Abstract
Background
The genetic composition of cynomolgus macaques used in biomedical research is not as well-characterized as that of rhesus macaques.
Methods
Populations of cynomolgus macaques from Sumatra, Corregidor, Mauritius, Singapore, Cambodia and Zamboanga were analyzed using 24 STRs.
Results
The Sumatran and Cambodian populations exhibited the highest allelic diversity while the Mauritian population exhibited the lowest. Sumatran cynomolgus macaques were the most genetically similar to all others, consistent with an Indonesian origin of the species. The high diversity among Cambodian animals may result from interbreeding with rhesus macaques. The Philippine and Mauritian samples were the most divergent from other populations, the former due to separation from the Sunda Shelf by deep water and the latter due to anthropogenic translocation and extreme founder effects.
Conclusions
Investigators should verify their research subjects’ origin, ancestry and pedigree to minimize risks to biomedical experimentation from genetic variance stemming from close kinship and mixed ancestry as these can obscure treatment effects.
Keywords: Population structure, founder effects, genetic isolation, inbreeding, drift
Introduction
Cynomolgus or long-tailed macaques (Macaca fascicularis) are used more frequently as non-human primate (NHP) models for biomedical research on human diseases than any other non-human primate species with the exception of rhesus macaques (M. mulatta). The steep increase in the number of cynomolgus macaques being imported to the US reflects their growing popularity as an alternative to rhesus macaques [5, 39]. According to a 2009 US Fish and Wildlife Department report, approximately 126,000 cynomolgus macaques were imported to the US during the period 2000–2005 [35], compared to only 20,000 rhesus macaques during the same time period. While their importance for biomedical studies, especially outside of the US National Primate Research Center (NPRC) system, rivals that of rhesus macaques, the genetic, genomic and bioinformatics resources currently available for rhesus macaques have not been developed for cynomolgus macaques. Consequently, the relevant genetic parameters for selecting cynomolgus macaques employed in biomedical research, including parentage, kinship and ancestry, remain poorly characterized.
The high demand for cynomolgus macaques for use in biomedical research has fueled the capture of large numbers of wild founders from Indonesia, the Philippines and Indochina (including Cambodia, Laos and Vietnam) for breeding in farms, particularly in China. Chinese breeders exported over 65% of the cynomolgus macaques imported to the US in 2009 [3]. Cynomolgus macaques imported directly from Mauritius and southern Sumatra/western Java as well as those of unknown origin imported from Chinese breeding farms, most of which probably originated in Indochina, have dominated US imports. In addition, animals originating in different countries are typically interbred in Chinese breeding farms [Dr. Hongli Du, South China Technology University, Guangzhou, pers. comm., 2012]. Thus, the points of origin and precise ancestry of cynomolgus macaques whose genetic characteristics have been reported in previous studies are not typically known.
Genetic evidence of natural interspecies admixture between cynomolgus and rhesus macaques in Indochina, leading to gene flow well beyond the hybrid zone, is well documented [4, 11, 22, 41, 43, 48, 56]. Thus, the majority of cynomolgus macaques employed in biomedical research in the US also exhibit variable but unknown levels of admixture with rhesus macaques [23].
Several studies of the mtDNA of relatively small samples of cynomolgus macaques indicate a fundamental genetic difference between animals from insular (e.g., Indonesia, Sarawak, and Philippines) and mainland (e.g., Indochina, Thailand, Burma and Peninsular Malaysia, north of the Isthmus of Kra) Southeast Asia [15, 54] and a paucity of genetic diversity in those from Mauritius [27, 29] and the Philippines [47]. Estimates of genetic differences reported among regional populations of cynomolgus macaques [4, 21, 22, 48] equal or surpass that between Indian and Chinese rhesus macaques [24, 33, 45, 46] which differ from each other as much as some species and are not always appropriate for use as the same animal model.
If genetic differences among some cynomolgus macaque populations are as high as that between Indian and Chinese rhesus macaques, then they, like Indian and Chinese rhesus macaques, should be separately bred and not necessarily considered appropriate as research models for the same diseases. The potential for genetic diversity among island populations should also be considered in experimental designs [62]. If some groups of cynomolgus macaques exhibit more genetic heterogeneity influencing the phenotype than other groups, their genetic background can affect their suitability for some types of research.
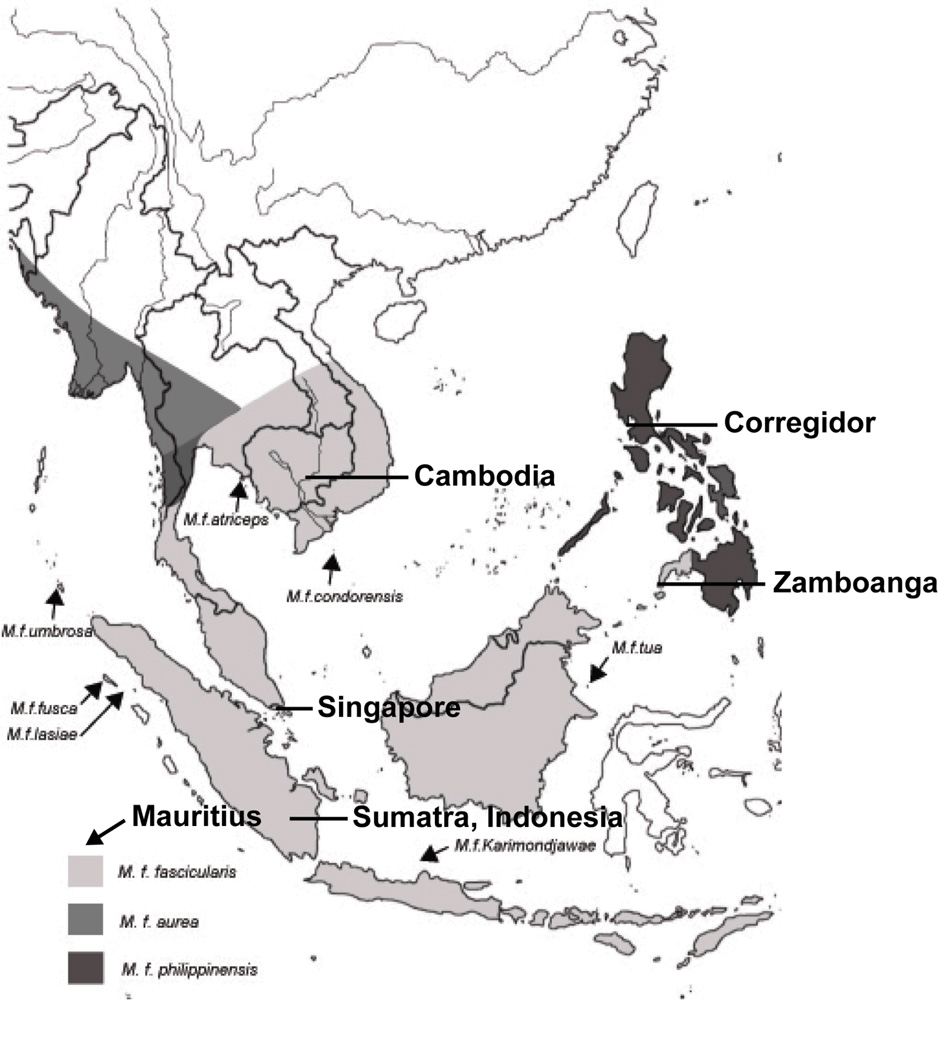
In the present study, we infer the population structure of several cynomolgus populations derived from well documented localities that represent major sources for wild-caught breeding stock for biomedical research in the US. We analyzed DNA samples from wild caught animals and their descendants known to have originated from the following locations, shown in Fig. 1: 1) Corregidor, in Manilla Bay on the island of Luzon in the northern Philippines, 2) Zamboanga, in the northwestern part of the island of Mindanao in the southern Philippines, 3) Singapore, off the southern coast of the Malay Peninsula, 4) Cambodia in mainland Southeast Asia, 5) Palembang in southern Sumatra and 6) five different localities on the island of Mauritius.
Figure 1.
Geographic range of cynomolgus macaques based on Groves (2001). Study samples were derived from the islands of Sumatra, Indonesia (1), Mindanao [Zamboanga (2) and Corregidor (4)], Mauritius (3), Singapore (5) and from Cambodia (6). Mauritius is approximately 3200 km southwest of Sumatra.
Our southern Sumatran population represents the alleged original homeland of cynomolgus macaques [6, 44]. However, the data of Tosi et al. [55], along with those of Street et al. [49] and Bonhomme et al. [3], imply that the cynomolgus macaques in Indochina represent a very old population because of the large number of private alleles it retains, supporting the alternative hypothesis that cynomolgus macaques instead originated in Indochina and later dispersed to insular Southeast Asia. An alternative hypothesis for the origin of private alleles in Indochinese cynomolgus macaques is admixture with rhesus macaques. The mtDNA haplotype network in Smith et al [42] demonstrates that the haplotypes from which all Mauritian, Philippine and Indochinese haplotypes derive originated on the Sunda Shelf. The cynomolgus macaque population with the greatest diversity and least genetic distance to all other populations of the species should be that population inhabiting the original homeland of the species. If the Cambodian population represents the descendants of immigrants from Indonesia that first reached the mainland nearly one million years ago [10] and has also experienced admixture with rhesus macaques, these animals should exhibit relatively high genetic distances from all other cynomolgus populations and relatively high genetic diversity due to admixture with rhesus macaques.
Mauritian cynomolgus macaques are believed to have originated in Indonesia and had been introduced to Mauritius several centuries ago by sailors [51, 52]. An Indonesian homeland for the Mauritian cynomolgus macaques is supported by the presence of the Indonesian MHC class I Mafa-B*011:01 and Mafa-B*075:01 alleles in the Mauritian macaques [7, 63]. Tosi and Coke [54] specifically suggest Sumatra as a source for Mauritian cynomolgus macaques because both exhibit the otherwise uniquely Sumatran characteristic of sharing their mtDNA with insular populations and their Y-chromosome DNA with mainland populations. The Mauritian population should exhibit a relatively low level of genetic diversity and share the lowest genetic distance with the population occupying its homeland.
Two different subspecies of cynomolgus macaques often employed in biomedical research inhabit the Philippines, M. f. fascicularis, in western Mindanao (e.g., Zamboanga) and M.f philippinensis, in eastern Mindanao and most of the remaining islands. The latter subspecies is believed to have dispersed to Luzon, probably via Palawan, long before the former, and has evolved markedly different morphology than the former. The former subspecies dispersed to Mindanao, probably via the Sulu archipelago, much later and should share much closer genetic similarity to all other cynomolgus macaques in our study than to M.f philippinensis. Our cynomolgus macaques from Corregidor are believed to be descendants of animals introduced by an exporter of NHPs who collected animals of unknown subspecies from unknown locations on the island of Mindanao in the Philippines and released 400–600 of them after the collapse of his business in the mid-1980s [R. Razalo, pers. comm., 2011 and J. Nazareno, pers. comm. 2012]. If this scenario is correct and the animals were collected from western Mindanao, our samples from Corregidor and Zamboanga should exhibit similar population genetic parameters and share a low genetic distance and approximately equal genetic distance to our other cynomolgus populations.
While Singapore is not presently connected to Peninsular Malaysia, it was connected by a land bridge throughout most of the Quaternary Period [1]. For this reason, we believe that our Singapore samples should resemble cynomolgus macaques of the Malaysian Peninsula at least as closely as the Chinese rhesus macaques from Hainan Island resemble those from the mainland of Guangdong province from which they were last separated by the flooding of the Qiongzhou Strait approximately 8,500 years ago [64]. The single mitochondrial DNA haplotype represented on Hainan Island is a single mutational step removed from another haplotype that is common in the mainland provinces of Guangxi and Guangdong [30]. Thus, we anticipate that the cynomolgus macaques from Singapore will resemble those from Cambodia, its closest neighbor in our study, more closely than does any other population. The genetic distances among and genetic diversity of the six regional populations studied might also suggest the route of dispersal of the species, whether from the mainland or Indonesia. As peninsular Malaysia is separated from Sumatra by a short and very shallow length of water that was among the earliest to disappear during glacial events [62], we expect the Sumatran population to be the insular cynomolgus population most similar to the mainland populations of Cambodia and Singapore.
Methods
The research reported in this manuscript complied with the protocols approved by the UC Davis Animal Care and Use Committee and adhered to the legal requirements of the US, where the research took place. In addition to the 13 STRs used by Kanthaswamy et al. [22] in an earlier study of cynomolgus and rhesus macaque hybridization, DNA samples from the six regional populations of cynomolgus macaques discussed above were genotyped for 11 additional STRs (Table 1). Table 2 lists the number of animals (N) sampled from each of these six study populations. Cumulatively, these 24 loci represent a two-fold wider coverage of the cynomolgus macaque genome than reported previously [20, 22]. Procedures for DNA extraction and PCR amplification of all 24 STR loci were those described by Satkoski et al. [40] and Kanthaswamy et al. [22]. Only individuals with at least 90% complete genotypes were used in subsequent analyses.
Table 1.
The 24 STRs used in the present study and per locus estimates of allele numbers (na), and observed (HO) and expected (HE) heterozygosities averaged across populations are provided below. Estimates of na, HO and He averaged across 10 iterations of analyses involving the random sampling with replacement of 18 individuals are in brackets. Only D5s1457 was in HWE (p < 0.05) when data from all populations were pooled.
| Locus | Allele number | Observed heterozygosity | Expected heterozygosity |
|---|---|---|---|
| D1s548 | 4 [4] | 0.548 [0.563] | 0.611 [0.607] |
| D3s1768 | 10 [8] | 0.656 [0.663] | 0.772 [0.771] |
| D4s1626 | 7 [6] | 0.718 [0.727] | 0.742 [0.747] |
| D5s1457 | 5 [4] | 0.571 [0.574] | 0.611 [0.613] |
| D6s501 | 7 [6] | 0.727 [0.725] | 0.74 [0.739] |
| D7s794 | 8 [6] | 0.582 [0.584] | 0.681 [0.679] |
| D7s1826 | 9 [7] | 0.704 [0.707] | 0.719 [0.72] |
| D8s1106 | 8 [7] | 0.743 [0.759] | 0.79 [0.796] |
| D8s1466 | 7 [6] | 0.581 [0.585] | 0.724 [0.722] |
| D9s921 | 8 [7] | 0.771 [0.774] | 0.808 [0.809] |
| D9s934 | 8 [7] | 0.71 [0.704] | 0.786 [0.784] |
| D10s1432 | 6 [6] | 0.595 [0.591] | 0.69 [0.683] |
| D11s1975 | 5 [5] | 0.687 [0.669] | 0.715 [0.713] |
| D11s2002 | 7 [6] | 0.725 [0.734] | 0.762 [0.77] |
| D13s765 | 10 [7] | 0.661 [0.654] | 0.772 [0.769] |
| D14s306 | 12 [9] | 0.807 [0.813] | 0.843 [0.84] |
| D16s750 | 3 [3] | 0.528 [0.533] | 0.513 [0.513] |
| D18s536 | 6 [5] | 0.549 [0.548] | 0.593 [0.593] |
| D18s537 | 5 [4] | 0.48 [0.491] | 0.631 [0.618] |
| AGAT007 | 6 [5] | 0.666 [0.668] | 0.706 [0.714] |
| 27oe8 | 6 [5] | 0.685 [0.687] | 0.717 [0.716] |
| 270o7 | 4 [4] | 0.466 [0.46] | 0.538 [0.543] |
| 271j8 | 9 [8] | 0.856 [0.841] | 0.815 [0.807] |
| 272o12 | 7 [6] | 0.588 [0.591] | 0.702 [0.699] |
Table 2.
Sample numbers (N), mean allele numbers (na), mean observed (HO) and mean expected heterozygosity (HE) averaged across all 24 STRs for each study population. The number of loci per population that were not in HWE is given in parentheses. Pairwise FST values between populations are given below the diagonal. Population-specific inbreeding coefficients (FIS) and genetic differentiation (FST) are the average of pairwise values for each population (below diagonal). All population-specific FIS and FST values and all pairwise FST values were statistically significant at the 0.05 level of probability. Overall FIS, FIT and FST estimates among the six populations are 0.083 [0.077], 0.185 [0.172], and 0.110 [0.102], respectively. Averaged estimates of parameters for a random sampling of 18 individuals across 10 iterations of sampling are given in brackets.
| S. Sumatra (5) | Zamboanga (4) | Mauritius (7) | Corregidor (1) | Singapore (4) | Cambodia (1) | |
|---|---|---|---|---|---|---|
| N | 98 [18] | 35 [18] | 79 [18] | 34 [18] | 26 [18] | 18 |
| na | 9.5 [6.7] | 6.3 [5.6] | 5.0 [4.2] | 6.6 [6.0] | 6.8 [6.3] | 6.9 [6.9] |
| HO | 0.693 [0.701] | 0.642 [0.639] | 0.540 [0.539] | 0.675 [0.683] | 0.641 [0.639] | 0.725 [0.725] |
| HE | 0.743 [0.743] | 0.700 [0.696] | 0.620 [0.618] | 0.683 [0.686] | 0.745 [0.745] | 0.759 [0.759] |
| Population-specific FIS | 0.066 [0.055] | 0.079 [0.076] | 0.120 [0.126] | 0.016 [0.004] | 0.149 [0.153] | 0.043 [0.043] |
| Population-specific FST | 0.079 [0.082] | 0.105 [0.105] | 0.154 [0.149] | 0.110 [0.109] | 0.089 [0.089] | 0.090 [0.088] |
| Zamboanga | 0.092 [0.099] | |||||
| Mauritius | 0.109 [0.110] | 0.185 [0.176] | ||||
| Corregidor | 0.100 [0.105] | 0.062 [0.059] | 0.181 [0.177] | |||
| Singapore | 0.049 [0.047] | 0.103 [0.105] | 0.136 [0.130] | 0.108 [0.108] | ||
| Cambodia | 0.049 [0.050] | 0.084 [0.086] | 0.160 [0.153] | 0.100 [0.097] | 0.058 [0.057] |
Genepop 3.4 was used to detect pairwise linkage disequilibrium (LD) among the 24 STRs [38] and to determine if the genotypes at each locus segregated independently at the 0.05 level of probability. Unbiased estimates were made through randomization (1,000 iterations) using the Markov-chain method to generate a contingency table representing the random association of genotypes at their observed frequencies at all possible pairs of loci. The same program was used to test the data for deviations from Hardy-Weinberg Equilibrium (HWE) at the 0.05 level of probability.
To determine the extent of genetic variation within and among the populations studied, the number of alleles (na), observed heterozygosity (HO) and gene diversity (HE: heterozygosity expected under equilibrium conditions) for each locus and population were computed using Arlequin [9]. FSTAT 1.2 [12] was used to compute F-statistics (FIS (inbreeding coefficient), FST (population subdivision coefficient) and FIT (the total reduction in observed heterozygosity due to non-random mating) [37]) to test for non-random mating among the populations. The statistical significance of the deviation of per locus and overall FIS values from zero was tested using the FSTAT program, and the 95% Confidence Intervals (CI) for each computation were estimated by bootstrapping over loci. Positive F-statistics imply an excess of homozygosity due to inbreeding (FIS) and/or genetic subdivision (FST). Values of FIT ranging from 0 to 0.15 and from 0.15 to more than 0.25 suggest negligible/moderate and moderate/high levels of homozygosity, respectively. Arlequin was also used to compute pairwise FST [61] and conduct an analysis of molecular variance (AMOVA). In the AMOVA, genetic variation within populations corresponds to the within-population inbreeding coefficient or FIS, variation among populations corresponds to FST and the net effect of both factors corresponds to FIT. The significance of the pairwise FST estimates was tested using a probability distribution constructed from permutation tests (N = 1000) with Bonferroni corrections for multiple comparisons. A population-specific FST, the mean pairwise FST estimate generated by comparing a particular population with all others, was also calculated for each regional population. Values of locus-specific and population-specific FIS were tested for normal distribution and pairwise FST values were tested for linear correlation with values of gene diversity (expected heterozygosity) that are expected under the neutral hypothesis [38].
We used van Oosterhout et al.’s [57] Micro-Checker STR data checking software to evaluate the presence of null alleles at each locus among and within study populations, and we used Wang’s [60] method featured in the SPAGeDi program [14] to assess the degree of pairwise relatedness among the study animals within each study population. To insure that variability in sample sizes of the six population samples, which varied from 18 to 98, did not influence our estimates of the parameters described above, the parameters were also computed based on genotypes of 18 sample members of each population selected with replacement using random number generation and averaged over 10 permutations.
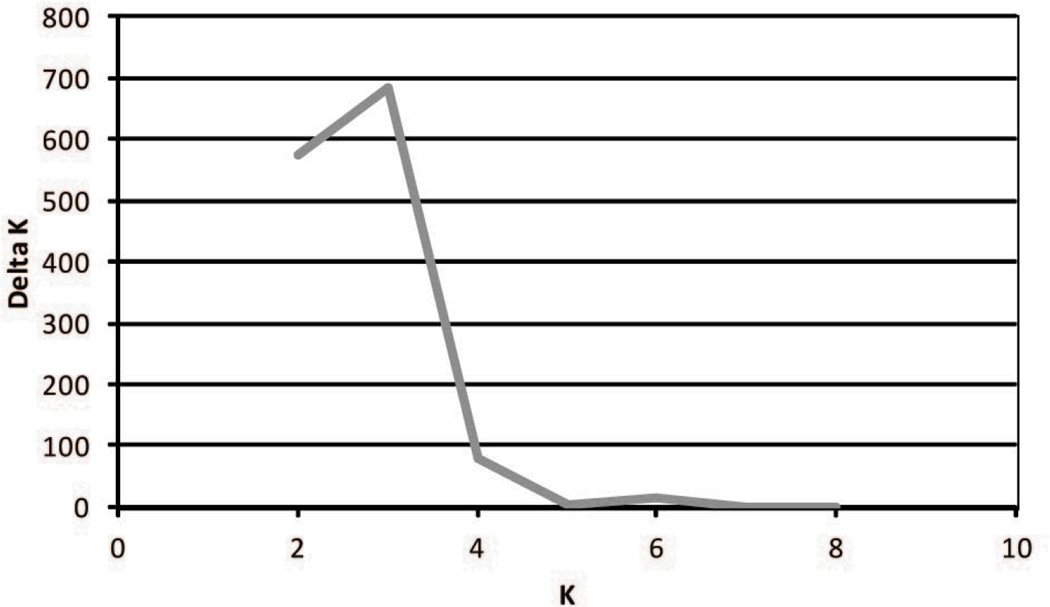
We used the program STRUCTURE 2.3.3 [16, 37] to characterize the population structure among cynomolgus macaque populations derived from six different mainland and insular populations. To determine if the cynomolgus macaques’ nuclear genetic variation at the 24 STRs follows a geographic pattern, the software program utilizes a Markov Chain Monte Carlo (MCMC) method to compute L(K), the posterior probability that the data fits the hypothesis of K geographically/genetically distinct groups and to estimate the fractional membership of each animal in each group. Often the most realistic value of K is established using the maximum value of L(K) that STRUCTURE generates [13]. However, Pritchard and Wen [36] observed that once the “true” value of K is reached, estimates of L(K) for higher values of K plateau because of increasing variance. Thus, we computed ΔK, a measure of second order rate of change of the STRUCTURE likelihood function [L(K)] as described by Evanno et al. [8], who demonstrated that the height of the modal value of ΔK is correlated with the strength of the genetic subdivision among the study populations.
We conducted STRUCTURE runs assuming that between two and eight (2≤K≤8) genetically distinct groups of individuals exist among the study populations. Simulations were performed with 5X105 iterations, after a burn-in period of 105 using prior population information. It was assumed that allele frequencies among populations are correlated, and that despite a priori assignment of an individual to a particular population, there is a high probability that it has ancestors in other populations. The STRUCTURE runs were replicated 10 times with each set of assumptions, to assure that group assignments with the greatest probabilities were detected. We also performed discriminant analysis of principal components (DAPC) using the adegenet 1.3 package for R [18]. The DAPC provides a visual and quantitative method for identifying genetic clusters [19] by partitioning within- and between-group variance and maximizing the latter.
Results
Table 1 presents estimates of allele number (na), and observed (HO) and expected (HE) heterozygosities averaged across study populations for each of the 24 loci analyzed. Up to seven loci departed from HWE at the 0.05 level of probability when individual populations were examined separately (Table 2). All 24 markers were statistically unlinked (p > 0.05) when data from the different populations were pooled, and according to the Micro-Checker program, none of them presented any evidence for null alleles at statistically significant levels (p < 0.05). The average number of STR alleles (na) and the average observed (HO) and expected (HE) proportion of heterozygous genotypes in each population, as well as the estimates (in brackets) standardized for a sample size of 18, are presented in Table 2.
The Sumatran population exhibited the highest average number of alleles per locus (na = 9.5), the Mauritian population the lowest (5.0) while other populations exhibited 6.3–6.9 alleles per locus. Sample size clearly influenced na as the number of alleles in the largest sample, Sumatra (N=98), was especially overestimated compared to its size-adjusted value of na = 6.7. Based on size-adjusted values, Sumatra and Cambodia exhibited the highest allele numbers (6.7 and 6.9, respectively) while Mauritius exhibited the lowest (4.2). A lower average number of alleles was generally accompanied by lower values of HO and HE, but these values were not influenced by sample size, probably because alleles lost by the Mauritian and Philippine populations were those of low frequency. Estimates of HO ranged from 0.54 (Mauritius) to 0.73 (Cambodia) while HE ranged from 0.62 (Mauritius) to 0.76 (Cambodia). The Singapore sample exhibited the greatest discrepancy between HE and HO (0.74 versus 0.64) while the animals from Corregidor exhibited no difference between HE and HO.
Pairwise estimates of coefficients of relationship did not exceed 0.01, confirming that no pair of the animals in any group was closely related, and none of the F-statistics were influenced by variation in sample size by more than 0.01. Contrary to the locus-specific FIS estimates, the variability in FIS estimates among the six populations of cynomolgus macaques did not fit a normal distribution (data not shown) due to an excess of both very high (e.g., Mauritius) and very low (Corregidor) values. However, the negative correlation (r = −0.89) between genetic differentiation (FST) and gene diversity (HE) expected under the neutral hypothesis was observed. All pairwise FST comparisons showed statistically significant differentiation among the six populations (Table 2) at the 0.05 level of probability. Genetic differentiation was lowest (pairwise FST = 0.05–0.06) among Sumatra, Singapore and Cambodia and highest (pairwise FST = 0.14–0.19) between Mauritius and four of the other five populations, except Sumatra, their presumed ancestral homeland [6, 10]. Thus, Mauritius was the most divergent of the six populations despite its recent derivation from Indonesian cynomolgus macaques (pairwise FST = 0.11).
Population-specific FIS estimates for Mauritius and Singapore, the two small island populations, were the highest (FIS = 0.12 and 0.15, respectively), those for Sumatra and Zamboanga were intermediate (0.07–0.08, respectively) while that for Corregidor was the lowest (0.016). Population-specific (average pairwise) FST values ranged from approximately 0.08 (Sumatra) to 0.15 (Mauritius) with the other populations ranging between 0.09 and 0.11, reflecting the lowest and highest average divergence of Sumatra and Mauritius from all other populations, respectively. The corresponding population-specific values of FIT (0.19) suggest a relatively high excess of homozygosity in the small island populations of Mauritius (0.26) and Singapore (0.22) exhibiting the greatest excess and Corregidor (0.12), Cambodia (0.13) and Sumatra (0.14) exhibiting the least excess.
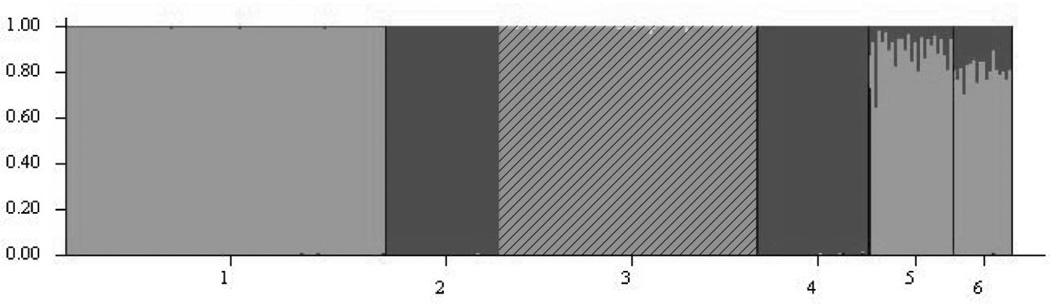
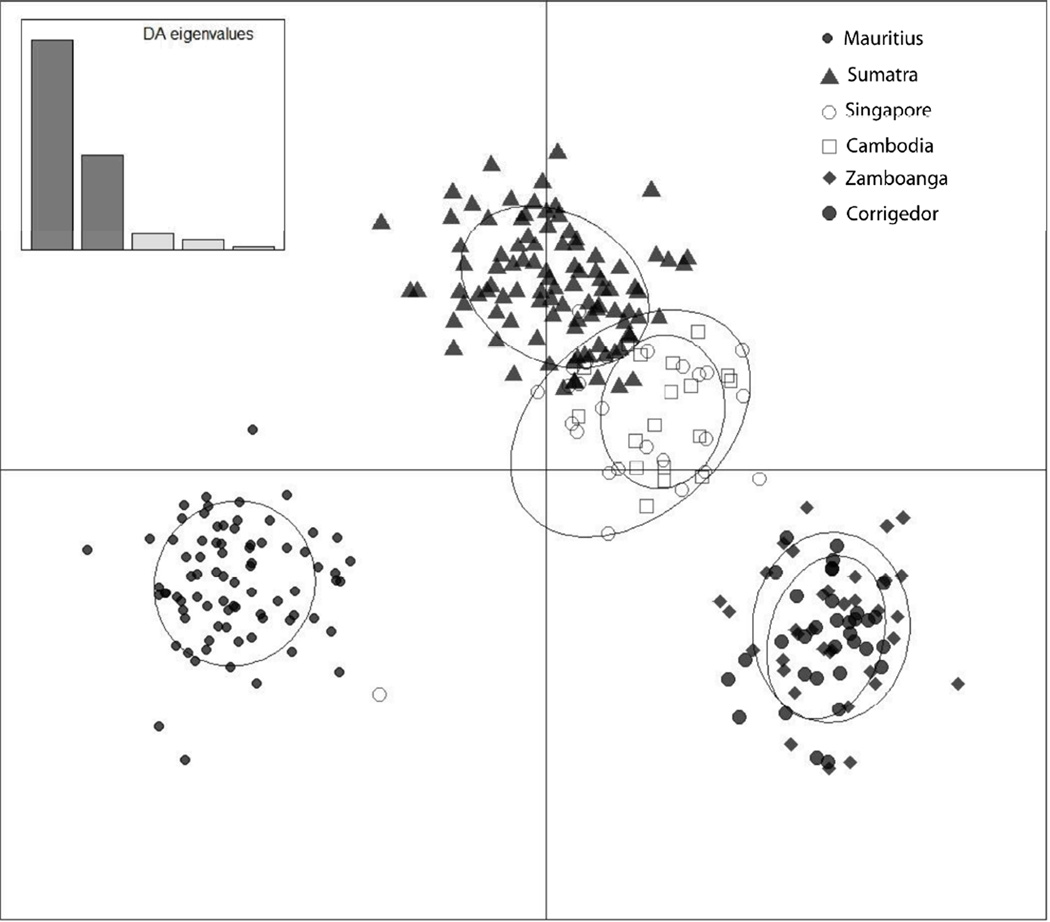
The use of prior population information greatly assisted the Bayesian clustering process of the STRUCTURE program (K = 3; Fig. 2a) and assigned the cynomolgus study populations to inferred structure clusters based on robust posterior probabilities. Examination of Ln P(X|K) values suggested that the highest value of ΔK was at K = 3 (Fig. 2b) indicating that there are three highly differentiated genetic clusters of cynomolgus populations. Posterior assignment probabilities generated by the STRUCTURE analysis maintained the Mauritian group, and assigned all animals in the two Philippine populations, Zamboanga and Corregidor, to the same cluster. The island populations of Singapore and Sumatra clustered with the mainland population, Cambodia. This result was also reflected in the DAPC (Fig. 3) and the pairwise FST values (Table 2). When subjected to discriminant analysis, the DAPC values were found to have discriminant functions producing a high ratio of explained variance (eigenvalues; 90.5%) to the error term, accounted for by the first two principal components and the K value of 3.
Figure 2.
a. Graphical representation of the STRUCTURE analysis for the most likely value of K (K = 3), where each individual is represented by a vertical bar and each shade corresponds to a genetically distinct population cluster. The numbers on the X-axis correspond to a specific regional population of cynomolgous macaques: Sumatra, Indonesia (1), Zamboanga, Mindanao (2), Mauritius (3), Corregidor (4), Singapore (5), Cambodia (6). The Y-axis represents the probability of assignment of an individual to each population cluster.
b. The modal value of Ln P(X|K) identifies the most realistic value of K, the uppermost level of structure that represent only three genetic clusters (i.e., K = 3).
Figure 3.
Results from the DAPC of the samples support the presence of the same three genetic clusters identified in the STRUCTURE analysis.
When all sampling localities were pooled according to their genetically distinct groups with K=3 as determined by the STRUCTURE and DAPC analyses (i.e., Sumatra/Singapore/Vietnam, Zamboanga/Corregidor and Mauritius, Table 3), the Mauritian sample, whose geographic separation from the Indonesian population is only a few hundred years old, emerged as the most genetically isolated and homogenous population of cynomolgus macaques. Of the three clusters, Mauritius exhibited the highest group-specific FIS (0.12), FIT (0.24) and FST (0.14), respectively and highest pairwise FST (0.10–0.17) values. The majority (80%) of variation in the AMOVA analysis for the combined dataset is within each of the three genetically distinct groups (Table 4). The FST value of 0.11 among the three DAPC clusters (when K=3) was identical to that for all six populations (K=6), confirming the relationships implied by the three clusters.
Table 3.
N, na, HO, HE and cluster-specific FIS and FST values for each population cluster in the STRUCTURE analysis. Group-specific FIS and FST values are the average pairwise values for each of the three population clusters, given above the diagonal, and all pairwise FST values among the three clusters are given below the diagonal. All group-specific FIS and FST values and all pairwise FST values are statistically different from zero (p<0.05). FIS, FIT and FST estimates among all six populations are 0.102 [0.104], 0.196 [0.188], and 0.105 [0.094], respectively. The same parameters for 18 randomly selected individuals, averaged over 10 iterations of sampling, from each population (to match the population size of the Cambodian population) are in brackets.
| S. Sumatra/Singapore/Cambodia | Zamboanga/Corregidor | Mauritius | |
|---|---|---|---|
| N | 142 [54] | 69 [36] | 79 [18] |
| na | 11.6 [9.8] | 7.5 [7.0] | 5.0 [4.2] |
| HO | 0.668 [0.689] | 0.658 [0.661] | 0.540 [0.539] |
| HE | 0.764 [0.776] | 0.715 [0.713] | 0.620 [0.618] |
| Group-specific FIS | 0.100 [0.115] | 0.077 [0.070] | 0.120 [0.126] |
| Group-specific FST | 0.088 [0.087] | 0.120 [0.114] | 0.136 [0.133] |
| Zamboanga/Corregidor | 0.072 [0.068] | ||
| Mauritius | 0.104 [0.107] | 0.167 [0.160] |
Table 4.
Analysis of molecular variance (AMOVA) using the combined dataset of study populations and groups.
| Populations (K = 6) | Structure Groups (K = 3) | |||
|---|---|---|---|---|
| Source of Variation | Degrees of Freedom | Variation % | Degrees of Freedom | Variation % |
| Among populations | 5 | 18.47 | 2 | 19.61 |
| Within populations | 289 | 81.53 | 289 | 80.39 |
| Total | 294 | 100 | 291 | 100 |
| FST | 0.110 | 0.105 | ||
Discussion
The uniqueness of Mauritius in the STRUCTURE and DAPC does not reflect the very recent derivation of this population from Indonesia [51, 52], nor does the clustering of Sumatra with the two mainland populations (Cambodia and Singapore) reflect either the fundamental genetic division between insular and mainland cynomolgus macaques that others have reported [e.g., [15, 54] or the hypothesis that the Isthmus of Kra represents a geographic barrier to gene flow [56]. The high genetic diversity and lowest divergence of the Indonesian cymomolgus macaques from all other cynomolgus macaque populations is consistent with an Indonesian, rather than Indochinese, homeland for the species, as concluded by Smith et al [44], but is not consistent with an earlier report based on a smaller subset of the STR loci employed in this study [22]. The difference between our study and others based on mtDNA and Y-chromosomes might result from the lineage sorting effects of these genomes/chromosomes based on their low effective population sizes. The influence of sampling subjects and loci on the results of such studies may also be substantial, as suggested by the difference between the results of the present study and that of many of the same samples based on a smaller subset of loci. The clustering of southern Sumatra with Singapore and Cambodia might reflect the route that cynomolgus macaques followed during dispersal from southern Indonesia to the mainland, since Sumatran and peninsular Malaysian long-tailed macaques are separated by one of the narrowest and shallowest waterways on the Sunda Shelf [58]. When K=4 (not shown), Sumatra was removed from its cluster with Singapore and Cambodia, leaving the latter two populations in their own cluster, as suggested by the DAPC analysis, and underscoring the mainland affiliation with Singapore.
The greatest component of the genetic variance (axis 1, encompassing 62.2% of the variance) separates the Philippine (both Corregidor and Zamboanga) and Mauritian samples from the mainland and shallow water populations, as previously cited. The second greatest component (axis 2, encompassing 28.3% of the variance) does not distinguish the Mauritian and Philippine samples, but rather illustrates differentiation of the mainland populations, Cambodia and Singapore from both Sumatra and the Mauritius-Philippines group. Thus, the first and second DAPC axes correspond with the K=3 clusters emerging from the STRUCTURE analysis. Samples from all mainland populations (those that are contiguous or separated by shallow water) are genetically equidistant from the Mauritius and Philippines populations. The Philippine and Mauritian populations are most divergent from the other populations of cynomolgus macaques because animals in the Philippines, an island that is separated from the Sunda Shelf by deep water, and those from Mauritius, an island off the coast of Africa, have become significantly genetically homogeneous and distinct from the other populations. The remarkable divergence of Mauritian cynomolgus macaques from those from Indonesia in fewer than 500 years underscores the speed with which genetic divergence can occur in an isolated population with a short generation length [43] expanding rapidly from a very small number of founders. To a lesser extent, evolutionary forces have led to genetic differentiation among all insular populations of cynomolgus macaques while those on the mainland remain relatively undifferentiated [42].
Patterns of pairwise FST estimates suggest that population differentiation occurred at varying rates not proportional to their corresponding geographic distances and probably accounts for the lack of a normal distribution of variability in FIS estimates. The increased divergence among and genetic homogeneity within isolated populations of limited size reflects the Wahlund effect [59]. The higher estimates of HE compared to those of HO stem from the relatively high F statistics (higher estimates of FIS and FST are often correlated with lower observed heterozygosity) for populations of a single species [34] and might suggest further subdivision within at least some of the populations studied. Although the individual population clustering patterns created by STRUCTURE, DAPC, and pairwise FST revealed three genetically differentiated groups; the Zamboanga-Corregidor, the Indonesia-Singapore-Cambodia and the Mauritius clusters, estimates of FIS calculated in this study are 9 to 18 times greater than those among rhesus macaques used for biomedical research at the California NPRC. Since none of the 24 loci analyzed showed significant and systematic effects from null alleles across populations, the high degree of allele sharing and excess homozygosity among animals in this study probably resulted from many generations of genetic isolation and random inbreeding. The FIS values were higher for small island populations (Mauritius and Singapore) than for the larger island population, the mainland population of Cambodia or the potentially artificially heterogeneous population of Corregidor. The lower estimates of FIS for the Corregidor population might reflect a wide breadth of sampling from the island of Mindanao or even from other islands of the Philippines notwithstanding its close clustering with the Zamboanga population. In the smaller populations of Mauritius, Zamboanga and, perhaps Singapore, founder effects, small effective population size and genetic drift probably played a significant role in shaping these populations’ genetic structure. The especially low level of genetic heterogeneity of Mauritian cynomolgus macaques reported for both mtDNA [2, 25, 29], STRs [22] and MHC alleles [63] is confirmed by the present study.
The Philippine population may represent the remnant of ancient dispersals from the Sunda Shelf that is presently restricted to the outer limits of the cynomolgus macaques’ natural geographic distribution. The modest level of genetic differentiation between cynomolgus macaques from the islands of Corregidor and Zamboanga (FST = 0.06) and those from Cambodia and Singapore (FST = 0.06) compared to other pairs of Southeast Asian populations corresponds with their close geographic proximity. Because the animals from Corregidor and Zamboanga are equally differentiated from all other cynomolgus macaque populations, we conclude that both populations represent the subspecies M. f. fascicularis and that the former were collected predominantly from the western part of Mindanao.
While the level of divergence of the Mauritian population from their Indonesian relatives was unexpected, the former resembled the latter more closely than any other population. The pairwise FST value of 0.11 implies a closer genetic relationship between Mauritian and South Sumatran animals than reported by Kanthaswamy et al. [22]. In that study, estimates of genetic differences between these populations (0.14) were slightly greater than in the present study. The use of more STRs than in previous studies also led to better detection of genetic similarity between the Mauritian and Indonesian cynomolgus populations, but the value is still unexpectedly high and underscores how strong stochastic processes can mask phylogenetic relationships. Cynomolgus macaques purportedly arrived in Mauritius from Indonesia as shipboard companions of sailors in the seventeenth or eighteenth centuries [51, 52] and are thought to have all originated from a small founder population originating in Java or Sumatra. The drastic founder effect and subsequent rapid expansion in their population size [3] undoubtedly resulted in the loss of alleles, especially rarer ones, high levels of inbreeding, and a unique and genetically homogeneous set of genetic traits, including remarkably low levels of genetic variation at the MHC loci [28, 63]. Founder effects, isolation, inbreeding and drift must have been exceedingly profound to so obscure the recent derivation of these animals from their Indonesian ancestors.
Kanthaswamy et al. [21, 22] showed that the rhesus and cynomolgus macaques from Indochina exhibited the greatest genetic diversity based on STRs and SNPs, respectively, perhaps reflecting the natural zone of inter-species hybridization in Indochina. This is consistent with an introgression of rhesus alleles that is both unique to cynomolgus macaques in Indochina [22, 41, 43, 54] and extends much broader than suggested by the current zone of hybridization described by Tosi et al. [56]. Moreover, a much more extensive overlap of troops of rhesus and cynomolgus macaque in Indochina, including Vietnam [32], Cambodia [53], Laos [26] and Thailand [17] than previously suspected has also been reported.
Our observations of a lack of close consanguinity of cynomolgus macaques in each population is in agreement with Melnick’s [31] conclusion that cynomolgus macaques generally avoid mating among close relatives. However, our estimates of FIS were much higher than his, a conflict that may have stemmed from the different sampling approaches employed by the two studies. While Melnick [31] focused on social groups in the wild, our sampling scheme was based on samples obtained opportunistically from animals trapped for biomedical research.
Because the effect of inbreeding was not considered in STRUCTURE analyses reported elsewhere [20, 21], the presence of remotely related individuals in the sample-set from Singapore and/or Mauritius, suggested by high values of FIS, could have led to an overestimation of the real value of K [36], further compounding the effects from the correlated allele frequency model. However, the overall FST values among all six populations exceeded 0.11, suggesting that if anything, K is more likely to have been under-estimated than over-estimated in this study.
Our results suggest that animals from different source populations vary substantially and should not be included in the same experiments as models for heritable human diseases because they may not be ideal for valid comparisons. Combining information on quantitative risk factors for disease from different populations of cynomolgus macaques could obscure risk factor-disease associations or create spurious or artificial associations that are biologically irrelevant [50].
Though Indonesia and the Philippines consist of thousands of islands, little attention has been given to the island of origin of cynomolgus macaques chosen as models for research. The potential effects of the distribution of genetic diversity of cynomolgus macaques within and among islands should be adequately described in future studies. Since many cynomolgus macaques have been imported to the US from breeding farms in China that mix and breed cynomolgus macaques originating in Indochina, Indonesia and elsewhere, both intra-specific and inter-specific hybrid animals might persist in the domestic supply of cynomolgus macaques in the US [21]. This should be considered by investigators when selecting animals as subjects in biomedical research. To maximize resolution of experimental treatment effects, animals of unmixed ancestry with paired coefficients of relationship below that of first cousins (r<0.125) should be employed in biomedical research. Relatively inexpensive genetic testing can ensure that these conditions are met. By requiring accurate information on their animals’ origin, ancestry and pedigree, investigators colony managers can 1) prevent admixture among animals from different geographic origins that can inflate genetic variability among experimental subjects, and 2) prevent mating among related animals that can increase inbreeding coefficients among their breeding stock.
Acknowledgement
The authors are grateful to our former and current staff at the Molecular Anthropology Laboratory, Department of Anthropology, UC Davis for their contributions to this study. We wish to thank the various facilities listed in Smith et al. [45, 47], Satkoski et al. [40] and Kanthaswamy et al. [20, 21] that contributed samples used in this study. This study was supported by a National Institutes of Health grant (No. RR05090 awarded to DGS).
References
- 1.Bird MI, Pang WC, Lambeck K. The age and origin of the Straits of Singapore. Palaeogeography, Palaeoclimatology, Palaeoecology. 2006;241:531–538. [Google Scholar]
- 2.Blancher A, Bonhomme M, Crouau-Roy B, Terao K, Kitano T, Saitou N. Mitochondrial DNA Sequence Phylogeny of 4 Populations of the Widely Distributed Cynomolgus Macaque (Macaca fascicularis fascicularis) Journal of Heredity. 2008;99:254–264. doi: 10.1093/jhered/esn003. [DOI] [PubMed] [Google Scholar]
- 3.Bonhomme M, Blancher A, Cuartero S, Chikhi L, Crouau-Roy B. Origin and number of founders in an introduced insular primate: estimation from nuclear genetic data. Molecular Ecology. 2008;17:1009–1019. doi: 10.1111/j.1365-294X.2007.03645.x. [DOI] [PubMed] [Google Scholar]
- 4.Bonhomme M, Cuartero S, Blancher A, Crouau-roy B. Assessing Natural Introgression in 2 Biomedical Model Species, the Rhesus Macaque (Macaca mulatta) and the Long-Tailed Macaque (Macaca fascicularis) Journal of Heredity. 2009;100:158–169. doi: 10.1093/jhered/esn093. [DOI] [PubMed] [Google Scholar]
- 5.Bowden D, Smith O. Conservationally sound assurance of primate supply and diversity. ILAR Journal. 1992;34:53–55. [Google Scholar]
- 6.Delson E. Fossil macaques, phyletic relationships and a scenario of deployment. New York: Van Nostrand-Reinhold; 1980. [Google Scholar]
- 7.Ellis S, Bontrop R, Antczak D, Ballingall K, Davies C, Kaufman J, Kennedy L, Robinson J, Smith D, Stear M, Stet RM, Waller M, Walter L, Marsh SE. ISAG/IUIS-VIC Comparative MHC Nomenclature Committee report, 2005. Immunogenetics. 2006;57:953–958. doi: 10.1007/s00251-005-0071-4. [DOI] [PubMed] [Google Scholar]
- 8.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 9.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary Bioinformatics. 2007;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 10.Fooden J. Comparative Review of Fascicularis-group Species of Macaques (primates:Macaca) Fieldiana Zoology. 2006:1–43. [Google Scholar]
- 11.Fooden J. Systematic review of Southeast Asian longtail macaques, Macaca fascicularis (Raffles 1821) Fieldiana Zoology. 1995;81:1–206. [Google Scholar]
- 12.Goudet J. FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- 13.Hampton JO, Spencer PBS, Alpers DL, Twigg LE, Woolnough AP, Doust J, Higgs T, Pluske J. Molecular techniques, wildlife management and the importance of genetic population structure and dispersal: a case study with feral pigs. Journal of Applied Ecology. 2004;41:735–743. [Google Scholar]
- 14.Hardy OJ, Vekemans X. spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2:618–620. [Google Scholar]
- 15.Harihara S, Saitou N, Hirai M, Aoto N, Terao K, Cho F, Honjo S, Omoto K. Differentiation of mitochondrial DNA types in Macaca fascicularis. Primates. 1988;29:117–127. [Google Scholar]
- 16.Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jadejaroen J, Malaivijitnond S, Hamada Y. Genetic survey of hybrids in a semi-wild population of longtailed (Macaca fascicularis) and rhesus macaques (Macaca mulatta): Human ABO Blood Groups; 23rd Congress of the International Primatological Society; 2010. [Google Scholar]
- 18.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 19.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanthaswamy S, Capitanio JP, Dubay CJ, Ferguson B, Folks T, Ha JC, Hotchkiss CE, Johnson ZP, Katze MG, Kean LS, Michael Kubisch H, Lank S, Lyons LA, Miller GM, Nylander J, O’Connor DH, Palermo RE, Smith DG, Vallender EJ, Wiseman RW, Rogers J. Resources for genetic management and genomics research on non-human primates at the National Primate Research Centers (NPRCs) Journal of Medical Primatology. 2009;38:17–23. doi: 10.1111/j.1600-0684.2009.00371.x. [DOI] [PubMed] [Google Scholar]
- 21.Kanthaswamy S, Kou A, Smith DG. Population Genetic Statistics from Rhesus Macaques (Macaca mulatta) in Three Different Housing Configurations at the California National Primate Research Center. Journal of the American Association for Laboratory Animal Science. 2010;49:598–609. [PMC free article] [PubMed] [Google Scholar]
- 22.Kanthaswamy S, Satkoski J, George D, Kou A, Erickson B, Smith D. Hybridization and Stratification of Nuclear Genetic Variation in Macaca mulatta and M. fascicularis. International Journal of Primatology. 2008;29:1295–1311. doi: 10.1007/s10764-008-9295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanthaswamy S, Satkoski J, Kou A, Malladi V, Glenn Smith D. Detecting signatures of inter-regional and inter-specific hybridization among the Chinese rhesus macaque specific pathogen-free (SPF) population using single nucleotide polymorphic (SNP) markers. Journal of Medical Primatology. 2010;39:252–265. doi: 10.1111/j.1600-0684.2010.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanthaswamy S, Smith DG. Use of microsatellite polymorphisms for paternity exclusion in Rhesus macaques (Macaca mulatta) Primates. 1998;39:135–145. [Google Scholar]
- 25.Kawamoto Y, Kawamoto S, Matsubayashi K, Nozawa K, Watanabe T, Stanley MA, Perwitasari-Farajallah D. Genetic diversity of longtail macaques (Macaca fascicularis) on the island of Mauritius: an assessment of nuclear and mitochondrial DNA polymorphisms. Journal of Medical Primatology. 2008;37:45–54. doi: 10.1111/j.1600-0684.2007.00225.x. [DOI] [PubMed] [Google Scholar]
- 26.Kingsada P, Pathomthong S, Praxaysombath B, Malaivijitnond S, Hamada Y. Distribution pattern of macaque species in the southern Laos PDR: Examining the ecological segregation among macaque species; 23rd Congress of the International Primatological Society; 2010. [Google Scholar]
- 27.Kondo M, Kawamoto Y, Nozawa K, Matsubayashi K, Watanabe T, Griffiths O, Stanley M-A. Population genetics of crab-eating macaques (Macaca fascicularis) on the island of mauritius. American Journal of Primatology. 1993;29:167–182. doi: 10.1002/ajp.1350290303. [DOI] [PubMed] [Google Scholar]
- 28.Krebs KC, Jin Z, Rudersdorf R, Hughes AL, O’Connor DH. Unusually High Frequency MHC Class I Alleles in Mauritian Origin Cynomolgus Macaques. The Journal of Immunology. 2005;175:5230–5239. doi: 10.4049/jimmunol.175.8.5230. [DOI] [PubMed] [Google Scholar]
- 29.Lawler SH, Sussman RW, Taylor LL. Mitochondrial DNA of the Mauritian macaques (Macaca fascicularis): An example of the founder effect. American Journal of Physical Anthropology. 1995;96:133–141. doi: 10.1002/ajpa.1330960203. [DOI] [PubMed] [Google Scholar]
- 30.Li DY, Xu HL, Smith DG, Cheng AC, Trask JS, Zhu Q, Yao YF, Du DD, Ni QY. Phylogenetic analysis of chinese rhesus macaques (Macaca mulatta) based on mitochondrial control region sequences. American Journal of Primatology. 2011;73:883–895. doi: 10.1002/ajp.20956. [DOI] [PubMed] [Google Scholar]
- 31.Melnick DJ. The genetic consequences of primate social organization: a review of macaques, baboons and vervet monkeys. Genetica. 1987;73:117–135. doi: 10.1007/BF00057443. [DOI] [PubMed] [Google Scholar]
- 32.Minh NV, Van NH, Hamada Y. Distribution and present status of non-human primates in central Vietnam; 23rd Congress of the International Primatological Society; 2010. [Google Scholar]
- 33.Morin PA, Kanthaswamy S, Smith DG. Simple sequence repeat (SSR) polymorphisms for colony management and population genetics in rhesus macaques (Macaca mulatta) American Journal of Primatology. 1997;42:199–213. doi: 10.1002/(SICI)1098-2345(1997)42:3<199::AID-AJP3>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 34.Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- 35.Pavlin BI, Schloegel LM, Daszak P. Risk of importing zoonotic diseases through wildlife trade, United States. Emerging Infectious Diseases. 2009 doi: 10.3201/eid1511.090467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pritchard J, Wen W. Documentation for STRUCTURE software: version 2. Chicago: Department of Human Genetics, University of Chicago; 2003. [Google Scholar]
- 37.Pritchard JK, Stephens M, Donnelly P. Inference of Population Structure Using Multilocus Genotype Data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raymond M, Rousset F. GENEPOP (Version 1.2): Population Genetics Software for Exact Tests and Ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- 39.Research Resources Information Center (RRIC) Demands for rhesus macaques in biomedical research: a workshop report. Institute of Laboratory Animal Research (ILAR) Journal. 2003;44:222–238. [Google Scholar]
- 40.Satkoski J, George D, Smith DG, Kanthaswamy S. Genetic characterization of wild and captive rhesus macaques in China. Journal of Medical Primatology. 2008;37:67–80. doi: 10.1111/j.1600-0684.2007.00228.x. [DOI] [PubMed] [Google Scholar]
- 41.Satkoski Trask JA, Garnica WT, Smith DG, Houghton P, Lerche N, Kanthaswamy S. Single-Nucleotide Polymorphisms Reveal Patterns of Allele Sharing Across the Species Boundary Between Rhesus (Macaca mulatta) and Cynomolgus (M. fascicularis) Macaques. American Journal of Primatology. 2013;75:135–144. doi: 10.1002/ajp.22091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiina T, Tanaka K, Katsuyama Y, Otabe K, Sakamoto K, Kurata M, Nomura M, Yamanaka H, Nakagawa H, Inoko H, Ota M. Mitochondrial DNA Diversity among Three Subpopulations of Cynomolgus Macaques (<i>Macaca fascicularis</i>) Originating from the Indochinese Region. Experimental Animals. 2010;59:567–578. doi: 10.1538/expanim.59.567. [DOI] [PubMed] [Google Scholar]
- 43.Smith D. Potential for cumulative inbreeding and its effects upon survival in captive groups of non-human primates. Primates. 1980;21:430–436. [Google Scholar]
- 44.Smith D, Trask J, George D, Kanthaswamy S, Houghton P. Rhesus macaque (Macaca mulatta) ancestry in Indochinese cynomolgus macaques (Macaca fascicularis) ILAR Journal. 2013 (in press) [Google Scholar]
- 45.Smith DG, George D, Kanthaswamy S, McDonough J. Identification of country of origin and admixture between Indian and Chinese rhesus macaques. International Journal of Primatology. 2006 [Google Scholar]
- 46.Smith DG, Kanthaswamy S, Viray J, Cody L. Additional highly polymorphic microsatellite (STR) loci for estimating kinship in rhesus macaques (Macaca mulatta) American Journal of Primatology. 2000;50:1–7. doi: 10.1002/(SICI)1098-2345(200001)50:1<1::AID-AJP1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 47.Smith DG, McDonough JW, George DA. Mitochondrial DNA variation within and among regional populations of longtail macaques (Macaca fascicularis) in relation to other species of the fascicularis group of macaques. American Journal of Primatology. 2007;69:182–198. doi: 10.1002/ajp.20337. [DOI] [PubMed] [Google Scholar]
- 48.Stevison LS, Kohn MH. Divergence population genetic analysis of hybridization between rhesus and cynomolgus macaques. Molecular Ecology. 2009;18:2457–2475. doi: 10.1111/j.1365-294X.2009.04212.x. [DOI] [PubMed] [Google Scholar]
- 49.Street S, Kyes R, Grant R, Ferguson B. Single nucleotide polymorphisms (SNPs) are highly conserved in rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) macaques. BMC Genomics. 2007;8:480. doi: 10.1186/1471-2164-8-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sturt E. Analysis of linkage and association for diseases of genetic aetiology. Statistics in Medicine. 1984;3:57–72. doi: 10.1002/sim.4780030108. [DOI] [PubMed] [Google Scholar]
- 51.Sussman R, Tattersall I. Behavior and ecology of…lt;i…gt;macaca fascicularis in Mauritius: A preliminary study. Primates. 1981;22:192–205. [Google Scholar]
- 52.Sussman RW, Tattersall I. Distribution, abundance, and putative ecological strategy of Macaca fascicularis on the island of Mauritius, southwestern Indian Ocean. Folia Primatologica. 1986;46:28–43. [Google Scholar]
- 53.Thao S, Meas S, Oi T. Recorded species of non-human primates in Cambodia and their present status; 23rd Congress of the International Primatological Society; 2010. [Google Scholar]
- 54.Tosi AJ, Coke CS. Comparative phylogenetics offer new insights into the biogeographic history of Macaca fascicularis and the origin of the Mauritian macaques. Molecular Phylogenetics and Evolution. 2007;42:498–504. doi: 10.1016/j.ympev.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Tosi AJ, Morales JC, Melnick DJ. Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution. 2003;57:1419–1435. doi: 10.1111/j.0014-3820.2003.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 56.Tosi AJ, Morales JC, Melnick DJ. Y-Chromosome and Mitochondrial Markers in Macaca fascicularis Indicate Introgression with Indochinese M. mulatta and a Biogeographic Barrier in the Isthmus of Kra. International Journal of Primatology. 2002;23:161–178. [Google Scholar]
- 57.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- 58.Voris HK. Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. Journal of Biogeography. 2000;27:1153–1167. [Google Scholar]
- 59.Wahlund S. ZUSAMMENSETZUNG VON POPULATIONEN UND KORRELATIONSERSCHEINUNGEN VOM STANDPUNKT DER VERERBUNGSLEHRE AUS BETRACHTET. Hereditas. 1928;11:65–106. [Google Scholar]
- 60.Wang J. An Estimator for Pairwise Relatedness Using Molecular Markers. Genetics. 2002;160:1203–1215. doi: 10.1093/genetics/160.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weir BS, Cockerham CC. Estimating F-Statistics for the Analysis of Population Structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 62.Williams-Blangero S, Vandeberg JL, Blangero J, Konigsberg L, Dyke B. Genetic differentiation between baboon subspecies: Relevance for biomedical research. American Journal of Primatology. 1990;20:67–81. doi: 10.1002/ajp.1350200202. [DOI] [PubMed] [Google Scholar]
- 63.Wiseman RW, Wojcechowskyj JA, Greene JM, Blasky AJ, Gopon T, Soma T, Friedrich TC, O'Connor SL, O'Connor DH. Simian Immunodeficiency Virus SIVmac239 Infection of Major Histocompatibility Complex-Identical Cynomolgus Macaques from Mauritius. Journal of Virology. 2007;81:349–361. doi: 10.1128/JVI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yao YT, Harff J, Meyer M, Zhan WH. Reconstruction of the paleocoastlines for the northwestern South China Sea since the last glacial maximum. Science in China, Series D-Earth Sciences. 2009;52:1127–1136. [Google Scholar]