two weeks prior to initiating treatment, and treatment with 1 was performed twice per week for a total of four weeks. We observed no significant change in tumor size at the experimental endpoint, although we found a sustained suppression of luciferase output in the polyamide treated arm as compared to vehicle treated, consistent with our initial observations. The IC50 for cytotoxicity of 1 in cell culture is 0.47 µM, which we believe to be higher than the concentrations achieved within the tumor tissues in this study.
Tissue distribution of FITC conjugate 5 in mice after repeated subcutaneous injections
Fixed, frozen sections through multiple internal organs harvested from T47D-KBLUC engrafted mice treated with 5 reveal widespread organ distribution of fluorescent signal, but with differing patterns of fluorescence between tissues, and with little obvious systemic toxicity. The tumor sections show nuclear fluorescence in a subcellular pattern that is similar to what is observed in cell culture (Figure 6A). The liver and kidneys also demonstrate strong nuclear fluorescence (Figure 6D–E), while sections through the intestinal epithelium and cardiac muscle show predominantly cytoplasmic, and both cytoplasmic and nuclear fluorescence, respectively. A difference in the cellular uptake of polyamide-FITC conjugates between cell types has also been observed in vitro (27). Recent work has shown that polyamides can form aggregates in solution (49). Whether polyamide aggregation influences distribution in vivo is unknown. Tissue-specific targeting of small molecule drugs is an area of current investigation that may become relevant for this class of molecules as additional in vivo experiments are planned.
Conclusion
Polyamide 1 delivered by subcutaneous injection in a simple DMSO/saline vehicle distributed widely in host and tumor tissue and demonstrated adequate bioavailability to affect luciferase expression in xenografted tumor tissue, with acceptable toxicity. Future investigations will include optimization of polyamides for lower systemic toxicity without a compromise in efficacy.
Supplementary Material
Acknowledgments
We thank Dr. Janet Baer, Dr. Karen L. Lencioni, and Gwen E. Williams for helpful discussions and technical assistance with animal experiments. Sequencing was conducted at the Millard and Muriel Jacobs Genetics and Genomics Laboratory at California Institute of Technology.
Grant support
This work was supported in large part by the National Institutes of Health Grant GM-51747. N.G. Nickols received support from the Jonsson Cancer Center Foundation at UCLA. J.O. Szablowski and B.C. Li were supported by NIH GM-51747. J.A. Raskatov received postdoctoral support from the Alexander von Humboldt foundation. A.E. Hargrove received postdoctoral support from the California Tobacco-Related Disease Research Program (19FT-0105) and the NIH (NRSA number 1F32CA156833).
Footnotes
Conflict of interest:
The authors have no conflict of interest to declare.
References cited
- 1.Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 2.Manni A, Arafah B, Pearson OH. Estrogen and progesterone receptors in the prediction of response of breast-cancer to endocrine therapy. Cancer. 1980;46:2838–2841. doi: 10.1002/1097-0142(19801215)46:12+<2838::aid-cncr2820461421>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 3.Burstein HJ, Prestrud AA, Seidenfeld J, Anderson H, Buchholz TA, Davidson NE, et al. American Society of Clinical Oncology Clinical Practice Guideline: Update on Adjuvant Endocrine Therapy for Women With Hormone Receptor-Positive Breast Cancer. J Clin Oncol. 2010;28:3784–3796. doi: 10.1200/JCO.2009.26.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dervan PB. Molecular recognition of DNA by small molecules. Biorg Med Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 5.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr Opin Struct Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 6.Kielkopf CL, White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB, et al. A structural basis for recognition of A•T and T•A base pairs in the minor groove of B-DNA. Science. 1998;282:111–115. doi: 10.1126/science.282.5386.111. [DOI] [PubMed] [Google Scholar]
- 7.White S, Baird EE, Dervan PB. On the pairing rules for recognition in the minor groove of DNA by pyrrole-imidazole polyamides. Chem Biol. 1997;4:569–578. doi: 10.1016/s1074-5521(97)90243-x. [DOI] [PubMed] [Google Scholar]
- 8.Chenoweth DM, Dervan PB. Allosteric modulation of DNA by small molecules. Proc Natl Acad Sci U S A. 2009;106:13175–13179. doi: 10.1073/pnas.0906532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chenoweth DM, Dervan PB. Structural Basis for Cyclic Py-Im Polyamide Allosteric Inhibition of Nuclear Receptor Binding. J Am Chem Soc. 2010;132:14521–14529. doi: 10.1021/ja105068b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muzikar KA, Nickols NG, Dervan PB. Repression of DNA-binding dependent glucocorticoid receptor-mediated gene expression. Proc Natl Acad Sci U S A. 2009;106:16598–16603. doi: 10.1073/pnas.0909192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nickols NG, Dervan PB. Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc Natl Acad Sci U S A. 2007;104:10418–10423. doi: 10.1073/pnas.0704217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raskatov JA, Meier JL, Puckett JW, Yang F, Ramakrishnan P, Dervan PB. Modulation of NF-kappa B-dependent gene transcription using programmable DNA minor groove binders. Proc Natl Acad Sci U S A. 2012;109:1023–1028. doi: 10.1073/pnas.1118506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Modulating hypoxia-inducible transcription by disrupting the HIF-1-DNA interface. ACS Chem Biol. 2007;2:561–571. doi: 10.1021/cb700110z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olenyuk BZ, Zhang GJ, Klco JM, Nickols NG, Kaelin WG, Dervan PB. Inhibition of vascular endothelial growth factor with a sequence-specific hypoxia response element antagonist. Proc Natl Acad Sci U S A. 2004;101:16768–16773. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang F, Nickols NG, Li BC, Marinov GK, Said JW, Dervan PB. Antitumor activity of a pyrrole-imidazole polyamide. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1222035110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukasawa A, Aoyama T, Nagashima T, Fukuda N, Ueno T, Sugiyama H, et al. Pharmacokinetics of Pyrrole-imidazole Polyamides after Intravenous Administration in Rat. Biopharm Drug Disposition. 2009;30:81–89. doi: 10.1002/bdd.648. [DOI] [PubMed] [Google Scholar]
- 17.Nagashima T, Aoyama T, Fukasawa A, Watabe S, Fukuda N, Ueno T, et al. Determination of pyrrole-imidazole polyamide in rat plasma by liquid chromatography-tandem mass spectrometry. J Chromat B: Biomed Sci Appl. 2009;877:1070–1076. doi: 10.1016/j.jchromb.2009.02.041. [DOI] [PubMed] [Google Scholar]
- 18.Nagashima T, Aoyama T, Yokoe T, Fukasawa A, Fukuda N, Ueno T, et al. Pharmacokinetic Modeling and Prediction of Plasma Pyrrole-Imidazole Polyamide Concentration in Rats Using Simultaneous Urinary and Biliary Excretion Data. Biol Pharm Bull. 2009;32:921–927. doi: 10.1248/bpb.32.921. [DOI] [PubMed] [Google Scholar]
- 19.Matsuda H, Fukuda N, Ueno T, Katakawa M, Wang X, Watanabe T, et al. Transcriptional inhibition of progressive renal disease by gene silencing pyrrole-imidazole polyamide targeting of the transforming growth factor-beta 1 promoter. Kidney Int. 2011;79:46–56. doi: 10.1038/ki.2010.330. [DOI] [PubMed] [Google Scholar]
- 20.Raskatov JA, Nickols NG, Hargrove AE, Marinov GK, Wold B, Dervan PB. Gene expression changes in a tumor xenograft by a pyrrole-imidazole polyamide. Proc Natl Acad Sci U S A. 2012;109:16041–16045. doi: 10.1073/pnas.1214267109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gearhart MD, Dickinson L, Ehley J, Melander C, Dervan PB, Wright PE, et al. Inhibition of DNA binding by human estrogen-related receptor 2 and estrogen receptor alpha with minor groove binding polyamides. Biochemistry (Mosc) 2005;44:4196–4203. doi: 10.1021/bi047872o. [DOI] [PubMed] [Google Scholar]
- 22.White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature. 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 23.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Improved nuclear localization of DNA-binding polyamides. Nucleic Acids Res. 2007;35:363–370. doi: 10.1093/nar/gkl1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Synold TW, Xi B, Wu J, Yen Y, Li BC, Yang F, et al. Single-dose pharmacokinetic and toxicity analysis of pyrrole-imidazole polyamides in mice. Cancer Chemother Pharmacol. 2012;70:617–625. doi: 10.1007/s00280-012-1954-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson VS, Bobseine K, Gray LE. Development and characterization of a cell line that stably expresses an estrogen-responsive luciferase reporter for the detection of estrogen receptor agonist and antagonists. Toxicol Sci. 2004;81:69–77. doi: 10.1093/toxsci/kfh180. [DOI] [PubMed] [Google Scholar]
- 26.Puckett JW, Green JT, Dervan PB. Microwave Assisted Synthesis of Py-Im Polyamides. Org Lett. 2012;14:2774–2777. doi: 10.1021/ol3010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Best TP, Edelson BS, Nickols NG, Dervan PB. Nuclear localization of pyrrole-imidazole polyamide-fluorescein conjugates in cell culture. Proc Natl Acad Sci U S A. 2003;100:12063–12068. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edelson BS, Best TP, Olenyuk B, Nickols NG, Doss RM, Foister S, et al. Influence of structural variation on nuclear localization of DNA-binding polyamide-fluorophore conjugates. Nucleic Acids Res. 2004;32:2802–2818. doi: 10.1093/nar/gkh609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappa B gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 30.Raskatov JA, Hargrove AE, So AY, Dervan PB. Pharmacokinetics of Py-Im Polyamides Depend on Architecture: Cyclic versus Linear. J Am Chem Soc. 2012;134:7995–7999. doi: 10.1021/ja302588v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10 doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11 doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FCGJ, Span PN, et al. ChIP-Seq of ER alpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stabile LP, Lyker JS, Gubish CT, Zhang W, Grandis JR, Siegfried JM. Combined targeting of the estrogen receptor and the epidermal growth factor receptor in non-small cell lung cancer shows enhanced antiproliferative effects. Cancer Res. 2005;65:1459–1470. doi: 10.1158/0008-5472.CAN-04-1872. [DOI] [PubMed] [Google Scholar]
- 35.Lau KM, LaSpina M, Long J, Ho SM. Expression of estrogen receptor (ER)-alpha and ER-beta in normal and malignant prostatic epithelial cells: regulation by methylation and involvement in growth regulation. Cancer Res. 2000;60:3175–3182. [PubMed] [Google Scholar]
- 36.Le Page Y, Scholze M, Kah O, Pakdel F. Assessment of xenoestrogens using three distinct estrogen receptors and the zebrafish brain aromatase gene in a highly responsive glial cell system. Environ Health Perspect. 2006;114:752–758. doi: 10.1289/ehp.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devidze N, Fujimori K, Urade Y, Pfaff DW, Mong JA. Estradiol regulation of lipocalin-type prostaglandin D synthase promoter activity: evidence for direct and indirect mechanisms. Neurosci Lett. 2010;474:17–21. doi: 10.1016/j.neulet.2010.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madak-Erdogan Z, Kieser KJ, Kim SH, Komm B, Katzenellenbogen JA, Katzenellenbogen BS. Nuclear and extranuclear pathway inputs in the regulation of global gene expression by estrogen receptors. Mol Endocrinol. 2008;22:2116–2127. doi: 10.1210/me.2008-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Little M, Wells C. A clinical overview of WT1 gene mutations. Hum Mutat. 1997;9:209–225. doi: 10.1002/(SICI)1098-1004(1997)9:3<209::AID-HUMU2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 41.Yang L, Han Y, Saurez Saiz F, Minden MD. A tumor suppressor and oncogene: the WT1 story. Leukemia. 2007;21:868–876. doi: 10.1038/sj.leu.2404624. [DOI] [PubMed] [Google Scholar]
- 42.Loeb DM, Evron E, Patel CB, Sharma PM, Niranjan B, Buluwela L, et al. Wilms' tumor suppressor gene (WT1) is expressed in primary breast tumors despite tumor-specific promoter methylation. Cancer Res. 2001;61:921–925. [PubMed] [Google Scholar]
- 43.Miyoshi Y, Ando A, Egawa C, Taguchi T, Tamaki Y, Tamaki H, et al. High expression of Wilms' tumor suppressor gene predicts poor prognosis in breast cancer patients. Clin Cancer Res. 2002;8:1167–1171. [PubMed] [Google Scholar]
- 44.Foekens JA, Rio MC, Seguin P, Vanputten WLJ, Fauque J, Nap M, et al. Prediction of relapse and survival in breast-cancer patients by PS2 protein status. Cancer Res. 1990;50:3832–3837. [PubMed] [Google Scholar]
- 45.Stender JD, Kim K, Charn TH, Komm B, Chang KC, Kraus WL, et al. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol Cell Biol. 2010;30:3943–3955. doi: 10.1128/MCB.00118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 47.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishii Y, Waxman S, Germain D. Tamoxifen stimulates the growth of cyclin D1 - Overexpressing breast cancer cells by promoting the activation of signal transducer and activator of transcription 3. Cancer Res. 2008;68:852–860. doi: 10.1158/0008-5472.CAN-07-2879. [DOI] [PubMed] [Google Scholar]
- 49.Hargrove AE, Raskatov JA, Meier JL, Montgomery DC, Dervan PB. Characterization and solubilization of pyrrole-imidazole polyamide aggregates. J Med Chem. 2012;55:5425–5432. doi: 10.1021/jm300380a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.