Abstract
The forkhead box transcription factor FOXM1 is considered to be a promising target for cancer therapy. However, the significance of FOXM1 in tumors harboring mutation in p53, which is very common, is unclear. In this study, we investigated the efficacy of FoxM1-targeting in spontaneous p53-null tumors using genetic ablation as well as using a peptide-inhibitor of FOXM1. We show that conditional deletion of FoxM1 inhibits growth of the p53 null thymic lymphoma and sarcoma cells. In addition, deletion of FoxM1 induces apoptotic cell death of the p53 null tumors, accompanied by reduced expression of the FOXM1 target genes Survivin and Bmi1. An ARF-derived peptide that inhibits the activity of FOXM1, by targeting it to the nucleolus, also induces apoptosis in the p53 null sarcoma and lymphoma, leading to a strong inhibition of their metastatic colonization. Together, our observations suggest that FOXM1 is critical for survival and growth of the p53-null lymphoma and sarcoma, and provide proof-of-principle that FOXM1 is an effective therapeutic target for sarcoma and lymphoma carrying loss of function mutation in p53.
Introduction
The tumor suppressor p53, encoded by the TP53 gene, is a short-lived transcription factor involved in a wide range of cellular processes that are critical for tumor suppression (1-3). Though p53 is expressed at a low level in normal cells, it serves as a protective barrier against development of many types of cancers mainly through preventing proliferation of the incipient cancer cells, induction of apoptosis, as well as through its role in the maintenance of genome integrity (3). Mice deficient in p53 develop spontaneous tumors including thymic lymphoma and sarcoma (3, 4). The essential role of p53 as a tumor suppressor is further manifested by the fact that the p53 gene is mutated in approximately half of the human cancers (1). Given the high prevalence of p53 inactivation in human cancers, it is important to validate therapeutic strategies targeting cancer cells with loss of p53 function.
The forkhead box transcriptional factor FOXM1 is aberrantly overexpressed in many types of human malignancies including liver, prostate, breast, colon, neural tissue and others (5-8). FOXM1 facilitates development of cancers in several ways. First, it transcriptionally activates genes involved in cell-proliferation, and promotes progression through G1-S and G2-M phases of the cell cycle (9, 10). It stimulates expression of DNA repair genes, ensuring the chromosome stability (11, 12). In addition, FoxM1 has been implicated in alleviating oxidative stress in tumor cells by activating ROS scavenger proteins (13) and mediating resistance (14, 15). A recent study in a mouse hepatocellular carcinoma (HCC) model demonstrated that FOXM1 also functions as a potent activator of tumor metastasis through promoting the epithelial to mesenchymal transition (EMT), increased motility of the tumor cells, and establishment of the pre-metastatic niches in the distal target organ (16, 17). Two recent studies in neuroblastoma and embryonic carcinoma cells indicated a role of FOXM1 in the maintenance of the undifferentiated state of the tumor cells by activating pluripotency-associated genes (8, 18).
Given the multifaceted functions of FOXM1 in tumor progression, targeting FOXM1 represents a rational and promising anti-cancer therapeutic strategy. This is further supported by the fact that FOXM1 is a proliferative-specific transcriptional factor whose expression is unique to the proliferating cells (19, 20). Several strategies have been developed to target FoxM1 in cancer cells. Based on the fact that FoxM1 is an inhibitory target of mouse ARF tumor suppressor, a cell penetrating ARF 26-44 peptide which consists of 9 N-terminal D-arginine (D-Arg) residues and amino acid residues 26-44 of the mouse ARF protein was synthesized (6). The ARF 26-44 peptide, which inhibits FOXM1 by sequestering it to the nucleolus, is effective in diminishing tumor size in HCC by reducing tumor cell proliferation and inducing apoptosis (21). That ARF peptide also effectively prevents pulmonary metastasis of HCC cells (16). In addition, thiazole antibiotics have been shown to down-regulate FOXM1 and induce apoptosis in various cancer cells (22, 23).
In this study, we demonstrate that FOXM1 is critical for survival and growth of p53−/− tumor cells both in vitro and in vivo. The ARF 26-44 peptide, which inhibits the activity of FOXM1, induces apoptosis in p53 null tumors. These observations validate the therapeutic strategy of targeting FOXM1 in tumors with p53 loss of function.
Materials and Methods
Animals
The CreERT2 strain (Strain 01XAB) was obtained from Tyler Jacks’ laboratory (Massachusetts Institute of Technology, USA). Foxm1b fl/fl strain was generated as previously described(24). The C57Bl/6 p53 +/− strain was obtained from the Jackson Laboratories (Bar Harbor, ME). The triple transgenic CreERT2, Foxm1b fl/fl, p53−/− mice were generated by mating the three individual strains. NU/NU nude mice were obtained from Charles River Laboratories (Wilmington, MA). ICR SCID mice were obtained from Taconic Farms (Germantown, N.Y.)
Establishment of p53 null thymic lymphoma and sarcoma cell lines
Thymic lymphoma tissue was isolated from the thymus of mice and sarcoma was isolated from a tumor encompassing the rear leg of the mouse. Tumors were excised, minced and enzymatically dissociated with 0.25% trypsin or papain (10u/ml). Cells were then washed and replaced with fresh media. Thymic lymphoma cells grew in suspension and sarcoma cells were adherent and they were maintained in DMEM medium supplemented with 10% fetal bovine, L-glutamine and penicillin-streptomycin. No established cell lines were used in this study.
Antibodies and immunoblots
The following antibodies were also used: FoxM1 (Santa Cruz: sc-500), Survivin (Novus Biologicals: NB500-201), α-tubulin (Sigma:T6074), Cleaved-PARP (Asp214) (Cell Signaling : #9544), Bmi1 (Cell Signaling; #5856), Cleaved Caspase-3(Asp175) (Cell Signaling #9661). Horseradish peroxidase-conjugated secondary antibodies were used to amplify the signal from primary antibody (Bio-rad). Protein lysates were prepared in NP-40 lysis buffer consisted of 1% NP-40, 5% glycerol, 20nM β-glycerophosphate, 2mM NaF, 5mM EDTA, 5mM EGTA and freshly added protease inhibitor cocktail (Roche).
Cell viability assay
Cells were counted and seeded at a density of 2×103 cells per well in triplicate in 48-well plate (Corning). The growth of the cell was monitored by measuring the luminescent signal using the CellTiter-Glo kit (Promega) every other day following manufacture’s protocol.
Soft agar assays and foci formation assay
For soft agar assay, cells were counted and plated in six-well plates in 0.35% agarose on a 0.7% agarose bed in triplicate for 2 weeks. For foci formation assay, one thousand cells were plated in six-well plates for 2 weeks. In both assays, colonies were stained with crystal violet and counted after 3 weeks. Pictures were taken under dissecting microscope.
Allograft assay and intravenous tail vein injection
Cells were counted and suspended in cold PBS. For allograft model, 1×106 cells were injected subcutaneously into rear flank of the nude mice. After palpable tumor formation, mice were randomized into two groups. Either corn oil or tamoxifen (1mg/per injection) were injected into the nude mice intraperitoneally every other day. Tumor sizes were measured with a caliper and calculated by length*height*width*0.5. For tail vein injection, cells were stably transduced with pFU-L2G luciferase construct obtained from Sanjiv Sam Gambhir (M.D., Ph.D) of Stanford University and optimized by Dr. Huiping Liu (25). This construct enables the expression of both the bioluminescence and green fluorescence protein. eGFP positive cells were sorted by Beckman Coulter MoFlo. 3×106 cells were suspended in cold PBS and injected through tail vein. Live animal imaging was done on the IVIS Spectrum optical imaging machine (Caliper Life Sciences, Alameda, CA).
Peptide treatment
Both wild type ARF 26-44 (rrrrrrrrrKFVRSRRPRTASCALAFVN) and mutant ARF 37-44 rrrrrrrrrSCALAFVN peptides were synthesized by Genemed Synthesis Inc. (San Antonio ,Texas). The N-terminus of each peptide was modified with nine D-Arg(r) residues. The peptides were also blocked with amidation at the C terminus and acetylation at the N terminus. For sarcoma cells, mice were treated with 5mg/kg body weight of peptide every other day for 10 times. For lymphoma cells, mice were treated with 2.5mg/kg body weight of peptide every other day for 10 times.
Statistical analysis
Statistical significance was calculated by the Student’s t test (two tailed) with GraphPad Prism software, Microsoft Excel and R. Statistically significant changes were indicated with asterisks (* p<0.05, ** p<0.01).
Results
p53 null thymic lymphoma and sarcoma cells are addicted to FoxM1 for survival
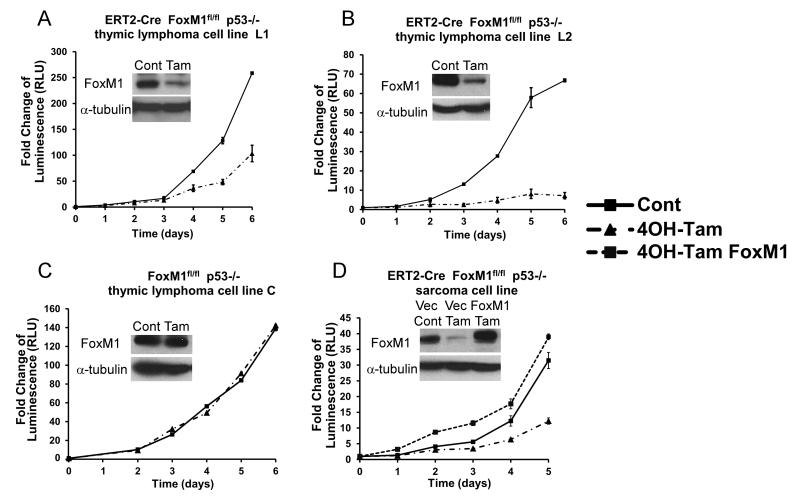
FoxM1 is a p53-regulated gene (26, 27). Our database analyses indicated that FoxM1-mRNA is upregulated in cancers harboring mutations in p53 (Supplementary Fig.1)(28-31). In this study, we analyzed the role of FoxM1 in p53 loss-of-function tumors. To investigate this, we generated a strain of triple transgenic mice harboring CreERT2, Foxm1 fl/fl and p53 −/− alleles by crossing the three individual strains. Mice developed a spectrum of spontaneous tumors, as expected from the p53 null background (4). The presence of CreERT2 allele in the triple transgenic strain permits Cre recombinase expression upon 4-OH tamoxifen treatment to excise flox flanked Foxm1 alleles and thus silencing FoxM1 expression. However, our attempts to study the effects of Foxm1 deletion on endogenous lymphomas/sarcomas were inconclusive mainly because the lymphomas/sarcomas developed at different times in the cohorts of mice used in the study. Also, since the Foxm1 alleles are deleted in most cell types in this system, it would be difficult to avoid the effects of Foxm1-deletion in the other cell types on the lymphoma/sarcoma development and progression. Therefore, we decided to isolate lymphoma/sarcoma cells from the triple transgenic and analyze them in host mice. Two thymic lymphoma (L1 and L2) and a sarcoma (S) triple transgenic cell lines were generated from the endogenous tumors. In addition, a control thymic lymphoma line (C) isolated from Foxm1 fl/fl p53−/− tumor was established in parallel. We tested the deletion efficiency of FoxM1 by immunoblot and confirmed that FoxM1 expression was significantly reduced in triple transgenic lines L1, L2 and S but not in control line C upon treatments with 4-OH tamoxifen (Fig.1A-D). A sarcoma line stably transduced with exogenous FoxM1 expression was generated. Treatments with 4-OH tamoxifen did not diminish the exogenous FoxM1 expression (Fig.1D).
Figure 1. FoxM1 is critical for the survival and tumorigenicity of p53 null thymic lymphoma and sarcoma.
A-C, CreERT2, Foxm1 fl/fl and p53 −/− thymic lymphoma (represented by “L1” and “L2”) and Foxm1 fl/fl and p53 −/− thymic lymphoma (represented by “C”) were treated with ethanol as vehicle or 800nM of 4OH-tamoxifen (Tam). D, CreERT2, Foxm1 fl/fl and p53 −/− sarcoma (represented by “S”) was treated with ethanol as vehicle or 800nM of 4OH-tamoxifen (Tam). Sarcoma line stably transduced with FoxM1 expression was constructed (S: FoxM1) and treated with 800nM of 4OH-tamoxifen. Cell viability was measured by proportional luminescence signal generated by celltiter-glo assay.
To examine the effect of FoxM1 ablation, growth curves were plotted following 4-OH tamoxifen treatment. FoxM1 deletion led to a profound decrease in the cell viability starting from early time point in all three of the triple transgenic lines L1, L2 and S (Fig.1A,B and D). The control lymphoma cell line C (Fig.1C) as well as the sarcoma cells stably expressing the exogenous FoxM1 (Fig. 1D) did not exhibit inhibition, demonstrating that the phenotype was caused by FoxM1 ablation. We also tested the tumorigenic properties of the sarcoma cells by performing soft agar assay. FoxM1 deletion significantly reduced the ability of cells to grow under anchorage-independent conditions (Supplemental Fig.2A). After FoxM1 deletion, cells formed about 60% less colonies on the soft agar plate compared to the control. In addition, cells without FoxM1 also formed about 50% less colonies on the adherent plate (Supplemental Fig.2B). These results indicate that FoxM1 function is important for the survival and tumorigenicity of tumor cells with p53 loss of function.
FoxM1 ablation diminishes expression of Survivin and Bmi1 in p53 null tumors accompanied by apoptosis
Several studies have suggested targeting FoxM1 could serve as a therapeutic strategy towards treatment of cancer (21, 32, 33). To validate this strategy in tumors harboring p53 loss of function, we utilized a nude mice allograft model. One million thymic lymphoma (L1) or sarcoma (S) triple transgenic cells were injected subcutaneously into nude mice. About one week after injection, when the tumors became palpable, we randomized animals into two treatment groups and started to administer either tamoxifen or vehicle for two weeks. For both p53 null tumor lines, the tumors in the vehicle-treated control group grew significantly faster than of the tumors treated with tamoxifen (Fig.2A and B). FoxM1 expression was examined by performing immunohistochemical staining. FoxM1 expression was largely reduced following two-weeks of tamoxifen treatment, while abundant FoxM1 staining was detected in the vehicle treated group, consistent with FoxM1 over-expression in tumor cells (Supplemental Fig.3A-D).
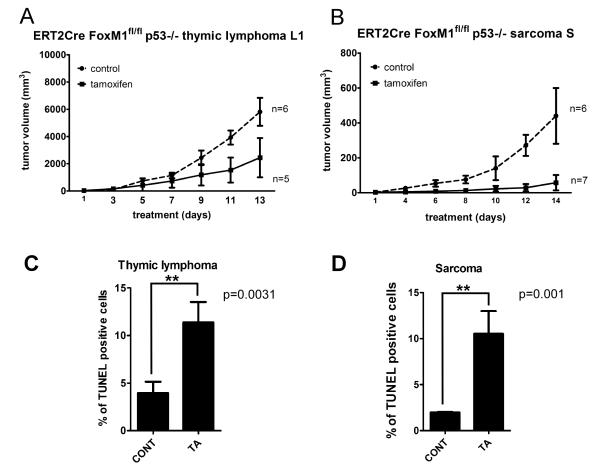
Figure 2. FoxM1 ablation retards growth and induces apoptosis of allografted p53 null lymphoma and sarcoma.
A, Tumor volumes of the subcutaneously inoculated CreERT2 Foxm1 fl/fl and p53 −/− thymic lymphoma cell L1 following FoxM1 ablation by tamoxifen and control treatment are indicated. B, Tumor volumes of the subcutaneously inoculated CreERT2 Foxm1 fl/fl and p53 −/− sarcoma cell S following FoxM1 ablation by tamoxifen and control treatment are shown. C, Quantification of percentage of TUNEL positive cell per field of sarcoma. D, Quantification of percentage of TUNEL positive cell per field of thymic lymphoma.
To investigate the basis for delayed tumor growth, we assayed for apoptosis of the tumor cells using TUNEL staining. In both lymphoma and sarcoma derived tumor sections, we observed an increased number of apoptotic cells following FoxM1 depletion, evidenced by increase number of TUNEL positive cells (Fig.2C-D, Supplemental Fig.3E-L). We also assayed for cleaved caspase-3 and cleaved PARP, two apoptosis markers. Significant increases in the number of cleaved caspase-3 and cleaved PARP positive cells were detected in FoxM1-depleted cells (Supplemental Fig.3M-N). These observations suggested that the inhibition of the p53−/− tumors following loss of FoxM1 resulted from enhanced apoptosis of the tumor cells.
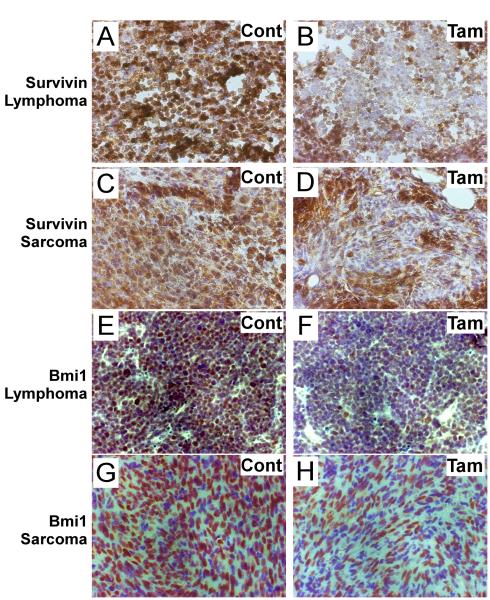
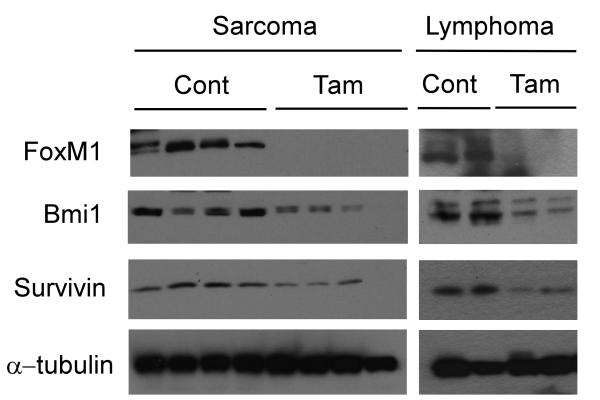
The increased apoptosis upon FoxM1-depletion was somewhat surprising because the p53−/− tumor cells are generally resistant to apoptosis (34). Survivin, which belongs to the inhibitor of apoptosis protein (IAP) family, is a known transcriptional target of FoxM1 that plays important roles in both cell cycle regulation and inhibition of apoptosis (9, 35). Previously, it was shown that reduced Survivin levels contributed to apoptosis of HCC cells (21). Consistent with this finding, we observed that expression of Survivin, which is abundant in control groups for both p53 null lymphoma and sarcoma, was down-regulated following depletion of FoxM1 (Fig.3A-D, Fig.4). Bmi1, another FoxM1-induced gene (8, 36), has been shown also to protect tumor cells from apoptotic stimuli (37). Therefore, we assayed for expression of Bmi1 in the tumor sections. We observed that the expression of Bmi1 was largely diminished in FoxM1-ablated tumors (Fig.3E-H, Fig.4). These observations suggest important roles of Bmi1 and Survivin in the survival of the p53−/− lymphoma and sarcoma. In Fig. 3I, the doublet for Bmi1 is not obvious in the Sarcoma samples because a higher percentage resolving gel was used. It is noteworthy that although the reduction of Survivin and Bmi1 was evident, it was not complete possibly due to the presence of other signaling pathways that control expression of these two proteins. In that regard, NF-κB/STAT3 and ERK/AMPK/p38MARK signaling pathways were shown to activate the expression of Survivin (38, 39). In addition, the expression of Bmi1 is regulated by microRNAs (40).
Figure 3. Immunohistochemistry Staining of Survivin and Bmi1 following FoxM1 ablation in p53 null tumor tissues.
A-D, Representative Survivin staining of subcutaneously inoculated CreERT2 Foxm1 fl/fl and p53 −/− lymphoma and sarcoma cells following tamoxifen and control treatment. E-H, Representative Bmi1 staining of subcutaneously inoculated CreERT2 Foxm1 fl/fl and p53 −/− lymphoma and sarcoma cells following tamoxifen and control treatment.
Figure 4. Reduced Survivin and Bmi1 expression following FoxM1 ablation in p53 null tumor lysates.
Western blot of protein lysates extracted from allografted tumors assayed for FoxM1, Bmi1 and Survivin. α–tubulin was used as a loading control. Lysates were collected from both control oil treated mice and tamoxifen treated mice.
ARF-derived peptide inhibitor of FoxM1 induces apoptosis in p53 null tumor cells
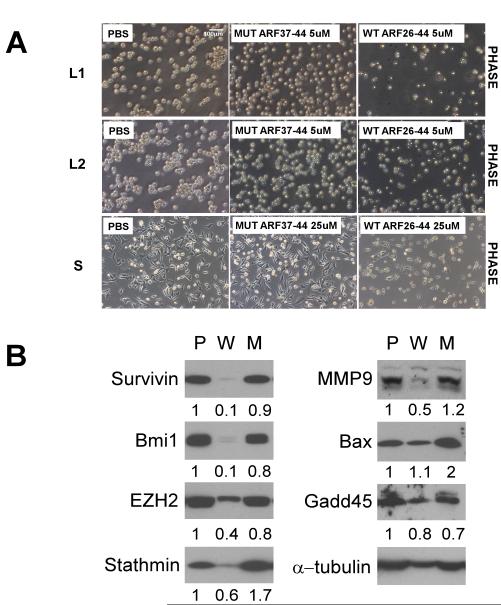
A peptide (ARF 26-44) derived from the mouse tumor suppressor ARF has been described that inhibits the activity of FoxM1 by re-localizing it to the nucleolus (6) (21). A cell-penetrating form of the peptide efficiently targets FoxM1 in liver tumors. In the DEN/PB induced mouse hepatocellular carcinoma model, the ARF-peptide is able to inhibit HCC progression by inducing apoptosis (21). The ARF-peptide induced apoptosis was observed mainly in the FoxM1-expressing cells (21). In addition, it has been shown to block the metastatic growth of the HCC cells (16). In order to see whether the ARF-peptide is able to inhibit the p53 null tumors, we first examined the effect of the peptide, in vitro. A mutant peptide (ARF 37-44), which lacks the interacting domain with FoxM1, was used as a control. One day after treatment the wild type ARF-peptide treated p53 null thymic lymphoma cell lines L1 and L2 underwent apoptosis. The number of viable cells was much less following treatment with wild type ARF peptide compared with cells treated with the mutant-peptide or PBS (Fig.5A). The induction of apoptosis by the wild type peptide was demonstrated by TUNEL staining (Fig.6A). A similar effect was observed in p53 null sarcoma cells (Fig.5A and Fig.6A). However, compared to the sarcoma lines, the p53−/− lymphoma cells are more sensitive to the ARF-peptide, where 5 μM of peptide was able to cause significant apoptosis (Fig.6A). Cell growth and foci formation assay, as well as cleaved caspase-3 staining were performed to confirm the finding (Supplemental Fig.4). Treatments with the ARF-peptide strongly inhibited expression of several FoxM1-induced genes, including Survivin, Bmi1, EZH2, Stathmin and MMP9 (Fig.6B). The ARF-peptide had only a marginal effect on expression of Bax and GADD45, which are not direct targets of FoxM1.
Figure 5. Cellular response of p53 null tumor cells following ARF 26-44 peptide treatment.
A, Phase contrast picture of CreERT2 Foxm1 fl/fl and p53 −/− thymic lymphoma and sarcoma cells treated with PBS, ARF 37-44 peptide (Mut) or ARF 26-44 peptide (WT) at 24 hours. B, Western blot of protein lysates extracted from thymic lymphoma cells treated with PBS, ARF 37-44 peptide (Mut) or ARF 26-44 peptide (WT). α–tubulin was used as a loading control. The band intensities were quantified by Image J program, and the relative intensities after adjusting for loading control (intensity of the tubulin bands) are shown below each panel.
Figure 6. ARF 26-44 peptide activates apoptotic response in the p53 null tumor cells.
A, TUNEL and DAPI staining of CreERT2 Foxm1 fl/fl and p53 −/− thymic lymphoma and sarcoma cells treated with PBS, ARF 37-44 peptide (Mut) or ARF 26-44 peptide (WT). B, Quantification of percentage of TUNEL positive cells per field.
ARF-peptide effectively reduces the colonization of p53 null tumor cells in vivo
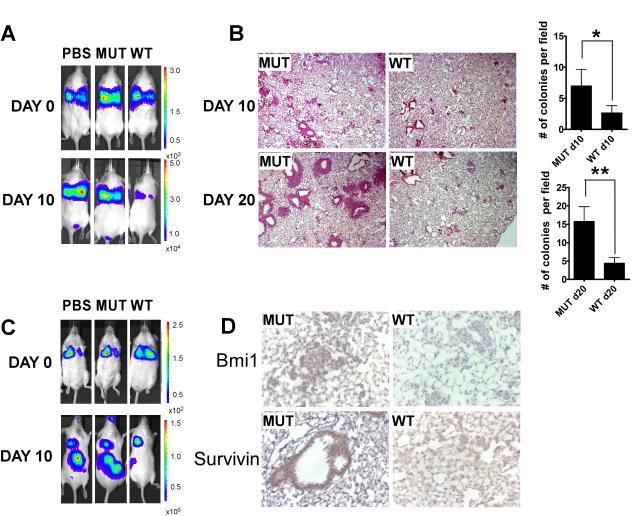
To test the therapeutic effect of the ARF-peptide on p53 null tumors in vivo, p53 null lymphoma/sarcoma cells were introduced into the circulation of SCID mice through intravenous injection. Both p53 null sarcoma and lymphoma cells were stably transduced with lentivirus carrying luciferase expression before injection. Shortly after injection, comparable fluorescence was detectable in the lung by injecting luciferin using Xenogen IVIS spectrum in vivo imaging machine (Fig.7A and C). Mice were randomized into three groups and treated with PBS, mutant-peptide or the wild type ARF-peptide for 10 injections every other day starting from day 0 by intraperitoneal injection. Ten days after tumor inoculation, p53 null sarcoma cells were found to colonize the lung (Fig.7A and B). After 20 days following the initial inoculation, compared to the PBS and the mutant peptide treated mice, the amount of luciferase signal from the wild type ARF-peptide treated mice was significantly reduced. The mice were sacrificed and lung sections were analyzed for tumor colonies. A reduced number of tumor colonies that were larger than 100 μm × 100 μm were detected in the lungs of the wild type ARF-peptide treated mice (Fig.7B). Moreover, Survivin and Bmi1 expression was inhibited in the colonized tumors from mice treated with the wild type ARF-peptide compared to those treated with the mutant peptide (Fig.7D).
Figure 7. ARF 26-44 peptide blocks colonization of intravenously inoculated p53 null tumors.
A, ICR SCID mice were intravenously inoculated with CreERT2 Foxm1 fl/fl and p53 −/− sarcoma cells. Luciferase intensity was monitored with IVIS image machine following peptide treatment at 10 days after initial injection and right after injection at day 0. B, H&E staining of the lung tissue section from MUT or WT peptide treated mice at day 10 and day 20 after initial sarcoma cell injection and quantification of the number of the colonies per field of the corresponding lung tissue section. C, ICR SCID mice were intravenously inoculated with CreERT2 Foxm1 fl/fl and p53 −/− thymic lymphoma cells. Luciferase intensity was monitored with IVIS image machine following peptide treatment at 10 days after initial injection and right after injection at day 0. D, Representative pictures of Bmi1 and Survivin IHC staining of colonized sarcoma cells in the lung after either MUT or WT peptide treatment at day 20.
The murine thymic lymphoma cells tended to colonize the kidney, liver and spleen (41). For the p53 null thymic lymphoma cells, we observed metastatic growth in kidney. Around 20 days after inoculation, PBS and the mutant ARF peptide treated mice displayed strong luciferase signals from the colonized lymphoma cells in the lower back region. On the other hand, the wild type ARF-peptide treated mice emitted very little fluorescence, indicating an inhibition of colonized tumors (Fig. 7C). When the mice were sacrificed, large tumor masses were found in the kidney by microscopic examination in the PBS and in the mutant peptide treated mice. Atypical pale coloration and enlargement of the kidney were observed in the mice, and the mice carried a large tumor mass that encompassed the two kidneys, the connective tissues and the spinal cords. On the other hand, kidneys from the wild type peptide treated mice still retained the original size and structure with only a small white mass started to build up on the surface of the kidney (Supplementary Fig.5A and B). These results clearly indicated that the wild type ARF-peptide was able to efficiently block the renal metastasis of the p53 null thymic lymphoma.
Discussion
The results presented here are significant in several ways. First, we show that p53-null lymphoma and sarcoma are addicted to FOXM1 for their survival, which might also be significant for other p53 loss-of-function tumors. Moreover, we show that the ARF-derived peptide inhibitor of FOXM1 effectively inhibits colonization and growth of the p53-null lymphoma and sarcoma, which will have strong implications with regards to a new therapeutic application of the ARF-peptide against lymphomas and sarcomas.
Loss-of-function of p53 confers resistance to apoptosis, because p53 stimulates expression of several pro-apoptotic genes, including Puma, Noxa, Bax, Bad, DR4, DR5, Apaf1, Caspase 6 and others (42). P53 also represses expression of anti-apoptotic genes, such as Survivin (43). The pro-apoptotic function of p53 is critical for elimination of cells harboring irreparable levels of DNA damage. It is noteworthy that p53 also stimulates several DNA repair genes (44). In the absence of p53, reduced DNA repair and apoptosis lead to the accumulation of mutant cells, which contribute to tumor development. For example, p53-null mice, used in this study, spontaneously develop lymphomas and sarcomas (4). P53 also stimulates expression of the cell cycle inhibitor p21 (45) and represses FOXM1 (26, 27), contributing to cell cycle arrest following DNA damage. Therefore it is not surprising that p53 mutation also leads to aggressive progression of already developed tumor cells because of increased survival and proliferation. Increased expression of FOXM1 in the p53 mutant tumors is expected to drive aggressive progression because of its role in cell proliferation and inhibition of apoptosis. FOXM1 has been shown to inhibit apoptosis by activating expression of Survivin, which is inhibited by p53.
FOXM1 is an important target for cancer therapy. It is expressed mainly in the proliferating cells and in tumors (19, 46). Based on available evidence, it appears that FoxM1 is dispensable for survival or function of the normal cells in a tissue. For example, deletion of FoxM1 in the adult mouse liver has not visible effect for at least one year (6). But, it blocks development of hepatocellular carcinoma (HCC). Moreover, conditional deletion of FoxM1 after HCC development causes inhibition of tumor progression (21). Moreover, a peptide inhibitor derived from the tumor suppressor ARF, the ARF-peptide used in this study, was shown to inhibit liver tumors, through increased apoptosis, without affecting the neighboring normal cells in the tumor bearing liver (21). Therefore, selective inhibition of FOXM1 would be effective in cancer treatment. Our observations with p53-null lymphoma and sarcoma are significant in that regard because over 50% of tumors harbor p53 mutations.
The observations that p53-null lymphoma and sarcoma cells depend upon FOXM1 and are highly sensitive to inhibition of FOXM1 suggest that the p53-null tumors are also candidates for therapeutic strategies that target FOXM1. Cre-recombinase mediated deletion of FoxM1 inhibited tumor growth at least partly by inducing apoptosis. Deletion of FoxM1 caused a reduction in the expression of Survivin, an anti-apoptotic protein. Moreover, there was a strong reduction of Bmi1, which was shown to support survival of tumor cells (47). It is therefore likely that these FOXM1 target genes are involved in the survival of the p53-null lymphoma and sarcoma. Moreover, a cell-penetrating form of the ARF-peptide, which inhibits FOXM1, also induced apoptosis and inhibited colonization of the p53-null lymphoma and sarcoma cells. The lymphoma cells were more sensitive to the peptide. It is possible that the entry of the peptide is more efficient in the less adherent lymphoma cells, raising the possibility that the ARF-peptide would be highly effective against the tumor cells in circulation. Consistent with that, there was a drastic inhibition of the lymphoma colonization to the kidney, a major site of colonization for the T-lymphoma cells (41). These observations indicate the possibility of a new application of the ARF-peptide in targeting the tumor cells in the circulation.
Supplementary Material
Acknowledgment
We thank Dr. Roberta Franks and Wanni Yu (University of Illinois at Chicago) for technical support. We dedicate this work to the memory of Dr. Robert H. Costa.
Grant support This work was supported by US Public Health Service (PHS) Grants CA 124488 and by a Merit Review Grant (IO1BX000131) from the Veteran’s Administration to P.Raychaudhuri. A.L.Tyner is supported by the PHS grants DK 44525 and DK068503. S.Bagchi is supported by the PHS grant CA156164.
Footnotes
Conflict of interest: There is no conflict of interest.
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 3.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 4.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 5.Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV, et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–1720. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshida Y, Wang IC, Yoder HM, Davidson NO, Costa RH. The forkhead box M1 transcription factor contributes to the development and growth of mouse colorectal cancer. Gastroenterology. 2007;132:1420–1431. doi: 10.1053/j.gastro.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Park HJ, Carr JR, Chen YJ, Zheng Y, Li J, et al. FoxM1 in tumorigenicity of the neuroblastoma cells and renewal of the neural progenitors. Cancer Res. 71:4292–4302. doi: 10.1158/0008-5472.CAN-10-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 11.Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007;27:1007–1016. doi: 10.1128/MCB.01068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–5189. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 13.Park HJ, Carr JR, Wang Z, Nogueira V, Hay N, Tyner AL, et al. FoxM1, a critical regulator of oxidative stress during oncogenesis. Embo J. 2009;28:2908–2918. doi: 10.1038/emboj.2009.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carr JR, Park HJ, Wang Z, Kiefer MM, Raychaudhuri P. FoxM1 mediates resistance to herceptin and paclitaxel. Cancer Res. 70:5054–5063. doi: 10.1158/0008-5472.CAN-10-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwok JM, Peck B, Monteiro LJ, Schwenen HD, Millour J, Coombes RC, et al. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res. 8:24–34. doi: 10.1158/1541-7786.MCR-09-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park HJ, Gusarova G, Wang Z, Carr JR, Li J, Kim KH, et al. Deregulation of FoxM1b leads to tumour metastasis. EMBO Mol Med. 3:21–34. doi: 10.1002/emmm.201000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raychaudhuri P, Park HJ. FoxM1: a master regulator of tumor metastasis. Cancer Res. 71:4329–4333. doi: 10.1158/0008-5472.CAN-11-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z, Tan G, Ding M, Dong D, Chen T, Meng X, et al. Foxm1 transcription factor is required for maintenance of pluripotency of P19 embryonal carcinoma cells. Nucleic Acids Res. 38:8027–8038. doi: 10.1093/nar/gkq715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25:1715–1719. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye H, Holterman AX, Yoo KW, Franks RR, Costa RH. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phase. Mol Cell Biol. 1999;19:8570–8580. doi: 10.1128/mcb.19.12.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gusarova GA, Wang IC, Major ML, Kalinichenko VV, Ackerson T, Petrovic V, et al. A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J Clin Invest. 2007;117:99–111. doi: 10.1172/JCI27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhat UG, Halasi M, Gartel AL. Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS One. 2009;4:e5592. doi: 10.1371/journal.pone.0005592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radhakrishnan SK, Bhat UG, Hughes DE, Wang IC, Costa RH, Gartel AL. Identification of a chemical inhibitor of the oncogenic transcription factor forkhead box M1. Cancer Res. 2006;66:9731–9735. doi: 10.1158/0008-5472.CAN-06-1576. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci U S A. 2002;99:16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci U S A. 107:18115–18120. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barsotti AM, Prives C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene. 2009;28:4295–4305. doi: 10.1038/onc.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandit B, Halasi M, Gartel AL. p53 negatively regulates expression of FoxM1. Cell Cycle. 2009;8:3425–3427. doi: 10.4161/cc.8.20.9628. [DOI] [PubMed] [Google Scholar]
- 28.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66:1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 30.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gartel AL. FoxM1 inhibitors as potential anticancer drugs. Expert Opin Ther Targets. 2008;12:663–665. doi: 10.1517/14728222.12.6.663. [DOI] [PubMed] [Google Scholar]
- 33.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 34.Lopes UG, Erhardt P, Yao R, Cooper GM. p53-dependent induction of apoptosis by proteasome inhibitors. J Biol Chem. 1997;272:12893–12896. doi: 10.1074/jbc.272.20.12893. [DOI] [PubMed] [Google Scholar]
- 35.Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22:8581–8589. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- 36.Li SK, Smith DK, Leung WY, Cheung AM, Lam EW, Dimri GP, et al. FoxM1c counteracts oxidative stress-induced senescence and stimulates Bmi-1 expression. J Biol Chem. 2008;283:16545–16553. doi: 10.1074/jbc.M709604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tracey L, Perez-Rosado A, Artiga MJ, Camacho FI, Rodriguez A, Martinez N, et al. Expression of the NF-kappaB targets BCL2 and BIRC5/Survivin characterizes small B-cell and aggressive B-cell lymphomas, respectively. J Pathol. 2005;206:123–134. doi: 10.1002/path.1768. [DOI] [PubMed] [Google Scholar]
- 39.Gritsko T, Williams A, Turkson J, Kaneko S, Bowman T, Huang M, et al. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12:11–19. doi: 10.1158/1078-0432.CCR-04-1752. [DOI] [PubMed] [Google Scholar]
- 40.Bhattacharya R, Nicoloso M, Arvizo R, Wang E, Cortez A, Rossi S, et al. MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res. 2009;69:9090–9095. doi: 10.1158/0008-5472.CAN-09-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aoudjit F, Potworowski EF, St-Pierre Y. The metastatic characteristics of murine lymphoma cell lines in vivo are manifested after target organ invasion. Blood. 1998;91:623–629. [PubMed] [Google Scholar]
- 42.Kuribayashi K, Finnberg N, Jeffers JR, Zambetti GP, El-Deiry WS. The relative contribution of pro-apoptotic p53-target genes in the triggering of apoptosis following DNA damage in vitro and in vivo. Cell Cycle. 10:2380–2389. doi: 10.4161/cc.10.14.16588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–2622. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- 44.Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 45.Agarwal ML, Agarwal A, Taylor WR, Stark GR. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci U S A. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Liu L, Andrews LG, Tollefsbol TO. Loss of the human polycomb group protein BMI1 promotes cancer-specific cell death. Oncogene. 2006;25:4370–4375. doi: 10.1038/sj.onc.1209454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.