Abstract
Mast cells differentiate from circulating pluripotent hematopoietic progenitors. During this differentiation, the progenitor cells are exposed to changes in oxygen availability. HIF1A is the major sensor of oxygen concentration in mammalian cells. We investigated the expression of HIF1A during the in vitro differentiation of peripheral blood-derived progenitors into human mast cells. In a series of experiments, we determined the changes in CD34 expression, selected mast cell markers, and HIF1A in human mast cell cultures. While the expression of CD34 dramatically decreased, the expression of mast cell-specific genes, including FCER1A, MS4A2, TPSB2, and CMA1, steadily increased. HIF1A expression similarly increased during mast cell differentiation, reaching its maximum level at five weeks of culture. The analysis of the promoter methylation status showed decreasing levels of methylation at the HIF1A promoter, increasing levels of methylation at the CD34 promoter, and no significant changes in other genes. In silico analysis of the promoter regions of these genes revealed large CpG islands in close proximity to the HIF1A and CD34 transcription initiation sites, but not in other investigated genes. In conclusion, in vitro mast cell differentiation was associated with decreased CD34 expression and increased HIF1A expression. These changes were paralleled with changes in the methylation status of the respective promoters, suggesting that DNA methylation-dependent epigenetic regulation mediates the gene expression changes involved in maintaining the phenotype of hematopoietic stem cells and mature mast cells. Therefore, the baseline expression of HIF1A is epigenetically regulated in a cell type- and differentiation stage-specific fashion.
Keywords: Human mast cells, Differentiation, Hif1a, Cd34, Epigenetic regulation
Introduction
Mast cells originate from pluripotent hematopoietic cells that are released from the bone marrow into the circulation and differentiate in the peripheral tissues (Metcalfe et al. 1997). One of the factors that differentially influences cellular physiology at different sites in the human body is the partial pressure of oxygen. Relatively low oxygen concentrations, often referred to as physiological hypoxia or physioxia, are characteristic of the bone marrow and the skin (Carreau et al. 2011; MacDougall and McCabe 1967). While the partial pressure of oxygen in various tissues ranges from 5 to 15 mmHg, the partial pressure of oxygen observed in blood plasma is significantly higher, reaching 40 mmHg (Carreau et al. 2011; MacDougall and McCabe 1967). Thus, during the differentiation of bone marrow progenitors into peripheral mature mast cells, the cells are exposed to a gradation of oxygen availability. The major sensor of oxygen concentration in mammalian cells is the dimeric transcription factor HIF-1, which consists of the protein product of HIF1A, known as HIF-1α, and the protein product of ARNT, known as HIF-1β (Semenza 2001). The amount of HIF-1α in the cell depends on the availability of oxygen, which induces HIF-1α degradation (Semenza 2001), and cell type-specific regulation of HIF1A expression at the transcriptional (Gorlach 2009; Lauzier et al. 2007; Walczak-Drzewiecka et al. 2008), and epigenetic levels (Koslowski et al. 2010; Walczak-Drzewiecka et al. 2010). The role of HIF-1α-mediated oxygen sensing is not limited to the physiological response to hypoxic stress, but is also important for other processes, such as astrocyte differentiation (Mutoh et al. 2012), and the maintenance of a quiescent state of bone marrow hematopoietic stem cells (Takubo et al. 2010). HIF-1α is also involved in innate immune responses to bacterial infection, where it upregulates antibacterial functions of granulocytes, macrophages, and keratinocytes (Peyssonnaux et al. 2008; Peyssonnaux et al. 2005; Zinkernagel et al. 2007).
We previously reported that CD34+ hematopoietic progenitors isolated from the peripheral blood and the bone marrow demonstrated significantly lower levels of HIF1A expression as compared to mature PBMC (Walczak-Drzewiecka et al. 2010). This, combined with the observation of epigenetic silencing of HIF1A in the HMC-1 human mast cell line (Walczak-Drzewiecka et al. 2010), prompted us to investigate the expression of HIF1A during in vitro differentiation of human mast cells from their peripheral blood-derived progenitors.
Materials and methods
Cytokines and reagents
Ficoll-Paque cell separation solution was purchased from GE Healthcare (Uppsala, Sweden). The AC133 cell isolation kit and LS+ cell separation columns were purchased from Miltenyi Biotech (Bergisch Gladbach, Germany). StemSpan culture medium was purchased from Stem Cell Technologies (Vancouver, Canada). Fetal calf serum (FCS) was bought from PAA (Pasching, Austria), and penicillin/streptomycin was acquired from Invitrogen/Gibco (Carlsbad, California). Human recombinant stem cell factor (rhSCF), human rhIL-6 and human rhIL-3 were purchased from R&D Systems (Abingdon, UK).
CD133+ cell isolation
The culture of primary human mast cells was initiated from a population of CD133+ cells that were obtained from buffy coat preparations as per Holm et al. (2008). Fresh buffy coats from a total of 35 anonymous blood donors (Table 1) were obtained from the Regional Center of Blood Donation and Blood Treatment in Lodz, Poland according to the center’s legal guidelines. Briefly, the 50 ml buffy coats were diluted in three volumes of PBS, gently layered on a Ficoll-Paque gradient, and centrifuged at room temperature for 30 min at 450×g. The interface layer of mononuclear cells was harvested and washed in PBS. The CD133+ cells were separated from the mononuclear cells using a magnetic separation column as follows: the mononuclear cells were resuspended in 300 μl of MACS buffer and were pre-treated with an FcR blocking reagent at 100 μl/108 cells. The cells were incubated with AC133 MicroBeads for 30 min at 4 °C. After incubation, the cells were washed with MACS buffer. The pellet was then resuspended in 500 μl of MACS buffer and passed through the separation columns. The LS+ column was washed three times with 3 ml of MACS buffer while in the magnetic field, and the CD133+ cells were retained in the column, while the lineage negative cells were eluted. The column was removed from the magnet, and the CD133+ cells were collected and counted in Trypan blue.
Table 1.
Source of cells employed for analysis of gene expression
| Donor no. | PBMC | CD133+ | Week of mast cell culture | ||||
|---|---|---|---|---|---|---|---|
| 1st | 2nd | 3rd | 4th | 5th | |||
| 1 | ■ | ■ | ■ | ■ | ■ | ■ | |
| 2 | ■ | ■ | |||||
| 3 | ■ | ■ | ■ | ||||
| 4 | ■ | ■ | ■ | ■ | ■ | ||
| 5 | ■ | ■ | ■ | ■ | ■ | ||
| 6 | ■ | ■ | ■ | ||||
| 7 | ■ | ■ | ■ | ■ | ■ | ■ | |
| 8 | ■ | ■ | ■ | ||||
| 9 | ■ | ■ | ■ | ■ | ■ | ■ | |
| 10 | ■ | ■ | ■ | ||||
| 11 | ■ | ■ | ■ | ||||
| 12 | ■ | ■ | ■ | ■ | ■ | ||
| 13 | ■ | ■ | ■ | ■ | |||
| 14 | ■ | ■ | ■ | ■ | ■ | ■ | |
| 15 | ■ | ■ | ■ | ■ | |||
| 16 | ■ | ■ | ■ | ■ | |||
| 17 | ■ | ■ | ■ | ■ | |||
| 18 | ■ | ■ | ■ | ■ | |||
| 19 | ■ | ■ | ■ | ||||
| 20 | ■ | ■ | ■ | ||||
| 21 | ■ | ■ | |||||
| 22 | ■ | ■ | ■ | ||||
| 23 | ■ | ■ | ■ | ||||
| 24 | ■ | ■ | ■ | ||||
| 25 | ■ | ■ | ■ | ||||
| 26 | ■ | ■ | ■ | ||||
| 27 | ■ | ■ | ■ | ||||
| 28 | ■ | ■ | ■ | ||||
| 29 | ■ | ■ | ■ | ||||
| 30 | ■ | ■ | ■ | ||||
| 31 | ■ | ■ | ■ | ■ | |||
| 32 | ■ | ■ | |||||
| 33 | ■ | ||||||
| 34 | ■ | ■ | ■ | ||||
| 35 | ■ | ■ | ■ | ■ | |||
Cell culture and differentiation of mast cells
The mast cell cultures were performed as described by Holm et al. (2008). Briefly, the cells were maintained at a density of 5 × 105/ml in StemSpan medium supplemented with 100 ng/ml rhSCF, 50 ng/ml rhIL-6, 1 ng/ml rhIL-3, and penicillin/streptomycin solution. IL-3 was removed from the medium after three weeks of culture, and 10 % FCS was introduced into the culture medium after six weeks. The medium was renewed once per week. For the gene expression analysis, 33 mast cell cultures were initiated from CD133+ cells that were obtained from individual donors. At the time points indicated in Table 1, aliquots of the cell suspensions were withdrawn from the culture for analysis. Seven cultures were continued until the seventh week. For the DNA methylation analysis, additional 8 cultures were initiated from CD133+ cells that were obtained from individual donors. At 3 and 5 weeks of culture, aliquots of equal numbers of cells from two of the cultures, or in one case four, were withdrawn, pooled, and analyzed.
Real-time RT-PCR
RNA was isolated using TRI Reagent® from Molecular Research Center. Aliquots of 1–5 μg of total RNA were reverse-transcribed using RevertAid™ H Minus M-MuLV Reverse Transcriptase (Fermentas, Vilnius, Lithuania) in the presence of oligo-dT18 primers in a 20 μl RT reaction. The level of cognate cDNA was measured by real-time PCR amplification performed on a LightCycler 480 (Roche, Basel, Switzerland) using SYBR Green I master mix (Roche). The cycling conditions were as follows: 95 °C 10 s, 55 °C 10 s, and 72 °C 20 s. The following intron spanning primers were used: 5′-GAAAGCGCAAGTCTTCAAAG-3′ and 5′-TGGGTAGGAGATGGAGATGC-3′ for HIF1A, 5′-CATCACAGAAACGACAGTCAA-3′ and 5′-ACTCCGCACAGCTGGAGG-3′ for CD34, 5′-TGTGGCAGCTGGACTATGAGTCT-3′ and 5′-ACTTCTCACGCGGAGCTTTTAT-3′ for FCER1A, 5′-ACAGGAGCCTTCCAGTGTGCCT-3′ and 5′-CAGTGGTGGGGATGAGGCCG-3′ for MS4A2, 5′- GGTTGAACAAGCAGCGGCCC-3′ and 5′-TTCCTGGTGCTCAGGCCCGT-3′ for FCER1G, 5′-GGGGAGGGGAGAGCCCACTG-3′ and 5′-CTTGACGTCCGGTCCCACGC-3′ for TPSAB1, 5′- TCATGCCAGGGCGACTCCG-3′ and 5′-ACCCAGGTGGACACCCCAGG-3′ for TPSAB2, and 5′-ACTTCCAACGGTCCCTCAAA-3′ and 5′- CCTCAAGCTTCTGCCATGTG-3′ for CMA1, 5′-ATTTTCTCTGCGTTCTGCTCCTAC-3′ and 5′-CGCCCACGCGGACTATTA for KIT, 5′-AGGATGAAAAACAAGCAGACATCA-3′ and 5'-CAGACTGGATGGCTTGGGATT-3′ for CPA3, 5'-GAATGACGCCCTCAATCAAAGT-3′ and 5′-TCATCTTGGGCAGTCACATACA for IL1A, 5′-ACAGATGAAGTGCTCCTTCCA-3′ and 5′-GTCGGAGATTCGTAGCTGGAT-3′ for IL1B, 5′-AAGAGACCTTGGCACTGCTTTC-3′ and 5′-GGAACAGGAATCCTCAGAGTCTCA for IL5, 5′-TGAGGAGCTGGTCAACATCA-3′ and 5′-CAGGTTGATGCTCCATACCAT-3′ for IL13, 5′-ATGCCCGACCTCAACTCCTCCACT-3′ and 5′-GCCACCCAGCTGCAAGATTTC-3′ for IL16, 5′-GACGCATGCCCTCAATCC-3′ and 5′-TAGAGCGCAATGGTGCAAT-3′ for IL18, 5′-CAAGGGCTACCATGCCAAC-3′ and 5′-AGGGCCAGGACCTTGCTG-3′ for TGFB1, 5′-AGGAGGAGGGCAGAATCATCA-3′ and 5′-CTCGATTGGATGGCAGTAGCT-3′ for VEGFA, 5′-GCAACCAGTTCTCTGCATCA-3′ and 5′-TGGCTGCTCGTCTCAAAGTA-3′ for CCL3, 5′-ACCACACCCTGCTGCTTTGC-3′ and 5′-CCGAACCCATTTCTTCTCTGG-3′ for CCL5, 5′-GGCCAGCCACTACAAGCAGCACT-3′ and 5′-CAAAGGGGATGACAAGCAGAAAG-3′ for CSF2, 5′-ACTTTGCAGCCAGGTACGTC-3′ and 5′-CTCAACCGAGAGATGCGAGC-3′ for TPSG1
HPRT1, HMBS, B2MG, RPL13A, TBP, and ACTB were chosen as the most reliable reference genes by a selection procedure that was developed by Vandesompele et al. (2002). The relative gene expression was calculated using ΔΔC T method with the geometric mean of selected reference genes.
Genomic DNA isolation and analysis of the in vivo methylation status of the human HIF1A and CD34 promoters
The genomic DNA from human cells was isolated using the Genomic DNA Extraction Kit (Panomics, Fremont, USA). The DNA was then sheared into 500–100 bp fragments by sonication (6 sets of 10-second pulses on wet ice) using a VCX130 sonicator (Sonics Inc., Newtown, USA). The methylated DNA was isolated based on the methylated CpG island recovery assay (MIRA) (Rauch and Pfeifer 2005) using a MethylCollectorTM Ultra Kit (Active Motif, Carlsbad, USA). The recovered methylated DNA fragments were purified by phenol/chloroform extraction, followed by ethanol precipitation, and then analyzed by RT-PCR with primers specific for the HIF1A promoter (forward: 5′-AGGATCACCCTCTTCGTCG-3′ and reverse: 5′-GCGGAGAAGAGAAGGAAAGG-3′; position: −259/−80) and the CD34 promoter (forward: 5′-ACTCGGTGCGTCTCTCTAGG-3′ and reverse: 5′-CCGTGAGACTCTGCTCTGC-3′; position: −35/+172). The relative amount of PCR product was calculated by transforming the Ct values according to the following formula: relative amount of PCR product = 1,000 × (2ΔCt), where ΔCt = Ctsample − Ctinput.
Bioinformatics
The promoter sequences of all of the analyzed genes were identified and downloaded from the UCSC genome browser (Kent et al. 2002). For the analysis of potential methylation sites in the selected gene promoters, the CpG Island Searcher was applied (Takai and Jones 2003). The putative HIF1α binding motifs were identified using MatInspector Software (Cartharius et al. 2005).
Statistics
The statistical significance of the data was evaluated with the Student parametric test and the Mann–Whitney and Kruskal–Wallis nonparametric tests using the SPSS 10 software. The statistical analysis of the in vivo methylation data was performed using a one-way ANOVA followed by the Dunn’s post hoc test. A p value of 0.05 or lower was considered statistically significant.
Results
In this study, we employed published experimental protocols for the culture of human mast cells. To verify the cell culture protocol, CD133+ cells were purified from the buffy coat preparations of five independent donors (>85 % purity), and 1–2 × 106 cells were cultured for 6 weeks. The cells were then analyzed for the presence of metachromatically stained granules and for surface expression of c-Kit and FcεRI. Greater than 92 % of the cells stained positive for toluidine blue, c-Kit, and FcεRI (data not shown). Next, the cells were harvested at the indicated time points and were analyzed by real-time PCR for the expression of several genes that are preferentially expressed either in hematopoietic progenitors or in mature mast cells. As seen in Fig. 1, CD34 (the marker of hematopoietic progenitors) was hardly detectable in PBMC but showed a high level of expression in the purified CD133+ cells 48 h post-magnetic bead separation. This expression level decreased dramatically after the first week of culture and reached a level comparable to that observed in the PBMC by the second week of culture. We next analyzed the expression of a series of genes that are known to be specifically expressed in mature human mast cells: FCER1A, which codes for the alpha polypeptide of the Fc fragment of the high affinity IgE receptor; MS4A2, which codes for the beta chain of the Fc fragment of the high affinity IgE receptor; TPSB2, which codes for the mast cell granule-specific tryptase; and CMA1, which codes for the mast cell granule-specific chymase, and KIT which codes for c-Kit receptor. Expression of KIT has steadily increased from the low level observed in CD133+ cells (median 0.02) to the significantly higher expression at the third (median 11.56) and the fourth (median 19.66) week of culture (data not shown). As seen in Fig. 2a, the expression of CMA1 in the PBMC and CD133+ cells was below the level of detection. However, it increased significantly following two weeks of culture and showed the highest level of expression in the third week of the differentiating culture. The expression of FCER1A in the PBMC was above the level of detection, likely due to the presence of a small fraction of FcεRI positive cells, but was below the limit of detection in the purified CD133+ cells. FCER1A expression significantly increased within the first week of the differentiating culture and further increased, reaching the maximum expression level, in the fourth week of culture (Fig. 2b). Similar kinetics were observed for the expression of MS4A2, which was also undetectable in CD133+ cells: the expression significantly increased in the first week of culture and reached the maximum expression in the third week of culture (Fig. 2c). The expression of TPSB2 was initially undetectable in the CD133+ cells. However, it increased significantly by the second week of culture and steadily increased until the fifth week, when it reached the maximum expression level (Fig. 2d) Continuation of culture until the seventh week previously reported as optimal time for development mast cell-specific phenotype (Holm et al. 2008) did not result in significant differences in FCER1A and TPSB2 expression as compared to the fifth week (data not shown). Thus, we observed increased expression of mast cell-specific genes upon culturing CD133+ cells under mast cell differentiating conditions. We next investigated the changes in HIF1A expression during culture. As seen in Fig. 3, the purified CD133+ cells revealed lower expression of HIF1A as compared to the PBMC. During in vitro differentiation, the expression of HIF1A significantly increased following three weeks of culture as compared to the CD133+ cells, and remained at this level until the seventh week of culture (data not shown). Thus, the expression of HIF1A paralleled the expression of several genetic markers of mast cell differentiation. We then analyzed the status of DNA methylation at the promoters of all the analyzed genes. We first performed a bioinformatics search for putative CpG islands in the upstream region of the analyzed genes. As seen in Table 2, only the upstream regions of CD34 and HIF1A unambiguously demonstrated the presence of CpG islands. There were no potential CpG islands detected in the upstream regions of the TPSB2, CMA1, MS4A2, and FCER1A genes. We experimentally determined the level of DNA methylation and found that the methylation status of the promoter regions of FCER1A, MS4A2, TPSB2, and CMA1 did not change during the differentiating culture (data not shown). In contrast, the methylation of the HIF1A and CD34 promoters changed during cell differentiation. Upon three weeks of culture under differentiation conditions, the level of DNA methylation at the CD34 promoter significantly increased (Fig. 4a), and the level of DNA methylation at the HIF1A promoter significantly decreased (Fig. 4b). Thus, we observed that during mast cell differentiation, both the expression and the methylation levels of the CD34 and HIF1A promoter sequences were inversely related. It has been previously shown that HIF1α is engaged in the regulation of mast cell cytokine expression (Sumbayev et al. 2012). To further assess the potential role of HIF1A in mast cell function, we analyzed genes of known function that are preferentially expressed in mast cells (Metcalfe 2008) for potential HIF-1 binding to hypoxia responsive elements (HRE). As seen in Table 3, this analysis identified several cytokine, chemokine and protease genes that are potentially regulated by HIF-1A. We next used the established human mast cell line LAD-2 to investigate the effects of hypoxia on the expression of genes that are potentially regulated by HIF-1α. We observed significant increases in the expression of several genes important for mast cell function, such as: c-Kit, IL-1, and RANTES (Table 4).
Fig. 1.
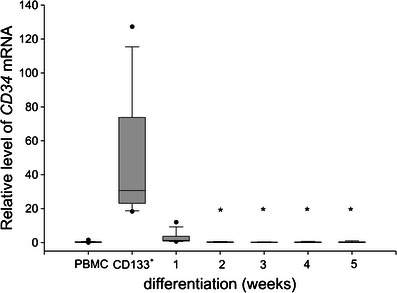
CD34 expression in differentiating human mast cells. Aliquots of PBMC obtained from anonymous blood donors were lysed for RNA isolation (PBMC), and the remaining cells were used for the purification of CD133+ cells by magnetic separation. Aliquots of the purified CD133+ cells were lysed for RNA isolation (CD133+), and the remaining cells were cultured under mast cell differentiating conditions for up to five weeks. At the indicated time points, the cells were lysed for RNA purification. The RNA was used to determine CD34 expression by real-time RT-PCR. The data obtained from a total of 35 donors and 33 mast cell cultures, each initiated from a single donor, are presented as median, lower and upper quartile, and minimum and maximum for each experimental condition from at least 11 cultures. * indicates significant differences at p < 0.05 as compared to the CD133+ cells
Fig. 2.
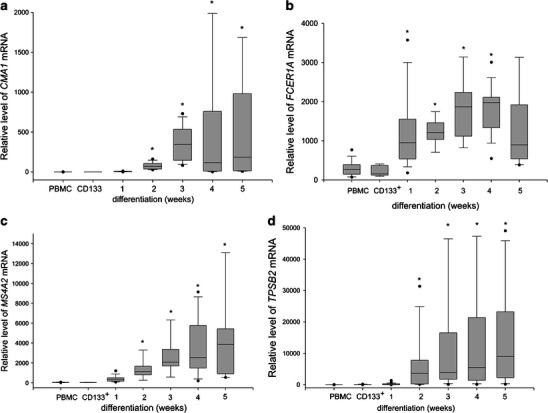
Mast cell-specific gene expression in differentiating human mast cells. CD133+ cells obtained from PBMC of individual blood donors by magnetic separation were cultured under mast cell differentiating conditions for up to five weeks. Aliquots of PBMC (PBMC), purified CD133+ (CD133+), and cell cultured for indicated time were lysed for RNA purification and the RNA was used to determine CMA1 (a), FCER1A (b), MS4A2 (c), and TPSB2 (d) expression by real-time RT-PCR. The data obtained from a total of 35 donors and 33 mast cell cultures, are presented as median, lower and upper quartile, and minimum and maximum for each experimental condition from at least 11 cultures. * indicates significant differences at p < 0.05 as compared to the CD133+ cells
Fig. 3.
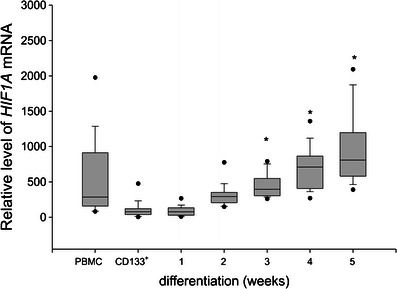
HIF1A and CD34 expression in differentiating human mast cells. CD133+ cells were obtained from PBMC of individual blood donors by magnetic separation and were cultured under mast cell differentiating conditions for up to five weeks. Aliquots of PBMC (PBMC), purified CD133+ (CD133+), and cell cultured for indicated time were lysed and the lysates were used for RNA purification. The RNA obtained from the different cell populations was used to determine HIF1A expression by real-time RT-PCR. The data obtained from a total of 35 donors and 33 mast cell cultures, are presented as median, lower and upper quartile, and minimum and maximum for each experimental condition from at least 11 cultures. * indicates significant differences at p < 0.05 as compared to the CD133+ cells
Table 2.
Summary of bioinformatics-based analysis of promoter regions of investigated genes and experimental analysis of their methylation status during mast cell differentiation
| Gene | CpG island | %GC | ObsCpG/ExpCpG | Length [bp] | Observed change in methylation upon differentiation |
|---|---|---|---|---|---|
| FCER1A | No | – | – | – | None |
| TPSAB1 | No | – | – | – | None |
| TPSB2 | No | – | – | – | None |
| CMA1 | No | – | – | – | None |
| CD34 | −197/+200 | 66.8 | 0.782 | 397 | Increase |
| HIF1A | −775/+200 | 68.2 | 0.809 | 978 | Decrease |
| MS4A2 | No | – | – | – | None |
Fig. 4.
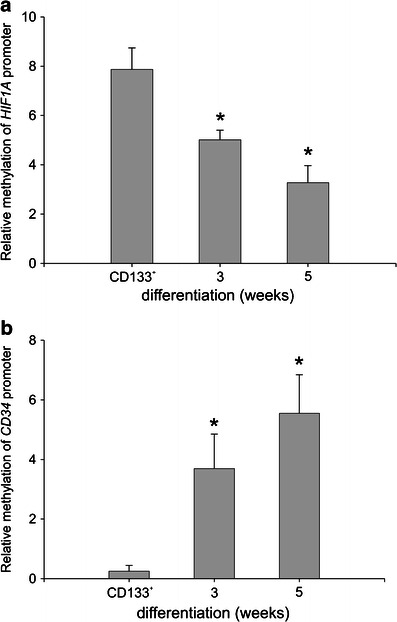
The methylation status of the human HIF1A and CD34 promoters in differentiating human mast cells. Aliquots containing equal numbers of CD133+ cells purified from the PBMC of two to four anonymous blood donors were pooled and lysed for the extraction of genomic DNA (CD133+). The remaining CD133+ cells were seeded into separate flasks and cultured under mast cell differentiating conditions. After 3 and 5 weeks of culture, aliquots containing equal numbers of cells originating from two to four blood donors were pooled and lysed for the extraction of genomic DNA. The DNA was analyzed by a methylated CpG island recovery assay followed by real-time PCR to determine the number of recovered HIF1A (a) promoter and CD34 (b) promoter sequences. The data represent the mean ± SEM of three DNA pools representing a total of eight donors
Table 3.
HIF1α binding motifs identified in the promoters of genes relevant for mast cell function. −2000/+100 sequences were scanned for Putative HIF1α binding sites for each gene. Locations of putative binding sites are relative to the ATG initiation codon
| Class | Gene symbol | Number of HRE | Matrix similarity | Sequence | Strand |
|---|---|---|---|---|---|
| Preformed mediators | CMA1 | 1 | 0.894 | −124gaagaaaaCGTGaccat−108 | (+) |
| CPA3 | 2 | 0.974 | −1763catggaCGTGttttt−1749 | (+) | |
| 0.698 | −603ttcacCACGttggccag−587 | (−) | |||
| CTSG | 0 | ||||
| TPSAB1 | 3 | 0.973 | −776gggtctcaCGTGttggccag−760 | (+) | |
| 0.946 | −731cccaccccCGTGgattc−715 | (+) | |||
| 0.976 | −368cgggggcaCGTGttaca−352 | (+) | |||
| TPSB2 | 2 | 0.946 | −732gccaccccCGTGgattc−716 | (+) | |
| 0.976 | −368gggggCGTGttaca−716 | (+) | |||
| TPSG1 | 5 | 0.982 | −1678tcaggaCGTGgcagg−1664 | (+) | |
| 1.000 | −1598cacccaCGTGgctg−1585 | (+) | |||
| 0.980 | −839ctccccaCGTGctggc−823 | (+) | |||
| 0.976 | −816gcggcCACGtctgtgcc−800 | (−) | |||
| 0.989 | −515ctgccggaCGTGgggac−499 | (+) | |||
| Cytokines | INFG | 0 | |||
| IL1A | 1 | 0.931 | −786gtagcCACGTagccacg−770 | (−) | |
| IL1B | 1 | 0.948 | −857tcttctaaCGTGggaaa−841 | (+) | |
| IL4 | 0 | ||||
| IL5 | 2 | 0.945 | −1502accaaaCGTGgtaca−1488 | (+) | |
| 0.955 | +40gctgcctaCGTGtatgc+56 | (+) | |||
| IL6 | 0 | ||||
| IL8 | 0 | ||||
| IL13 | 2 | 0.975 | −1998cggCAAGTgcgcgc−1985 | (−) | |
| 0.945 | −809gccagacaCGTGcacac−793 | (+) | |||
| IL16 | 1 | 0.973 | −1725gagCACGTagaccc−1712 | (−) | |
| IL18 | 1 | 0.892 | −1600gggcaaCGTGgtgaa−1586 | (+) | |
| TGFB1 | 2 | 0.989 | −1855gccccaCGTGgcggc−1841 | (+) | |
| 0.871 | −1645ctccaACGTcaccac−1631 | (+) | |||
| TNF | 1 | 0.975 | +19atccgggaCGTGgagct+35 | (+) | |
| Chemokines | CCL2 | 0 | |||
| CCL3 | 1 | 0.955 | +65ctgcatCACGtgagtct+81 | (+) | |
| CCL5 | 1 | 0.973 | −41agacagcaCGTGgacct−25 | (+) | |
| Growth | CSF2 | 1 | 0.986 | −1141tgggcaCGTGtgggc−1127 | (+) |
| factors | VEGFA | 2 | 0.987 | −1552tgaggaCGTGtgtgt−1538 | (+) |
| 0.892 | −1353tccgaACGTaacctc−1339 | (+) | |||
| Receptors | FCER1A | 0 | |||
| FCER1G | 0 | ||||
| KIT | 4 | 0.916 | −814ggggaaaaCGTGtatga−798 | (+) | |
| 0.979 | −496cgcccgaaCGTGctcga−480 | (+) | |||
| 0.916 | −424tgccctaaCGTGtgcgt−408 | (+) | |||
| 0.897 | −45agctggaaCGTGgacca−29 | (+) | |||
| MS4A2 | 0 | ||||
Table 4.
Changes in gene expression in LAD-2 mast cells under hypoxia
| Gene symbol | Relative gene expression | |
|---|---|---|
| Normoxia | Hypoxia | |
| KIT | 112 ± 24 | 401 ± 118* |
| CPA3 | 4 914 ± 239 | 102 130 ± |
| IL1A | 0 | 50 ± 16* |
| IL1B | 0 | 1 ± 1 |
| IL5 | 0 | 4 ± 2* |
| IL13 | 0 | 4 ± 4 |
| IL16 | 29 ± 3 | 24 ± 10 |
| IL18 | 96 ± 8 | 139 ± 48 |
| TGFB1 | 58 ± 6 | 1 281 ± 1054 |
| VEGFA | 25 ± 4 | 444 ± 209* |
| CCL3 | 106 ± 5 | 91 ± 5 |
| CCL5 | 7 ± 2 | 21 ± 5* |
| CSF2 | 87 ± 23 | 65 ± 15 |
| CMA1 | 254 ± 83 | 346 ± 66 |
| TPSB2 | 62 ± 18 | 100 ± 42 |
| TPSG1 | 1 ± 0 | 2 ± 0 |
LAD-2 mast cells were cultured for 5 days under normoxia (21 % O2) or hypoxia (1 % O2) and the gene expression was determined by real-time RT-PCR using ΔΔC T method with the geometric mean of selected reference genes. Data are presented as the mean ± SEM from four independent experiments (n = 4)
p < 0.05 statistically significant difference
Discussion
We report here that HIF1A expression significantly increased during the differentiation of peripheral blood-derived CD133+ cells into mast cells. While the level of HIF1A expression in undifferentiated CD133+ cells was significantly lower than in the PBMC, the level observed in mast cells was comparable to the PBMC, which consist mainly of differentiated hematopoietic cells (Fig. 3). This is similar to our previously reported observations that CD34+ cells isolated from the bone marrow and the peripheral blood revealed lower levels of HIF1A expression than mature hematopoietic and non-hematopoietic cells (Walczak-Drzewiecka et al. 2010). Upon culture under differentiating conditions, the level of HIF1A expression increased in parallel to genes encoding characteristic mast cell proteins, such as the surface receptor for Fc IgE and granule associated proteases (Fig. 2). This suggested that the observed changes in HIF1A expression are related to the process of mast cell differentiation and that increased HIF1A expression relative to progenitor cells is a marker of mature mast cells. The level of HIF1A expression in cultures predominantly containing differentiated mast cells was similar to that observed in PBMC. This is consistent with the hypothesis that increased HIF1A expression is not necessarily a phenomenon specific to mast cell differentiation, but might represent a general phenomenon of the differentiation of hematopoietic progenitors into mature blood cells.
One possible explanation for the increased baseline HIF1A expression in resting cells during differentiation is an epigenetic regulatory process. This hypothesis is consistent with the presence of large CpG islands in the HIF1A promoter region (Table 2). It also agrees with the observed decrease in methylation at the HIF1A promoters of differentiated mast cells at three and five weeks of culture (Fig. 3), which paralleled a significant increase in HIF1A expression. Interestingly, the CD34 promoter, which also contains CpG islands (Table 2), simultaneously demonstrated increased methylation that paralleled decreased CD34 expression. The opposing methylation statuses of these two genes suggests that they represent components of a complex epigenetic mechanism, which mediates cell maturation by silencing the genes involved in maintaining the phenotype of hematopoietic stem cells and upregulating the genes necessary for mature mast cell functions.
There is experimental evidence that HIF1A is essential for the cell cycle quiescence of hematopoietic and other stem cells, possibly due to its role in Notch-dependent signaling (Bar et al. 2010; Mukherjee et al. 2011; Takubo et al. 2010). This implies that the amount of HIF-1α mRNA in hematopoietic stem cells is distinct from mature hematopoietic cells and is tightly regulated, at least in part, by methylation of the HIF1A promoter region. The change in the level of HIF1A promoter methylation upon mast cell differentiation, resulting in increased levels of HIF-1α mRNA, might be important for the regulation of multiple HIF1A regulated genes that are necessary to maintain a mature mast cell phenotype. This hypothesis is consistent with the previously reported observations that the HIF1A product, HIF-1α, plays a role in LPS-mediated TNF release in human mast cells (Sumbayev et al. 2012) and our observations that genes important for mast cell function, such as Kit and IL1A, contain promoter HRE motifs (Table 3) and are influenced by changes in oxygen concentrations (Table 4).
Previously reported epigenetic regulation of HIF1A expression has involved the methylation of DNA in the region overlapping the HIF1A proximal promoter (Koslowski et al. 2010; Walczak-Drzewiecka et al. 2010). Considering the sequence of the upstream region that showed decreased methylation levels, one possible mechanism for the upregulation of HIF1A is the demethylation of cytosines that neighbor the HRE binding sites. These cytosines have previously been shown to regulate the accessibility of the HRE binding motif, which has been shown to positively autoregulate HIF1A expression in colon cancer cells exposed to hypoxia (Koslowski et al. 2010). However, the observed increase in HIF1A expression in differentiating human mast cells occurred under high concentrations of oxygen, and the same HRE motif was also reported to mediate negative regulation of HIF1A in other cell types (Xu et al. 2012). Furthermore, there are other binding motifs at close proximity to the HRE motif, including SP1, that are necessary for HIF1A promoter activity (Iyer et al. 1998; Minet et al. 1999).
In summary, we present evidence that during in vitro mast cell differentiation, HIF1A expression is upregulated by an epigenetic mechanism involving DNA methylation. This suggests that the baseline expression of HIF1A is epigenetically regulated in a cell type- and differentiation stage-specific fashion. Such precise regulation of HIF1A expression may be important for the cellular response to varying oxygen availability in particular organs and tissues.
Acknowledgments
This work was supported by the National Science Center grant no. N N301 164335
References
- Bar EE, Lin A, Mahairaki V, Matsui W, Eberhart CG. Hypoxia increases the expression of stem-cell markers and promotes clonogenicity in glioblastoma neurospheres. Am J Pathol. 2010;177:1491–1502. doi: 10.2353/ajpath.2010.091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreau A, El Hafny-Rahbi B, Matejuk A, Grillon C, Kieda C. Why is the partial oxygen pressure of human tissues a crucial parameter. Small molecules and hypoxia J Cell Mol Med. 2011;15:1239–1253. doi: 10.1111/j.1582-4934.2011.01258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Gorlach A. Regulation of HIF-1alpha at the transcriptional level. Curr Pharm Des. 2009;15:3844–3852. doi: 10.2174/138161209789649420. [DOI] [PubMed] [Google Scholar]
- Holm M, Andersen HB, Hetland TE, Dahl C, Hoffmann HJ, Junker S, Schiotz PO. Seven week culture of functional human mast cells from buffy coat preparations. J Immunol Methods. 2008;336:213–221. doi: 10.1016/j.jim.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Iyer NV, Leung SW, Semenza GL. The human hypoxia-inducible factor 1alpha gene: HIF1A structure and evolutionary conservation. Genomics. 1998;52:159–165. doi: 10.1006/geno.1998.5416. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslowski M, Luxemburger U, Tureci O, Sahin U. Tumor-associated CpG demethylation augments hypoxia-induced effects by positive autoregulation of HIF-1alpha. Oncogene. 2010;30:876–882. doi: 10.1038/onc.2010.481. [DOI] [PubMed] [Google Scholar]
- Lauzier MC, Page EL, Michaud MD, Richard DE. Differential regulation of hypoxia-inducible factor-1 through receptor tyrosine kinase transactivation in vascular smooth muscle cells. Endocrinology. 2007;148:4023–4031. doi: 10.1210/en.2007-0285. [DOI] [PubMed] [Google Scholar]
- MacDougall JD, McCabe M. Diffusion coefficient of oxygen through tissues. Nature. 1967;215:1173–1174. doi: 10.1038/2151173a0. [DOI] [PubMed] [Google Scholar]
- Metcalfe DD. Mast cells and mastocytosis. Blood. 2008;112:946–956. doi: 10.1182/blood-2007-11-078097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe DD, Baram D, Mekori YA. Mast cells. Physiol Rev. 1997;77:1033–1079. doi: 10.1152/physrev.1997.77.4.1033. [DOI] [PubMed] [Google Scholar]
- Minet E, Ernest I, Michel G, Roland I, Remacle J, Raes M, Michiels C. HIF1A gene transcription is dependent on a core promoter sequence encompassing activating and inhibiting sequences located upstream from the transcription initiation site and cis elements located within the 5'UTR. Biochem Biophys Res Commun. 1999;261:534–540. doi: 10.1006/bbrc.1999.0995. [DOI] [PubMed] [Google Scholar]
- Mukherjee T, Kim WS, Mandal L, Banerjee U. Interaction between Notch and Hif-alpha in development and survival of Drosophila blood cells. Science. 2011;332:1210–1213. doi: 10.1126/science.1199643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh T, Sanosaka T, Ito K, Nakashima K. Oxygen levels epigenetically regulate fate switching of neural precursor cells via hypoxia-inducible factor 1alpha-notch signal interaction in the developing brain. Stem Cells. 2012;30:561–569. doi: 10.1002/stem.1019. [DOI] [PubMed] [Google Scholar]
- Peyssonnaux C, Boutin AT, Zinkernagel AS, Datta V, Nizet V, Johnson RS. Critical role of HIF-1alpha in keratinocyte defense against bacterial infection. J Invest Dermatol. 2008;128:1964–1968. doi: 10.1038/jid.2008.27. [DOI] [PubMed] [Google Scholar]
- Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis EA, Gallo RL, Hurtado-Ziola N, Nizet V, Johnson RS. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J Clin Invest. 2005;115:1806–1815. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab Invest. 2005;85:1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- Semenza GL. HIF-1 and mechanisms of hypoxia sensing. Curr Opin Cell Biol. 2001;13:167–171. doi: 10.1016/S0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- Sumbayev VV, Yasinska I, Oniku AE, Streatfield CL, Gibbs BF. Involvement of hypoxia-inducible factor-1 in the inflammatory responses of human LAD2 mast cells and basophils. PLoS One. 2012;7:e34259. doi: 10.1371/journal.pone.0034259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, Shima H, Johnson RS, Hirao A, Suematsu M, Suda T. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, and Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034 [DOI] [PMC free article] [PubMed]
- Walczak-Drzewiecka A, Ratajewski M, Pulaski L, Dastych J. DNA methylation-dependent suppression of HIF1A in an immature hematopoietic cell line HMC-1. Biochem Biophys Res Commun. 2010;391:1028–1032. doi: 10.1016/j.bbrc.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Walczak-Drzewiecka A, Ratajewski M, Wagner W, Dastych J. HIF-1alpha is up-regulated in activated mast cells by a process that involves calcineurin and NFAT. J Immunol. 2008;181:1665–1672. doi: 10.4049/jimmunol.181.3.1665. [DOI] [PubMed] [Google Scholar]
- Xu J, Wang B, Xu Y, Sun L, Tian W, Shukla D, Barod R, Grillari J, Grillari-Voglauer R, Maxwell PH, Esteban MA. Epigenetic regulation of HIF-1alpha in renal cancer cells involves HIF-1alpha/2alpha binding to a reverse hypoxia-response element. Oncogene. 2012;31:1065–1072. doi: 10.1038/onc.2011.305. [DOI] [PubMed] [Google Scholar]
- Zinkernagel AS, Johnson RS, Nizet V. Hypoxia inducible factor (HIF) function in innate immunity and infection. J Mol Med (Berl) 2007;85:1339–1346. doi: 10.1007/s00109-007-0282-2. [DOI] [PubMed] [Google Scholar]


