Abstract
Background
In addition to activities needed to catalyze integration, retroviral integrases exhibit nonspecific endonuclease activity that is enhanced by certain small compounds, suggesting that integrase could be stimulated to damage viral DNA before integration occurs.
Methods
A nonradioactive, plate-based, solution-phase, fluorescence assay was used to screen a library of 50,080 drug-like chemicals for stimulation of nonspecific DNA nicking by human immunodeficiency virus type 1 (HIV-1) integrase.
Results
A semi-automated workflow was established, and primary hits were readily identified from a graphic output. Overall, 0.6% of the chemicals caused a large increase in fluorescence (the primary hit rate) without also having visible color that could have artifactually caused this result. None of the potential stimulators from this moderate-size library, however, passed a secondary test that included an inactive integrase mutant that assessed whether the increased fluorescence depended on the endonuclease activity of integrase.
Conclusions
This first attempt at identifying integrase stimulator (IS) compounds establishes the necessary logistics and workflow, which should encourage larger scale high-throughput screening to advance the novel antiviral strategy of stimulating integrase to damage retroviral DNA.
Introduction
Treatment of human immunodeficiency virus type 1 (HIV-1) infection is a major success story in clinical medicine and provides a model for antiviral therapy. However, development of drug resistance necessitates the continuing identification of new antiretroviral drugs as well as exploration of novel antiviral strategies to prevent or treat the acquired immunodeficiency syndrome (AIDS). One important target for antiretroviral agents is the viral integrase protein, which is present in all infectious retroviruses and must be enzymatically active for virus transmission, replication, and pathogenesis [1]. Integrase is an endonuclease that acts by nicking the ends of viral DNA at a specific site to prepare the viral DNA for integration, and then inserting those ends nonspecifically into cellular DNA [2]. After years of research, blocking the action of integrase was shown to have significant clinical benefits [3–5], and an integrase inhibitor is now commonly included in combination antiretroviral drug regimens.
In addition to the activities needed to catalyze integration, retroviral integrases have a nonspecific endonuclease activity that can nick any DNA sequence [6], and this activity is markedly stimulated by certain small compounds, including 1,2-ethanediol (ED); 1,2-propanediol; 1,3-propanediol; and 1,2,3-propanetriol (glycerol) [7]. In the case of ED, kinetic studies showed that the mechanism of stimulation involves an increase in Vmax [8]. It has been suggested by several investigators that stimulating or unregulating integrase-mediated DNA nicking could cause this enzyme to damage viral DNA before integration occurs and abort the infection before it becomes established [1,6,9], thus supplementing the host-mediated degradation of viral DNA that protects cells from integration [10]. The demonstration that viral DNA in HIV-1 preintegration complexes (PICs) is susceptible to nucleases [11,12] supports this novel idea, and studies of preintegration latency suggest that viral DNA could be vulnerable for several days [13]. In fact, the documented antiviral effect of packaging nonspecific bacterial nucleases into retroviruses by linking them to the Gag protein [14–17] provides experimental precedent for an antiretroviral strategy that involves a strategically-located nuclease. Thus, the idea of stimulating integrase — which is packaged into every infectious retrovirion as part of the Gag-Pol polyprotein — can be considered a natural version of capsid-targeted viral inactivation [18]. If the increased nicking activity were restricted to the PIC while still in the cytoplasm, which is likely if dissolution of the PIC exposes integrase to protein degradation [19], then toxicity to cellular DNA would be avoided. On the other hand, any collateral damage to cellular DNA would likely also be clinically beneficial because it would be limited to newly infected cells that had just been entered by HIV [20].
Development of a pharmacologic agent that stimulates integrase to damage DNA is a logical extension of the theoretical and experimental foundation described above. Although the four integrase stimulator (IS) compounds listed earlier can cause integrase to nick 50% of a DNA substrate [7], high concentrations of those chemicals are required for stimulation. Thus, to assist efforts to develop more potent IS compounds for preclinical evaluation, we recently described a nonradioactive, plate-based, solution-phase assay for nonspecific DNA nicking [8]. We now report the first use of this assay to screen a library of drug-like chemicals for stimulators of the nonspecific DNA nicking activity of HIV-1 integrase.
Methods
HIV-1 integrase
HIV-1 integrase was expressed in bacteria and purified under native conditions as described previously [21,22]. The purified protein was dialyzed against storage buffer [8], diluted to 4 pmol/μl, and stored in aliquots at −70°C. An active-site mutant of HIV-1 integrase that contains a D116I amino-acid substitution and lacks enzyme activity [23] was purified and handled similarly.
Chemical library
The DIVERSet collection of compounds was purchased from ChemBridge Corporation (San Diego, CA). The 50,080 chemicals in this library have molecular weights ranging from 200 to 540 Da and stock concentrations of 5 μg/μl in dimethylsulfoxide (DMSO). The chemicals were supplied as sets of 80 compounds in 96-well plates, which were stored at −20°C. Each chemical was screened at a final concentration of 1 μg/μl.
Plate-based fluorescence assay for DNA nicking
The dual-tagged 49-mer oligodeoxynucleotide in Figure 1 was purchased from Integrated DNA Technologies, Inc. (Coralville, IA). The 5′ end of this DNA is labeled with a fluorophore (fluorescein, or FAM), and the 3′ end is linked to a quencher (Black Hole Quencher-1, or BHQ-1). The oligonucleotide was dissolved in 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA, quantified by spectrophotometry at 260 nm [8], and stored in aliquots at −70°C. Reactions were assembled in 384-well plates on a cooling block with the aid of an Eppendorf epMotion 5070 Workstation (Mississauga, ONT, Canada). Each reaction had a final volume of 10 μl and contained 2 μl of test compound (or water, DMSO, or ED as controls, with the final concentrations of DMSO or ED being 20%), 5 pmol of substrate DNA (final concentration 500 nM), 25 mM Tris-HCl (pH 8.0), 10 mM dithiothreitol (DTT), 10 mM MnCl2, and 1 μl (4 pmol) of HIV-1 integrase. ED was used as a control because it is as active as other available IS compounds [7] but does not cause pipeting difficulties from high viscosity. Reagents were delivered in the following order: 2 μl of test compound or controls; 6 μl of a master mix that contained substrate DNA, Tris-HCl, DTT, and integrase; and 2 μl of MnCl2 (the divalent metal was added last to delay the start of the reaction, and assembling the mixtures on a cooling block also helped synchronize all reactions). DNase I (1 μl of a 1 μg/μl stock solution, for a final concentration of 0.1 μg/μl) was then added manually to two control wells to assess the maximum signal for each plate. Plates were sealed, briefly mixed by placing them for 10 sec on a vibrating Pipet-Aid (Drummond Scientific Company, Broomall, PA), spun for 30 sec at 4°C, then placed in an incubator at 37°C for 90 min. Reactions were stopped by transferring the plates to a 65°C oven for 10 min to inactivate integrase, then spun again.
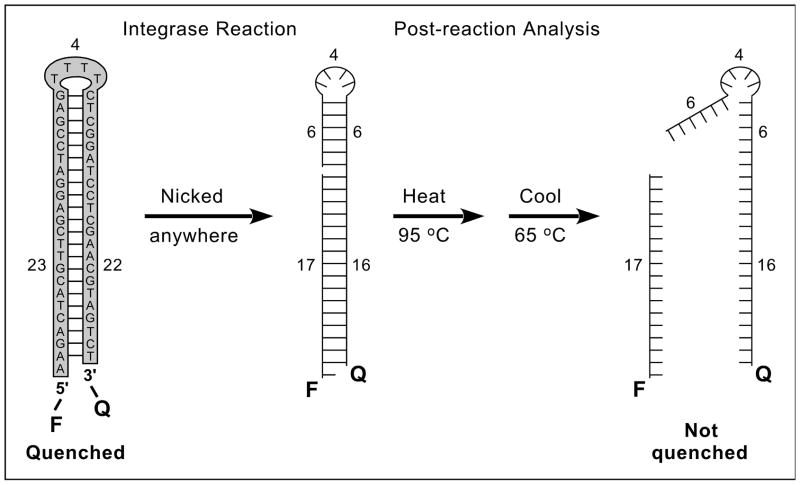
Figure 1. Nonradioactive, plate-based, solution-phase assay for nonspecific DNA nicking.
The sequence of the 49-mer oligonucleotide substrate is shown at the left, with base-pairing indicated by dashes. This DNA is 5′-labeled with a fluorophore (F), 3′-tagged with a quencher (Q), and designed to form a hairpin that brings these groups into proximity to quench fluorescence (numbers indicate lengths of structural features in nucleotides). During reactions with integrase, nicking on either side of the hairpin unlinks the F and Q groups (a nick at position 17 is used as an example, and the DNA is now shown without sequence). Completed reactions are analyzed by heating to 95°C to denature the DNA, then cooling to 65°C and measuring the fluorescence. 65°C is above the melting temperature of the nicked products (which will not be quenched, as shown at the right), but below the melting temperature of unnicked DNA (which will be quenched, as shown at the far left) [8]. The 5′ overhang in the initial substrate permits a nontagged version to be 32P-radiolabeled for gel-based assays.
Analysis of completed reactions
Fluorescence of completed reactions was measured using an Applied Biosystems (Foster City, CA) 7900HT Real-Time PCR System in the Absolute Quantitation mode for fluorescein (FAM) with the passive reference set to None. The machine was programmed for a single denaturation at 95°C for 1 min and renaturation at 65°C for 1 min, followed by 2 min 59 sec at 65°C during which fluorescence of each well was read 21 times (when we tried a data collection phase of exactly 3 min, the machine started another pass across the plate and read one of the columns of wells an extra time). Data were exported as a Microsoft Excel file, then copied and pasted into a pre-templated Excel file that automatically averages the final 10 reads for each well, calculates the control data, and displays the results on bar graphs [8].
Results
Nonspecific nicking assay for the primary screen
The development and validation of a solution-phase assay for nonspecific DNA nicking (Figure 1) was described previously [8]. Briefly, the assay uses a 49-mer oligonucleotide that is 5′-labeled with a fluorophore (F), 3′-tagged with a quencher (Q), and designed to form a hairpin that mimics the double-stranded radioactive substrates commonly used in gel-based nicking assays [6]. Reactions are conducted in 384-well plates during a 90-minute incubation at 37°C. Nicking anywhere in the sequence will unlink the F and Q groups and yield fluorescence after heat denaturation and subsequent cooling to a point above the melting temperatures of nicked products (Figure 1, right). In contrast, the unnicked hairpin should reform and quench fluorescence at temperatures below the melting temperature of the original stem (Figure 1, left). Post-reaction analysis is facilitated by programming a real-time PCR machine to perform a single heat denaturation followed by cooling to 65°C (which is between the melting temperatures of unnicked substrate and nicked products), and taking advantage of the ability of such machines to read fluorescence at a high temperature. No cycling is performed, and the PCR machine can process each plate and measure the fluorescence of every well in less than 10 minutes. The assay was shown to be linear with time, amount of integrase, and concentration of a known integrase stimulator, and the reaction conditions described in the Methods are within the linear portions of these curves; additionally, the presence of 20% DMSO did not unquench the fluorescence of unnicked substrate or mask the signal from nicked DNA [8].
Screening a chemical library: logistics and control data
We screened the 50,080-member DIVERSet collection of drug-like chemicals for stimulation of nonspecific DNA nicking in reactions with HIV-1 integrase. This library is supplied as sets of 80 compounds in 96-well plates, and a robotic workstation was used to deliver chemicals from 4 source plates to one 384-well assay plate. Thus, 320 chemicals were tested in each assay, and the entire library was screened on 157 assay plates (the last plate tested the final 160 compounds). Each assay plate also included 16 controls: 2 wells with water and 6 wells with DMSO as negative controls for baseline nicking, 6 wells with ED as an example of a known IS compound, and 2 wells to which DNase I was added as positive controls to indicate the maximum signal from extensive nicking of the DNA substrate.
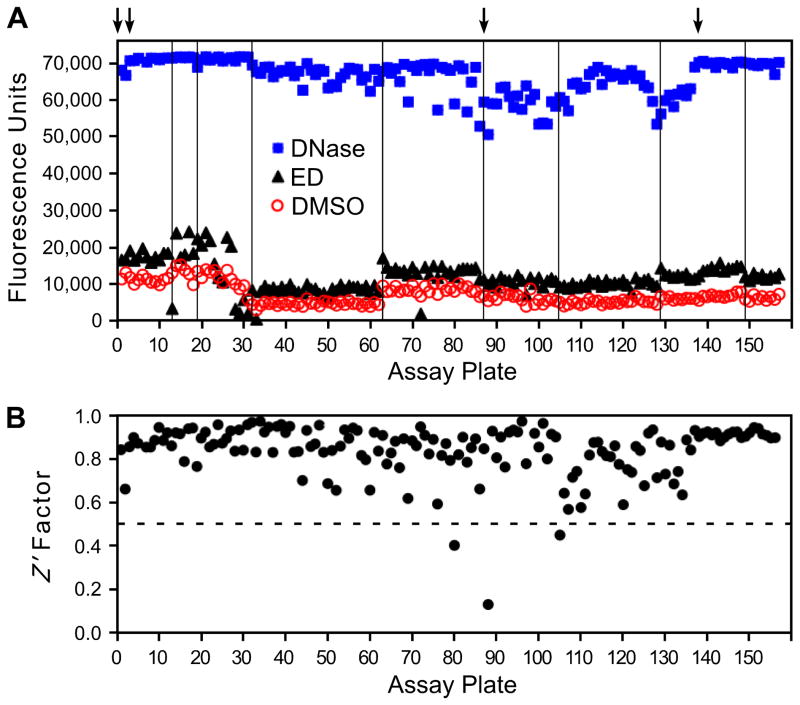
The mean fluorescence for the DMSO, ED, and DNase controls from each assay plate are shown in Figure 2A. As might be expected, the values for baseline nicking in reactions with DMSO (Figure 2A, circles) and stimulated nicking in reactions with the known integrase stimulator ED (triangles) varied slightly with each preparation of purified integrase (the vertical lines in Figure 2A indicate different integrase preparations). Note that baseline nicking in the absence of ED reflects unstimulated nicking plus a small amount of stimulation by the 1% final concentration of glycerol (another IS compound) that was provided by the integrase storage buffer [8,21]. Similarly, maximum signal after exposure of the substrate to DNase varied with each lot of fluorescent oligonucleotide, though these values were always very high (Figure 2A, squares, where the 4 arrows mark new stocks of the 49-mer substrate).
Figure 2. Quality control data from screening the full library.
The 50,080 chemicals in the library were tested on 157 plates. Panel A shows the mean values from each plate for reactions with integrase and DMSO (the negative control for baseline nicking, circles), reactions with integrase and ED (an example of a known stimulator, triangles), and reactions to which DNase I was added (the positive control for extensive nicking, squares). The arrows at the top indicate the use of new lots of the double-tagged oligonucleotide substrate, and the vertical lines denote the use of new preparations of purified integrase. Panel B shows the calculated Z′ factor for each plate. The dashed line is drawn at 0.5, and values above this level are considered optimal for high-throughput screening.
Stimulation of nicking by ED was observed on 145 (92%) of 157 plates, as indicated by clear separation (and an absolute difference of at least 2100 fluorescence units) between reactions with ED and concurrent reactions with DMSO (Figure 2A, compare triangles to circles; the 12 plates that did not pass this quality test [e.g., plate 13] were subsequently retested). Overall, stimulation by ED relative to the amount of nicking in the DMSO controls (omitting data for the 12 initially failed plates) averaged 1.8-fold, with an absolute difference that averaged 5,354 fluorescence units (these values were 1.9-fold and 5,400 units, respectively, when subsequent data from the 12 repeated plates were included).
We also calculated the Z′ factor for each assay; this parameter is a function of the range of an assay (the difference between positive and negative controls) and the variation of the controls, with values ≥0.5 considered optimal for high-throughput screening assays [24]. The Z′ factor for each assay plate (Figure 2B) averaged 0.84 (with or without inclusion of the repeated plates), and only 3 times was it < 0.5 (the low Z′ factors for plates 80, 88, and 105 were due to variation in the reactions with DNase, but even on these plates the DNase controls averaged > 50,000 fluorescence units). As in the original description of this assay [8], reactions with DNase were used as the positive controls for these calculations because a potent integrase stimulator is not yet available. In the absence of an optimal reference compound, others have also used reagents as positive controls (such as antibodies or other proteins) that were different from the test chemicals being screened [25].
Results from the primary screen
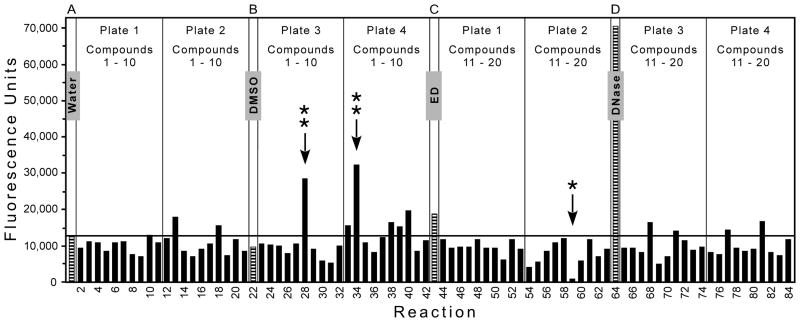
Each assay plate was processed as in the Methods to yield 4 bar graphs; Figure 3 shows an example of one of these graphs, which was modified slightly to include all 4 types of controls in the figure. Each graph presents data from one-fourth of an assay plate, representing 4 rows of 21 reactions (a total of 84 reactions). Each set of 21 reactions includes 1 of the controls (Figure 3, the striped bars) plus 20 assays with chemicals from the library (the solid bars, testing 10 compounds from each of 2 source plates). In the example shown, reactions 1 and 22 (with water and DMSO, respectively) yielded < 13,000 fluorescence units; reaction 43 (with ED) yielded approximately 19,000 units; and reaction 64 (with DNase I) had approximately 71,000 units. The fluorescence of the reaction that contained ED was greater than the mean + 3 standard deviations (SD) of the concurrent negative controls that contained DMSO (as indicated by the horizontal line in Figure 3); in fact, 84% of the almost 900 individual ED reactions across the 145 plates in Figure 2A (excluding the 12 plates that had to be repeated) had fluorescence above the mean + 3 SD of concurrent DMSO controls (and 95% were above the mean + 2 SD).
Figure 3. Example of output from the assay.
Each 384-well assay plate included 336 reactions (320 test chemicals plus 16 controls) using all 16 rows and 21 of the 24 columns. Each assay plate was processed as in the Methods to yield 4 bar graphs; an example of 1 graph is shown (slightly modified so that all 4 types of controls appear in this figure). Each graph presents data from 4 rows (A, B, C, D in this example) of 21 reactions. Each set of 21 reactions includes 1 control (the striped bar, representing a reaction with water or DMSO as negative controls, ED as a known stimulator, or DNase I as positive control for extensive nicking) plus 10 compounds (the solid bars) from each of 2 source plates from the library (Plates 1, 2, 3, 4 in this example, as demarcated by vertical lines). The horizontal line marks the mean + 3 SD for the DMSO negative controls. Of 80 reactions with test chemicals, the arrows indicate 2 with markedly increased fluorescence (**, primary hits) and 1 with greatly diminished signal (*).
Several of the test chemicals in Figure 3 also yielded fluorescence above the controls, sometimes in excess of that from the ED reaction (e.g., reaction 40). However, to have a manageable hit rate in the primary screen, we focused on chemicals that greatly exceeded the controls. Two such chemicals that yielded markedly increased fluorescence (reactions 28 and 34) are indicated by the double asterisks in Figure 3. Given the higher values for the DMSO control reactions on plates 1 to 31 compared to subsequent plates (which was related to the integrase preparation, as seen in Figure 2A), we defined potential stimulators as exceeding 25,000 fluorescence units on assay plates 1 – 31 or exceeding 20,000 units on plates 32 – 157. Overall, 292 of the 50,080 chemicals (a primary hit rate of 0.6%) exceeded these thresholds without also having a visible color that could have artifactually caused this result. We should note that 12.3% of the chemicals in the library caused discernible color on the assay plate (most often yellow or orange, which could increase or decrease fluorescence without a predictable pattern) and were excluded from further analysis; another 0.3% of the compounds yielded a negative value and also were excluded. We also observed that 0.7% of the chemicals in the library yielded markedly diminished fluorescence (< 1,000 fluorescence units), an example of which is indicated by the single asterisk in Figure 3 (reaction 59); although such chemicals might be worthy of further analysis, they were not the focus of this project.
Secondary assays for hits from the primary screen
Because many — if not most — hits from a chemical screen may be due to mechanisms other than the desired one, secondary (or counter) assays are required to validate primary hits. We initially used a gel-based assay with radioactive double-stranded oligonucleotide substrates [8] to retest potential stimulators identified in the primary screen. In fact, neither of the potential stimulators identified in Figure 3 was found to stimulate integrase in radioactive assays (data not shown). Similarly, no hits from any of the first 12 plates were validated as stimulators in radioactive assays. Given the large number of potential stimulators identified by the primary screen (and many others that were close to the defined threshold), the plate-based assay was adapted for an automated and more efficient counter screen, starting with assay plate 13. For this purpose, potential stimulators (primary hits) were retested in two parallel reactions, one using wild-type integrase (to confirm the initial results) and one using an inactive integrase mutant (to validate that the increased fluorescence depended on the action of integrase). Importantly, the known integrase stimulator ED increased the fluorescence of reactions with wild-type integrase but not reactions with the inactive integrase (data not shown).
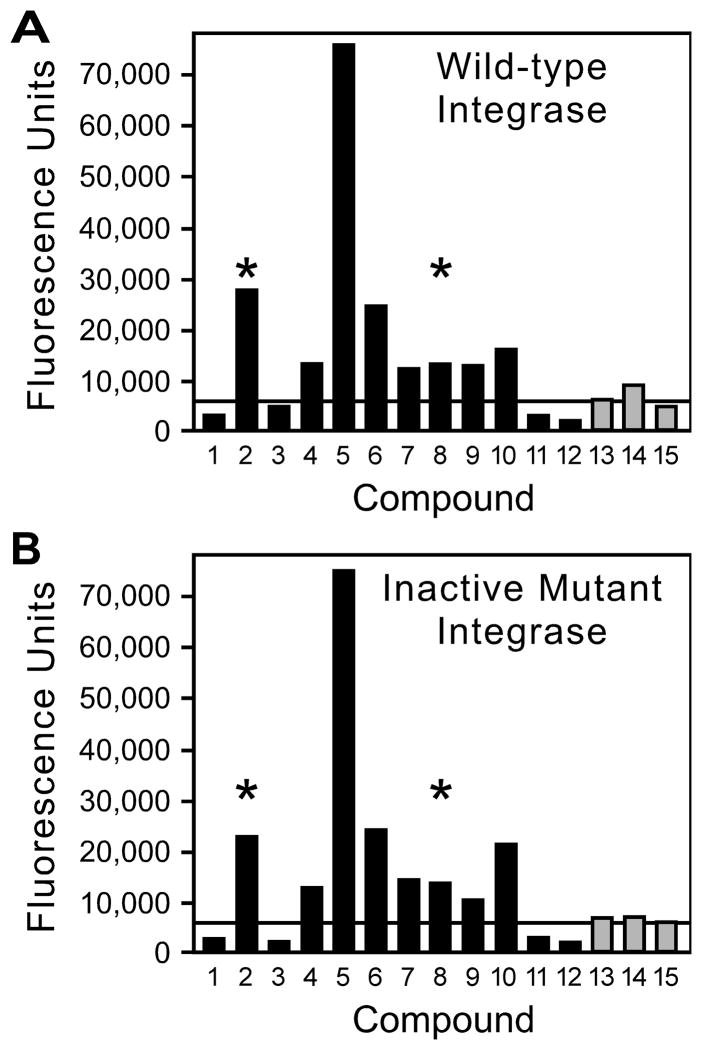
Many chemicals that increased fluorescence in the primary screen also yielded increased fluorescence upon retesting (e.g., Figure 4A, where the solid bars indicate primary hits from the initial screen). Overall, 38% of these chemicals when retested exceeded the mean + 3 SD of the concurrent controls (as indicated by the horizontal line in Figure 4A), 20% caused a signal above 15,000 fluorescence units, and 13% were above 20,000 units. Similar confirmation rates upon retesting have been reported in the literature for other high-throughput assays and underscore the importance of excluding false-positive hits from initial screens [26–29]. Moreover, none of the primary hits passed the predefined secondary test of causing greatly increased signal in reactions with wild-type integrase but not in reactions with an inactive mutant (i.e., the heights of the bars for each chemical were comparable between Figures 4A and 4B). Of note, we observed visible color on the source plate or assay plate for 18% of the primary hits during set up of the secondary assays (as indicated by the asterisks in Figure 4; that these colors had not been recorded during the primary screen reflects greater scrutiny during the counter screen, including examination of the source plates for any color). We also retested approximately 300 compounds that had caused more-moderately increased fluorescence in the primary screen (e.g., 15,000 to 20,000 units for assay plates 32 – 157). These chemicals often yielded results just above or below the mean + 3 SD of the concurrent DMSO controls upon retesting (e.g., Figure 4A, the shaded bars), but none clearly increased signal with wild-type but not with an inactive integrase (Figure 4A compared to Figure 4B).
Figure 4. Examples of secondary assay.
Compounds that caused stimulation of signal in the primary screen were retested in parallel reactions that used wild-type integrase (panel A) or an active-site D116I mutant of integrase (panel B). Solid bars indicate compounds that had caused greatly increased fluorescence in the primary screen (reactions 1 to 12), and shaded bars denote compounds that had caused moderately increased fluorescence (reactions 13 to 15). Asterisks mark compounds for which visible color was noted in the chemical source plate or in the well of the secondary assay plate.
Discussion
New strategies to attack human retrovirus infections are needed. There is precedent in clinical medicine for stimulating the function of other proteins, such as the cystic fibrosis transmembrane conductance regulator [30], and successful chemical screens have been reported for stimulators of other proteins and enzymes, including the human homologous recombination protein RAD51 [31] and glucokinase [32]. To date, four chemicals have been shown to stimulate HIV-1 integrase to nick DNA nonspecifically [6,7], but potent and relatively non-toxic IS compounds are needed for further preclinical development. Ultimately, a useful agent would have to perturb integrase in such a way that newly-synthesized viral DNA (or cellular DNA) is sufficiently damaged to overcome DNA repair mechanisms and prevent the permanent integration of viral DNA.
The recent development of a plate-based assay for nonspecific DNA nicking that is suitable for high-throughput screening (Figure 1) made it possible to begin efforts to identify IS compounds. In the current report, we describe the first attempt at such screening, using a moderate-sized library of approximately 50,000 chemicals. Importantly for future efforts, a semi-automated workflow was established for the primary and secondary screens, primary hits were readily identified from a graphic output, and a logical counter screen was developed. Thus, the approach and techniques were shown to be logistically feasible (Figures 2 and 3).
In this initial screening effort, none of the hits from the primary screen were validated using a secondary assay designed to exclude artifactual results that did not depend on the action of integrase (Figure 4). Increased fluorescence that was independent of integrase function may have been due to autofluorescence of the test compounds. Although pre-screening the chemical library for autofluorescence might have excluded such chemicals from the secondary screen (but not from the primary screen given the characteristics of our robotic workstation), it still would have been uncertain whether autofluorescence of stock compounds on the source plates would be relevant to diluted samples under reaction conditions; thus, testing all chemicals in the primary assay was appropriate. It also is possible that the increased signal from the false positives resulted from the compound directly damaging the DNA substrate or otherwise dissociating the hairpin, perhaps by affecting the pH or by chelating the divalent metal that we have found to be necessary to maintain the hairpin. Any of these possible explanations underscores the importance of performing an appropriate counter assay.
It should be noted that a reported screen for stimulation of another enzyme tested 120,000 compounds to identify one stimulator of glucokinase [32]. Thus, although the experiments described in the current report did not identify any new integrase stimulators from a much smaller chemical collection, the techniques and approaches described here can now be applied to larger scale high-throughput screening to advance the novel antiviral strategy of stimulating integrase to damage retroviral DNA.
Acknowledgments
We thank the Drug Discovery, Development, and Delivery Core of the Dept. of Pharmacology and the Penn State Hershey Cancer Institute for access to the chemical library, and Rob Brucklacher and Georgina Bixler of the Penn State Hershey Genome Sciences Core Facility for assistance with the PCR machine (all at the Penn State College of Medicine, Milton S. Hershey Medical Center); we also thank Fred Krebs (Drexel University College of Medicine) for support and encouragement. This work was supported by Public Health Service grant R21AI075929 from the Microbicide Innovation Program of the Office of Women’s Health and National Institute of Allergy and Infectious Diseases (to MK), and by an American Recovery and Reinvestment Act Administrative Supplement Providing Summer Research Experiences for Students and Science Educators (to MK to support JBE).
Footnotes
This is the author’s version of a work accepted for publication by International Medical Press. Changes resulting from the publishing process, including peer-review, editing and formatting, might not be reflected in this document. A definitive version was published in Antiviral Chemistry & Chemotherapy, (Vol.22, No.2), October 7, 2011. International Medical Press.
Disclosure Statement
The authors declare no competing interests.
References
- 1.Brown PO. Integration. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1997. pp. 161–203. [PubMed] [Google Scholar]
- 2.Craigie R. HIV integrase, a brief overview from chemistry to therapeutics. J Biol Chem. 2001;276:23213–6. doi: 10.1074/jbc.R100027200. [DOI] [PubMed] [Google Scholar]
- 3.Jegede O, Babu J, Di Santo R, et al. HIV type 1 integrase inhibitors: from basic research to clinical implications. AIDS Rev. 2008;10:172–89. [PubMed] [Google Scholar]
- 4.Havlir DV. HIV integrase inhibitors--out of the pipeline and into the clinic. N Engl J Med. 2008;359:416–8. doi: 10.1056/NEJMe0804289. [DOI] [PubMed] [Google Scholar]
- 5.Hicks C, Gulick RM. Raltegravir: the first HIV type 1 integrase inhibitor. Clin Infect Dis. 2009;48:931–9. doi: 10.1086/597290. [DOI] [PubMed] [Google Scholar]
- 6.Katzman M, Sudol M. Nonspecific alcoholysis, a novel endonuclease activity of human immunodeficiency virus type 1 and other retroviral integrases. J Virol. 1996;70:2598–2604. doi: 10.1128/jvi.70.4.2598-2604.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skinner LM, Sudol M, Harper AL, Katzman M. Nucleophile selection for the endonuclease activities of human, ovine, and avian retroviral integrases. J Biol Chem. 2001;276:114–124. doi: 10.1074/jbc.M007032200. [DOI] [PubMed] [Google Scholar]
- 8.Sudol M, Tran M, Nowak MG, et al. A nonradioactive plate-based assay for stimulators of nonspecific DNA nicking by HIV-1 integrase and other nucleases. Anal Biochem. 2010;396:223–30. doi: 10.1016/j.ab.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parissi V, Caumont A, de Soultrait VR, et al. The lethal phenotype observed after HIV-1 integrase expression in yeast cells is related to DNA repair and recombination events. Gene. 2003;322:157–68. doi: 10.1016/j.gene.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 10.Yoder KE, Roddick W, Hoellerbauer P, Fishel R. XPB mediated retroviral cDNA degradation coincides with entry to the nucleus. Virology. 2011;410:291–8. doi: 10.1016/j.virol.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Wei SQ, Engelman A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J Biol Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- 13.Pierson TC, Zhou Y, Kieffer TL, et al. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J Virol. 2002;76:8518–31. doi: 10.1128/JVI.76.17.8518-8531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natsoulis G, Seshaiah P, Federspiel MJ, et al. Targeting of a nuclease to murine leukemia virus capsids inhibits viral multiplication. Proc Natl Acad Sci U S A. 1995;93:364–368. doi: 10.1073/pnas.92.2.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schumann G, Qin L, Rein A, Natsoulis G, Boeke JD. Therapeutic effect of Gag-nuclease fusion protein on retrovirus-infected cell cultures. J Virol. 1996;70:4329–4337. doi: 10.1128/jvi.70.7.4329-4337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanBrocklin M, Federspiel MJ. Capsid-targeted viral inactivation can eliminate the production of infectious murine leukemia virus in vitro. Virology. 2000;267:111–23. doi: 10.1006/viro.1999.0113. [DOI] [PubMed] [Google Scholar]
- 17.Schumann G, Hermankova M, Cannon K, Mankowski JL, Boeke JD. Therapeutic effect of a Gag-nuclease fusion protein against retroviral infection in vivo. J Virol. 2001;75:7030–41. doi: 10.1128/JVI.75.15.7030-7041.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boeke JD, Hahn B. Destroying retroviruses from within. Trends Microbiol. 1996;4:421–6. doi: 10.1016/0966-842x(96)10065-2. [DOI] [PubMed] [Google Scholar]
- 19.Mulder LC, Muesing MA. Degradation of HIV-1 integrase by the N-end rule pathway. J Biol Chem. 2000;275:29749–53. doi: 10.1074/jbc.M004670200. [DOI] [PubMed] [Google Scholar]
- 20.Skalka AM, Katz RA. Retroviral DNA integration and the DNA damage response. Cell Death Differ. 2005;12 (Suppl 1):971–8. doi: 10.1038/sj.cdd.4401573. [DOI] [PubMed] [Google Scholar]
- 21.Katzman M, Sudol M. Mapping domains of retroviral integrase responsible for viral DNA specificity and target site selection by analysis of chimeras between human immunodeficiency virus type 1 and visna virus integrases. J Virol. 1995;69:5687–5696. doi: 10.1128/jvi.69.9.5687-5696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper AL, Sudol M, Katzman M. An amino acid in the central catalytic domain of three retroviral integrases that affects target site selection in nonviral DNA. J Virol. 2003;77:3838–3845. doi: 10.1128/JVI.77.6.3838-3845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harper AL, Skinner LM, Sudol M, Katzman M. Use of patient-derived human immunodeficiency virus type 1 integrases to identify a protein residue that affects target site selection. J Virol. 2001;75:7756–7762. doi: 10.1128/JVI.75.16.7756-7762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 25.Hou Y, McGuinness DE, Prongay AJ, et al. Screening for antiviral inhibitors of the HIV integrase-LEDGF/p75 interaction using the AlphaScreen luminescent proximity assay. J Biomol Screen. 2008;13:406–14. doi: 10.1177/1087057108317060. [DOI] [PubMed] [Google Scholar]
- 26.Sun C, Newbatt Y, Douglas L, et al. High-throughput screening assay for identification of small molecule inhibitors of Aurora2/STK15 kinase. J Biomol Screen. 2004;9:391–7. doi: 10.1177/1087057104264071. [DOI] [PubMed] [Google Scholar]
- 27.Delle Fratte S, Piubelli C, Domenici E. Development of a high-throughput scintillation proximity assay for the identification of C-domain translational initiation factor 2 inhibitors. J Biomol Screen. 2002;7:541–6. doi: 10.1177/1087057102238628. [DOI] [PubMed] [Google Scholar]
- 28.Zhang JH, Chung TD, Oldenburg KR. Confirmation of primary active substances from high throughput screening of chemical and biological populations: a statistical approach and practical considerations. J Comb Chem. 2000;2:258–65. doi: 10.1021/cc9900706. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Klock H, Yin H, et al. Homogeneous high-throughput screening assays for HIV-1 integrase 3′-processing and strand transfer activities. J Biomol Screen. 2005;10:456–62. doi: 10.1177/1087057105275212. [DOI] [PubMed] [Google Scholar]
- 30.Accurso FJ, Rowe SM, Clancy JP, et al. Effect of VX-770 in persons with cystic fibrosis and the G551D-CFTR mutation. N Engl J Med. 2010;363:1991–2003. doi: 10.1056/NEJMoa0909825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jayathilaka K, Sheridan SD, Bold TD, et al. A chemical compound that stimulates the human homologous recombination protein RAD51. Proc Natl Acad Sci U S A. 2008;105:15848–53. doi: 10.1073/pnas.0808046105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimsby J, Sarabu R, Corbett WL, et al. Allosteric activators of glucokinase: potential role in diabetes therapy. Science. 2003;301:370–3. doi: 10.1126/science.1084073. [DOI] [PubMed] [Google Scholar]