Abstract
Background
Olfactomedin-like is a polyfunctional polymeric glycoprotein. This family has at least four members. One member of this family is OLFML3, which is preferentially expressed in placenta but is also detected in other adult tissues including the liver and heart. However, the orthologous rat gene is expressed in the iris, sclera, trabecular meshwork, retina, and optic nerve.
Methods
OLFML3 amplification was performed by RT-PCR from human and baboon ocular tissues. The products were cloned and sequenced.
Results
We report OFML3 expression in human and baboon eye. The full CDS has 1221 bp, from which a OFR of 406 amino acid was obtained. The baboon OLFML3 gene nucleotidic sequence has 98%, and amino acidic 99% similarity with humans.
Conclusions
OLFML3 expression in human and baboon ocular tissues and its high similarity make the baboon a powerful model to deduce the physiological and/or metabolic function of this protein in the eye.
Keywords: Olfactomedin, eye, Old World Monkey, gene expression, animal model, corneal vascularization
INTRODUCTION
The Olfactomedin-family (OLF) is a group of extracellular glycoproteins that contain a domain in its C-terminal region [22]. This domain has about 250 amino acids and is named the OLF domain [6, 22]. The members of this family (seven diverse groups [26]) share different physiological functions between them. They seem to play a regulatory role in vertebrate embryogenesis [3] and other specific functions [26]. OLF includes noelins, implicated in neuronal differentiation during development, neurogenesis, and generation of the neural crest [3, 13–14]; tiarin, a dorsalizing factor of the neural tube [24]; amassin, a mediator of rapid intercellular adhesion [7]; myocilin, which has a role in trabecular meshwork inducible glucocorticoid response [10, 20] [mutations in the OLF domain of myocilin have been associated with primary open angle glaucoma (POAG) [1, 12]]; optimedin, which is expressed in the retina and the trabecular meshwork of the human eye and that has been suggested to be involved in disorders of the anterior segment of the eye and the retina[23]; and latrophilin, which is associated with cell adhesion and signal transduction[21].
Olfactomedin-like (OLFML) glycoproteins are other members of OLF and consist of at least five different molecules. OLFML1 regulates cell proliferation by increasing the percentage of cells in the S phase. OLFML2A and OLFML2B are known as photomedins. The former is expressed selectively in the outer segment of photoreceptor cells and the latter in all neurons of mice retina[6]. The OLFML3 gene, which has been suggested to have a physiological role as a matrix-related component involved in placental and embryonic development in human and mouse and that may be involved in eye disorders [22, 27].
In humans the OLFML3 gene encodes a 1852 nucleotide messenger RNA (mRNA), and contains a coding DNA sequence (CDS) of 1221 nucleotides flanked by two untranslated Regions (UTRs) (UTR-5′ with 119 nucleotides and UTR-3′ with 522 nucleotides). The gene has three exons (exon-1: 223 nucleotides, exon-2: 286 nucleotides and exon-3: 1352 nucleotides) separated by two introns and spans 2.9-kilo base pairs (kbp) in chromosome 1 at band p13.1. The Open Reading Frame (ORF) predicts a protein of 406 amino acids and 44 kilo Daltons (KDa)[27]. Expression of this gene has been reported in multiple human tissues, such as placenta, liver, heart, skeletal muscle, small intestine, lung, kidney, colon, thymus, spleen, and the brain [27], but not in the eye. On the other hand, OLFML3 has been identified by shotgun proteomics in rabbit corneal endothelium proteins [11].
The precise role of OLFML3 remains elusive. However, the orthologous rat gene of the human OLFML3 (HNOEL-iso) gene is expressed in the iris, sclera, trabecular meshwork, retina, and the optic nerve [2, 22].
We report for the first time, the expression of OFML genes in human eye tissues [endothelial layer of the cornea, uvea, lens, retina and retinal pigment epithelium (RPE)]. We also found expression of the OFML3 gene in cornea, lens, iris and RPE tissues of the baboon [Papio hamadryas (PHA)], an Old World Monkey (OWM).
MATERIALS AND METHODS
Animal model
Animal procedures were performed according to ethical guidelines and were reviewed by the Institutional Animal Care and Use Committee of the Texas Institute of Biomedical Research (TIBR). Animals were maintained at the Southwest National Primate Research Center in San Antonio, Texas at TIBR. All the animals shared the same diet and environmental conditions before sample collection. All baboons are gang-housed and fed ad libitum on a standard low-fat chow diet (Harlan Tecklad 15% Monkey Diet, 8715). Ocular pieces (n= 3) from three male baboons were collected at scheduled necropsy. Eyepieces were frozen in liquid nitrogen until dissection and nucleic acid extraction. The age of the baboons was 6, 9, and 11 years. Each ocular piece was dissected and the cornea, iris, lens, sclera, anterior chamber, posterior chamber, vitreous, retina-RPE and optic nerve were separated.
Human biological specimens
The study protocol was approved by the Health Research Ethics Board of the Medical School of the Universidad Autonoma de Nuevo Leon (UANL). Biopsies from five human eyes were collected at the Department of Ophthalmology of the Hospital Universitario “Dr. Jose Eleuterio Gonzalez” of the UANL. All patients voluntarily agreed to participate in the study and signed an informed consent. The specimens came from programmed enucleations or any other eye surgery procedure where ocular tissues were removed as part of the patient’s treatment. Biological samples were collected immediately after tissue removal and were immersed in RNAlater solution (Ambion Inc., Austin, TX). The samples were transported at 4°C to the laboratory and stored at −70°C until use. All tissues were from male patients. Characteristics of the tissues are shown in Table 1.
Table 1.
Characteristics of human eye tissues.
| Patient Age (yrs) | Diagnosis / Procedure | Eye Tissues collected |
|---|---|---|
| 47 | Enucleated eye for a traumatic penetrating injury | Retina, sclera and uvea |
| 82 | Penetrating keratoplasty and cataract extraction for a cornea stroma scar and dislocated cataract. | Lens, vitreous and corneal endothelium |
| 71 | Enucleated eye for an orbit carcinoma, infective keratitis and neovascular glaucoma | Optic nerve |
| 7 | Strabismus surgery (Medial Rectus Recession) | Conjunctiva |
| 13 | Strabismus surgery (Inferior oblique muscle myectomy) | Inferior oblique muscle, conjunctiva and tenon. |
RNA isolation from ocular tissues
Total RNA was extracted from the eye tissues samples with Trizol® reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). RNA was treated with RQ1 DNAse (Promega, Madison, WI) for 15 min at 37°C to remove traces of genomic DNA. For the assessment of RNA purity and integrity, we used standard methods of spectrophotometry and gel electrophoresis, respectively.
Reverse transcription and Polymerase Chain Reaction
Retrotranscription (RT) reactions were carried out with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA) and oligo (dT)12–18 Primer (Invitrogen) using 0.25 mg of total RNA from each tissue in 25 μL final volume. We used the online tool Primer3 (v. 0.4.0) to design primers that amplify OLFML3 related transcripts [8]. Primers were designed based on UTRs of OLFML3 mRNA sequences of Macaca mulatta (MMU) and Homo sapiens (HSA). Amplification reactions were carried out using 5 μL of cDNA (from RT reaction), 10 μM of each primer [sense hybridizing 13 bp before ATG initiation triplet (5′-CCTTCTACTCTGGCACCACT-3′) and antisense hybridizing 133 bp after TAG termination triplet (5′-GGAGCTGCAAGAATTTGATT-3′)] and the commercial kit PCR master mix from Qiagen (Valencia, CA). The amplification reaction design was for a final volume of 25 μL and it was carried out in a thermal cycler (Veriti 96-Well Thermal Cycler, Applied Biosystems). The amplification program used was as follows: an initial denaturation step of 4 min at 94°C, 40 cycles of 30 sec each at 94°C, 30 sec at 58°C, 90 seg at 72°C and finally an elongation step of 6 min at 72°C. The amplification products were visualized on 1% agarose gels stained with ethidium bromide and visualized under UV light (see Figure 1).
Figure 1. Electrophoresis analysis of the OLFML3 PCR amplicons.
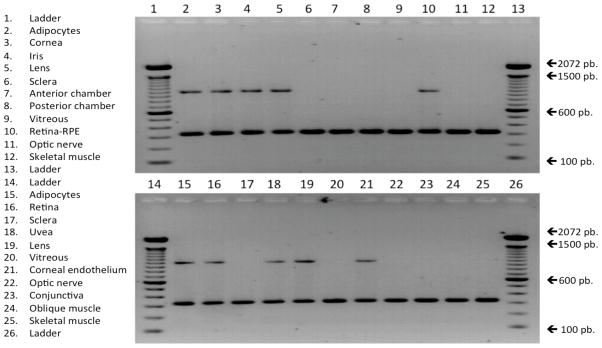
Lines 2 to 12 correspond to baboon cDNAs and lines 15 to 25 correspond to human cDNAs. The ladder consists of 15 blunt-ended fragments between 100 and 1500 bp in multiples of 100 bp and an additional fragment at 2072 bp (Invitrogen).
Molecular cloning, sequencing, and sequence analysis
The amplified products were cloned in the 3.5 kb-XL-TOPO vector and transformed into electrocompetent Escherichia coli bacteria strain Top 10 according to the manufacturer’s specifications (Invitrogen). Positive clones were sequenced using Big Dye terminator cycle sequencing kit v3.1 using specific primers and/or M13 universal primers. The reactions were analyzed in the ABI PRISM 3100 Genetic Analyzer using the Sequencing Analysis Software v5.3 (Applied Biosystems). The sequences obtained from clones were aligned with human orthologous gene [GenBank: NM_020190] using the CLUSTAL W program followed by manual corrections in case of need. Both, the nucleotidic and amino acid sequences were deposited in the GenBank database (PHA -OLFML3: KC237060 and HSA-OLFML3 KC237062) [16].
RESULTS
RT-PCR, molecular cloning and sequence analysis
Amplification of the OLFML3 nucleic acid species was performed by PCR using cDNA from different ocular tissues and OLFML3 specific primers (see Figure 1). The amplicon size was as expected (1405 bp) and no other bands were seen as possible isoforms. In HSA only the RT-PCR for corneal endothelium, uvea, lens and retina-RPE was positive. In baboon tissues, the RT-PCR for cornea, lens, iris and retina-RPE was positive. The PHA and HSA mRNA OLFML3 sequences were deposited in the GenBank database. The obtained amplified products were cloned and the sequences revealed the typical OLFML3 mRNA organization and a full CDS of 1221 bp, from which an ORF of 406 amino acids was predicted (see Figure 2-A). The PHA OLFML3 CDS nucleotidic sequence has 98% similarity with the HSA orthologous. The 23 nucleotidic changes for the 2% difference include: 19 synonymous and 4 non-synonymous changes. In the amino acidic sequence, the similarity between baboon and humans was 99% with only four amino acid changes: in amino acid position 10, there was a Leu in PHA and in HSA there was a Phe; in amino acid position 18, there was a Pro in PHA and a His in HSA, in amino acid position 232, there was a Gly in PHA and a Asp in HSA, and in amino acid position 235, there was a Met in PHA and a Leu in HSA.
Figure 2. Sequence analysis of the OLFML3 gene from HSA and PHA.
A. – OLFML3 gene anatomy. B. – Aligned aminoacidic sequences of PHA and HAS OLFML3. Lowercase sequence corresponds to signal peptide; italicized letter corresponds to coiled-coil domain and underlined letter to olfactomedin-like domain; highlighted in grey are potential sites susceptible to N-glycosylation and highlighted in black is the disulfide bond.
The same OLFML3 transcript was found expressed in the four analyzed tissues of both the PHA and HSA samples. The nucleotidic and amino acidic similarity was 98% and 99%, respectively. Based on these similarities and concordances we suggest that the baboon could be a good model to further explore and analyze the function of OLFML3 in human eyes [4, 18]. We identified OLFML3 expression in four human ocular tissues, which may provide a better understanding of this protein family.
DISCUSSION
Zeng et al., in 2004 reported OLFML3 gene expression in several human tissues but not in human eye [27]. Other analyses in rabbit corneas identified a peptide that shows high similarity to the peptide coded by the OLFML3 gene. In previous reports (human, baboon and rabbit), the metabolic function of OLFML3 remains unknown. A new finding in humans suggests that the OLFML3 gene may play a role in fetal development because this gene was found in the embryonic side of the placenta and not in the endometrial side [27]. However, there is no evidence suggesting its possible role in the eye thus far.
To our knowledge this is the first report of the expression of the OLFML3 gene in human ocular tissues (corneal endothelium, lens, uvea and retina-RPE). In ocular tissues (cornea, lens, iris and retina-RPE) of PHA primate, we found an OLFML3 transcript type, corresponding to the human isoform 1 (~1852 bp). It has been reported in humans that the ORF encoded by the OLFML3 gene contains a 21 amino acid α-signal peptide followed by a coiled-coil domain of 77 amino acids at its N-terminal, topped with an OLFML domain at the C-terminal of 268 amino acids. The ORFs predicted for the new baboon OFLML3 using the nucleotidic sequence coincide in all its components (see Figure 2-A). In the polypeptide sequence there are two potential sites susceptible to N-glycosylation and disulfide bonding, the latter being conserved in the PHA ORF but differing in two positions at the signal peptide and two positions in the OLFML domain. Sites susceptible to N-glycosylation and disulfide bridges predicted by Ling-Chun Zengand colleagues are conserved [27]. The OLF genes have a very diverse physiological role [22] and many of these genes are expression in the human eye [15]. Some mutations in these genes (e.g., myocilin and olfactomedin-2) are related with eye diseases, for instance, POAG [5, 9]. In mouse models (transgenic betaB1-crystallin-MYOC mice) overexpression of myocilin in the aqueous humor resulted in modification of the expression of multiple genes, including the OLFML3 gene, which showed a decrease of expression of 0.72 times. Changes in gene expression of these genes suggest that myocilin and other proteins from the OLF have a role in modulating cellular adhesion [17]. In this paper we show the OLFML3 gene structure and the presence of mRNAs in ocular tissues of baboons and humans. To confirm the presence of the OLFML3 protein in these tissues it is important to perform additional experiments such as immunohistochemistry and Western blot.
It is well known that several pro and anti-angiogenic mechanisms work together modulating eye vascularity. Likewise, maintaining avascularity of the cornea, regulating normal vascularization in the retina and choroid and regulating vascular proliferation are essential in maintaining homeostasis of the eye [19]. In animal models it was recently reported that some members of the OLF might have angiogenic effects on eye tissues. On June 10, 2010 some siRNA against OLFML3 transcripts and antibodies against OLFML3 and OLFML3 polypeptide were patented for use as mediators of angiogenesis. These siRNA and antibodies could be useful in the treatment of diseases with pathologic vascularity [25].
In this study, we demonstrate OLFML3 expression in humans and baboon eye tissue. Given these findings, and taking into consideration that some olfactomedins have a physiological role in angiogenesis [15] and with the new development of siRNA against OLFML3 transcripts to treat diseases with altered vascularity [25], we suggest that OLFML3 may play a possible angiogenic role in ocular tissues and might participate in anterior segment and retinal diseases.
It is important to point out that more work is needed to identify the biologic effect of OLFML3 on eye tissues. We could infer that blocking, modulating or stimulating the action of OLFML3 could be an innovative treatment for different eye diseases. But much more work is needed to clarify its actual role.
Finally, expression of the OLFML3 gene in human and baboon ocular tissues and the high similarity between them make the baboon a powerful model to deduce the physiological and/or metabolic function of this protein in the endothelial layer of the cornea, lens, iris/uvea and retina-RPE. Moreover, it would be important in the future to dig deeper into the role of these genes in normal and pathological physiology with tools such as antisense RNA assays and transgenic models.
Acknowledgments
We appreciate the donation of tissues by Dr. Peter W. Nathanielsz, from the Center for Pregnancy and Newborn Research, University of Texas Health Science Center Medical School, Department of Obstetrics and Gynecology, San Antonio, TX, USA. IPRS enjoyed an SNI assistantship and student fellowship to visit the SFBR. The authors gratefully acknowledge the critical reading of the manuscript by Dr. Sergio Lozano.
FUNDING: The present work was supported by grants from the Mexican Council of Science and Technology, CONACyT (U43987-Q), UANL’s PAICyT (SA972-04), Research Facilities Improvement Program (C06RR014578, C06 RR13556, C06 RR015456 and C06 RR017515) and from the NIH (PO1 HL028972 and P51 RR013986).
LIST OF ABBREVIATIONS
- OWM
Old word primates
- PHA
Papio hamadryas
- HSA
Homo sapiens
- b
base
- bp
base pairs
- PCR
Polymerase Chain Reaction
- RT
Reverse Transcription
- mRNA
messenger RNA
- CDS
coding DNA sequence
- UTR
untranslated region
References
- 1.Adam MF, Belmouden A, Binisti P, Brezin AP, Valtot F, Bechetoille A, Dascotte JC, Copin B, Gomez L, Chaventre A, Bach JF, Garchon HJ. Recurrent mutations in a single exon encoding the evolutionarily conserved olfactomedin-homology domain of TIGR in familial open-angle glaucoma. Hum Mol Genet. 1997;6:2091–2097. doi: 10.1093/hmg/6.12.2091. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed F, Torrado M, Zinovieva RD, Senatorov VV, Wistow G, Tomarev SI. Gene expression profile of the rat eye iridocorneal angle: NEIBank expressed sequence tag analysis. Invest Ophthalmol Vis Sci. 2004;45:3081–3090. doi: 10.1167/iovs.04-0302. [DOI] [PubMed] [Google Scholar]
- 3.Barembaum M, Moreno TA, LaBonne C, Sechrist J, Bronner-Fraser M. Noelin-1 is a secreted glycoprotein involved in generation of the neural crest. Nat Cell Biol. 2000;2:219–225. doi: 10.1038/35008643. [DOI] [PubMed] [Google Scholar]
- 4.Binder PS. Hydrogel implants for the correction of myopia. Curr Eye Res. 1982;2:435–441. doi: 10.3109/02713688208996346. [DOI] [PubMed] [Google Scholar]
- 5.Funayama T, Mashima Y, Ohtake Y, Ishikawa K, Fuse N, Yasuda N, Fukuchi T, Murakami A, Hotta Y, Shimada N. SNPs and interaction analyses of noelin 2, myocilin, and optineurin genes in Japanese patients with open-angle glaucoma. Invest Ophthalmol Vis Sci. 2006;47:5368–5375. doi: 10.1167/iovs.06-0196. [DOI] [PubMed] [Google Scholar]
- 6.Furutani Y, Manabe R, Tsutsui K, Yamada T, Sugimoto N, Fukuda S, Kawai J, Sugiura N, Kimata K, Hayashizaki Y, Sekiguchi K. Identification and characterization of photomedins: novel olfactomedin-domain-containing proteins with chondroitin sulphate-E-binding activity. Biochem J. 2005;389:675–684. doi: 10.1042/BJ20050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hillier BJ, Vacquier VD. Structural features and functional domains of amassin-1, a cell-binding olfactomedin protein. Biochem Cell Biol. 2007;85:552–562. doi: 10.1139/o07-055. [DOI] [PubMed] [Google Scholar]
- 8.Howard Hughes Medical Institute and by the National Institutes of Health NHGRI: Primer 3. In, 2011.
- 9.Kanagavalli J, Pandaranayaka E, Krishnadas SR, Krishnaswamy S, Sundaresan P. A review of genetic and structural understanding of the role of myocilin in primary open angle glaucoma. Indian J Ophthalmol. 2004;52:271–280. [PubMed] [Google Scholar]
- 10.Kubota R, Noda S, Wang Y, Minoshima S, Asakawa S, Kudoh J, Mashima Y, Oguchi Y, Shimizu N. A novel myosin-like protein (myocilin) expressed in the connecting cilium of the photoreceptor: molecular cloning, tissue expression, and chromosomal mapping. Genomics. 1997;41:360–369. doi: 10.1006/geno.1997.4682. [DOI] [PubMed] [Google Scholar]
- 11.Meade ML, Shiyanov P, Schlager JJ. Enhanced detection method for corneal protein identification using shotgun proteomics. Proteome Sci. 2009;7:23. doi: 10.1186/1477-5956-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menaa F, Braghini CA, Vasconcellos JP, Menaa B, Costa VP, Figueiredo ES, Melo MB. Keeping an eye on myocilin: a complex molecule associated with primary open-angle glaucoma susceptibility. Molecules. 2011;16:5402–5421. doi: 10.3390/molecules16075402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreno TA, Bronner-Fraser M. Noelins modulate the timing of neuronal differentiation during development. Dev Biol. 2005;288:434–447. doi: 10.1016/j.ydbio.2005.09.050. [DOI] [PubMed] [Google Scholar]
- 14.Moreno TA, Bronner-Fraser M. The secreted glycoprotein Noelin-1 promotes neurogenesis in Xenopus. Dev Biol. 2001;240:340–360. doi: 10.1006/dbio.2001.0472. [DOI] [PubMed] [Google Scholar]
- 15.Mukhopadhyay A, Talukdar S, Bhattacharjee A, Ray K. Bioinformatic approaches for identification and characterization of olfactomedin related genes with a potential role in pathogenesis of ocular disorders. Mol Vis. 2004;10:304–314. [PubMed] [Google Scholar]
- 16.NCBI. National Center for Biotechnology Information. 2011. [Google Scholar]
- 17.Paper W, Kroeber M, Heersink S, Stephan DA, Fuchshofer R, Russell P, Tamm ER. Elevated amounts of myocilin in the aqueous humor of transgenic mice cause significant changes in ocular gene expression. Exp Eye Res. 2008;87:257–267. doi: 10.1016/j.exer.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samples JR, Binder PS, Zavala EY, Deg JK, Baumgartner SD. Epikeratophakia: clinical evaluation and histopathology of a non-human primate model. Cornea. 1984;3:51–60. [PubMed] [Google Scholar]
- 19.Simo R, Carrasco E, Garcia-Ramirez M, Hernandez C. Angiogenic and antiangiogenic factors in proliferative diabetic retinopathy. Curr Diabetes Rev. 2006;2:71–98. doi: 10.2174/157339906775473671. [DOI] [PubMed] [Google Scholar]
- 20.Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, Sheffield VC. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 21.Sudhof TC. alpha-Latrotoxin and its receptors: neurexins and CIRL/latrophilins. Annu Rev Neurosci. 2001;24:933–962. doi: 10.1146/annurev.neuro.24.1.933. [DOI] [PubMed] [Google Scholar]
- 22.Tomarev SI, Nakaya N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol Neurobiol. 2009;40:122–138. doi: 10.1007/s12035-009-8076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torrado M, Trivedi R, Zinovieva R, Karavanova I, Tomarev SI. Optimedin: a novel olfactomedin-related protein that interacts with myocilin. Hum Mol Genet. 2002;11:1291–1301. doi: 10.1093/hmg/11.11.1291. [DOI] [PubMed] [Google Scholar]
- 24.Tsuda H, Sasai N, Matsuo-Takasaki M, Sakuragi M, Murakami Y, Sasai Y. Dorsalization of the neural tube by Xenopus tiarin, a novel patterning factor secreted by the flanking nonneural head ectoderm. Neuron. 2002;33:515–528. doi: 10.1016/s0896-6273(02)00590-1. [DOI] [PubMed] [Google Scholar]
- 25.WIPO. WO/2010/065437. WIPO Patent aplication. 2010
- 26.Zeng LC, Han ZG, Ma WJ. Elucidation of subfamily segregation and intramolecular coevolution of the olfactomedin-like proteins by comprehensive phylogenetic analysis and gene expression pattern assessment. FEBS Lett. 2005;579:5443–5453. doi: 10.1016/j.febslet.2005.08.064. [DOI] [PubMed] [Google Scholar]
- 27.Zeng LC, Liu F, Zhang X, Zhu ZD, Wang ZQ, Han ZG, Ma WJ. hOLF44, a secreted glycoprotein with distinct expression pattern, belongs to an uncharacterized olfactomedin-like subfamily newly identified by phylogenetic analysis. FEBS Lett. 2004;571:74–80. doi: 10.1016/j.febslet.2004.06.059. [DOI] [PubMed] [Google Scholar]



