Abstract
Colorectal cancer (CRC) is a classic example of a tumor that progresses through multiple distinct stages in its evolution. To understand the mechanisms regulating the transition from indolent to invasive disease, we profiled somatic copy number alterations in non-invasive adenomas and invasive adenocarcinomas from Apc and DNA mismatch repair (MMR) mutant mouse models. We identified a recurrent amplicon on mouse chromosome 8 that encodes microRNAs (miRs) 23a and 27a. miRs-23a and 27a levels are upregulated in mouse intestinal adenocarcinomas, primary tumors from stage I/II CRC patients, as well as in human CRC cell lines and cancer stem cells. Functionally, miR-23a promotes CRC cells and stem cells migration and invasion, while miR- 27a primarily promotes proliferation. We computationally and experimentally validated that Metastasis Suppressor 1 (MTSS1) is a direct miR-23a target and similarly validated that the ubiquitin ligase FBXW7 is a direct miR-27a target. Analyses of computationally predicted target genes in CRC patient microarray datasets are consistent with a role for miR-23a, but not miR-27a, specifically in invasive CRC.
Keywords: Colorectal cancer, microRNAs, tumor progression
INTRODUCTION
Colorectal cancer (CRC) is the 2nd leading cause of cancer death in the United States (1). CRC is a classic example of a tumor that progresses through multiple distinct stages in its evolution. Mutations activating WNT (most often APC) and KRAS pathways occur as an early event in cancer cells (2, 3). Subsequently, mutations in TGF-β, PI3-Kinase, TP53 pathways, DNA mismatch repair genes (MMR), FBXW7 and others accumulate in these long lived cells (4, 5). Morphologically, inappropriate proliferation causes formation of adenomas. Adenomas progress to carcinoma in situ and early stage CRCs. This process typically lasts several years (6, 7). Then, in a relatively short time early stage CRCs acquire the ability to invade through the colon wall, metastasize, and survive outside the colon niche microenvironment (6, 7). As 5 year survival for indolent CRC is ~90% vs. 10–15% for metastatic CRC, understanding the mechanisms that regulate the transition from indolent adenomas and carcinoma in situ to invasive and metastatic CRC is critical to improving patient outcomes (8).
MicroRNAs (miRs) are small, endogenous non-coding RNAs that simultaneously regulate levels of multiple proteins, primarily by binding to the 3’ UTR of targets and inhibiting protein translation(9). Important roles for miRs have been demonstrated in multiple types of cancer, including roles in tumor progression by modulating mechanisms of differentiation, proliferation, invasion and metastasis (10). Expression of the miR-23a~27a~24-2 cluster is altered in many cancers and has diverse effects (11). These include orchestration of target genes important for increasing proliferation, cell differentiation and growth (12, 13). The three miRs of this cluster are derived from a single primary transcript but the levels of each can vary because of post-transcriptional processing (i.e. levels of one or two of these miRs can increase while the third does not) (12).
Previously, we described a mouse model system to study the mechanisms regulating the transition from pre-invasive to invasive intestinal/colon tumors (14). Apc1638N and MMR-deficient (Mlh3−/−; Pms2−/−) mice develop almost exclusively adenomas and pre-invasive adenocarcinomas, while dual Apc1638N; MMR-deficient mice develop almost all invasive adenocarcinomas (14). Here, we use this model system to identify and characterize a novel mouse chromosome 8 locus that is amplified in invasive vs. pre-invasive intestinal adenocarcinomas and contains the miR-23a/24-2/27a cluster. Expression levels of miRs 23a and 27a are upregulated in Apc mutant/MMR-deficient invasive adenocarcinomas. miR-23a is upregulated specifically in invasive primary CRCs from stage I/II patients, while miR-27a levels are upregulated in primary CRCs from patients with disease that has spread beyond the colorectum (stage III/IV). Both miRs are also highly expressed in CRC cell lines and stem cells. Mechanistically, in CRC cell and cancer stem cell lines the ubiquitin ligase F-box protein FBXW7 (the 4th most commonly mutated gene in CRC) is a direct miR-27a target (15). In CRC stem cells, FBXW7 promotes proteasomal degradation of the transcription factors MYC and JUN, and downregulates NOTCH signaling components. Consequently, FBXW7 inhibition by miR-27a increases MYC, JUN and NOTCH signaling, promotes proliferation and prevents secretory lineage differentiation (16). Similarly, we show that Metastasis Suppressor 1 (MTSS1) is a direct miR-23a target; MTSS1 interacts directly with cortactin to promote filopodia formation and upregulates SRC signaling (17). Reduced MTSS1 levels promote CRC cell and cancer stem cell migration, invasion and metastasis. In vivo, miR-27a is required for subcutaneous CRC cell xenograft (18, 19) tumor growth, and both miR-23a and 27a are required for formation of hematogenous metastases. Computational analyses of publically available CRC gene expression profiling datasets are consistent with a role for miR-23a, but not miR-27a, specifically in invasive CRC. Overall, these data support a potential mechanistic role for miR-23a and its target genes in the transition from indolent to invasive CRC.
RESULTS
High Resolution Tiling Array Profiling of Mouse Intestinal Adenomas and Adenocarcinomas
To investigate the mechanisms that cause progression of intestinal adenomas to adenocarcinomas, we performed high-resolution tiling array based somatic copy number profiling of mouse chromosomes 6, 7,8 and 9 in Apc−/− ;MMR-deficient adenocarcinomas vs. MMR-deficient adenomas, which we had previously shown using array comparative genomic hybridization to contain amplified loci (14). This identified a recurrent 5MB amplicon on mouse chromosome 8 with increased copy number in Apc−/−;MMR-deficient invasive adenocarcinomas vs. normal mucosa (Fig. 1A; Supplemental Fig. S1). This amplicon contains both multiple coding genes and microRNAs. First, we evaluated the coding genes in the amplicon critical interval using quantitative real-time PCR. However, none of the protein coding genes in the amplicon had significantly or consistently increased mRNA expression levels in Apc−/−MMR-deficient invasive adenocarcinomas (Supplemental Fig. S2). Therefore, we next evaluated non-coding genes in this amplicon.
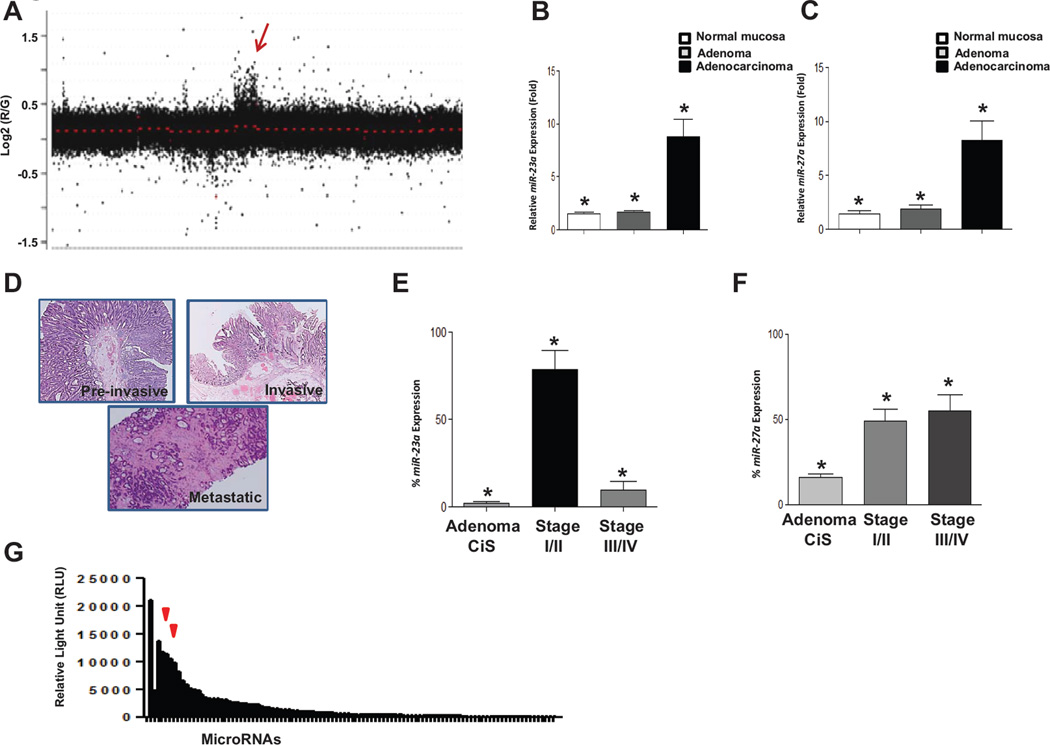
Figure 1.
Array-CGH analysis of Apc/MMR dual deficient Gl tumors. A, depiction of aCGH hybridization signal from a representative adenocarcinoma (additional tumors are shown in supplemental figure 1). The arrow indicates a region with gain of signal on chromosome 8. B,C, quantitative PCR of miR-23a and miR-27a levels in cDNA from MMR-deficient adenomas and Apc/MMR dual deficient adenocarcinomas. Levels are shown in fold relative to matched normal tissue. * denotes Student t test p value<0.001. D, representative images of a pre-invasive (adenoma, carcinoma in situ) (top), invasive (stage I, II) (middle) and metastatic (stage III and IV)(bottom) tumor stained with H&E. E, miR-23a level in pre-invasive tumors and primary CRCs from patients with invasive and metastatic disease. Pre-invasive (adenoma, carcinoma in situ), invasive (stage I, II) and metastatic (stage III and IV). * denotes Student t test p value<0.0001 in comparisons of pre-invasive vs. invasive levels or invasive vs. metastatic CRC expression level. Adenoma CiS n=31, stage I/II n=31, stage III/IV n=31. F, miR-27a levels in pre-invasive, invasive and metastatic human CRCs. Pre-invasive (adenoma, carcinoma in situ), invasive (stage I, II) and metastatic (stage III and IV). * denotes Student t test p value<0.005 in comparisons of pre-invasive vs. invasive levels or invasive vs. metastatic CRC expression levels. Adenoma CiS n=31, stage I/II n=31, stage III/IV n=31. G, array based microRNA profiling shows that miRs-23a and 27a are among the most highly expressed in colon cancer stem cells (left arrow miR-23a, right arrow miR-27a). The full list of microRNAs profiled is given in Supplemental Table 1.
miR-23a and miR-27a Expression Levels Are Increased in Mouse Intestinal Invasive Adenocarcinomas and Human Invasive/Metastatic CRCs
MicroRNAs 23a, 24-2, 27a and 181c are also contained in the critical interval for this amplicon. Using a stem-loop miR-qRT-PCR assay we confirmed that miR-23a and 27a levels were increased in Apc−/−; MMR-deficient adenocarcinomas as compared to MMR-deficient adenomas or normal mucosa (Fig. 1B and 1C), consistent with a potential role in tumor progression. In contrast, neither miR-24 nor miR-181c expression was significantly elevated (data not shown).
To understand whether microRNAs 23a and 27a are upregulated in human CRCs as well, we measured their expression levels in (a) pre-invasive tumors (adenomas and carcinoma in situ),(b) primary CRCs from patients with locally invasive disease (stage I/lI), or (c) primary CRCs from patients with tumor cells that had metastasized outside the colorectum (stage III/IV), each normalized to adjacent normal colon tissue from the same patient as control (Fig. 1D). Expression levels of miR-23a were upregulated in primary CRCs from stage I/II patients vs. pre-invasive adenomas and carcinoma in situ (p=0.0001). miR-23a upregulation was specific to CRCs from stage I/lI patients, as primary CRCs from stage III/IV patients had lower miR-23a levels vs. CRCs from stage I/II patients (p=0.0001) (Fig. 1E). miR-27a expression levels were higher in both comparisons of either stage I/II or III/IV CRCs vs. pre-invasive colon adenomas and carcinoma in situ (Fig. 1F). In summary, the human CRC and mouse intestinal tumor expression data are consistent with potential roles for miR-23a and 27a in CRC progression from pre-invasive to locally invasive and metastatic disease. Importantly, miR-23a expression is specifically higher in CRCs from patients with locally invasive (stage I/II) disease.
MicroRNA Profiling Reveals High Endogenous Levels of miRs 23a and 27a in Colon Cancer Stem Cells and Commonly Used Colon Cancer Cell Lines
Recent studies have highlighted the important role of colon cancer stem cells (CCSC) in CRC progression and metastasis (20). CCSC have high tumorigenicity and self-renewal capacity and are often found at the leading edge of invasive CRCs. CCSC are also proposed to play critical roles in seeding extra-colonic metastases. To understand the roles of miRs 23a and 27a in CCSC, we performed LNA-microarray based microRNA profiling of CCSC. MicroRNA profiling of CCSC showed that both miR-23a and miR-27a are among the most highly expressed miRs in CCSC, consistent with potential functional roles (Fig. 1G; Supplemental Table S1). We also tested several commonly used non-CCSC CRC cell lines for miR-23a and miR-27a expression and found that both microRNAs were highly expressed (Supplemental Fig. S3A). Overall, these data confirm mouse intestinal tumor and human CRC data and show that miRs 23a and 27a are highly expressed in both CCSC and non-CCSC CRC cell lines. There was no difference between expression levels of miR-23a or 27a in CRC patient MMR-proficient vs. deficient tumors (data not shown).
FBXW7 Is a Direct miR-27a Target
We used computational target prediction algorithms (21–24) to identify miR-27a target genes, with putative miR-27a binding motifs in their 3’UTRs. One of the targets shared by all algorithms tested included F-box and WD repeat domain-containing 7 (FBXW7), which is mutated in 6–9% of CRCs (25–27). FBXW7 encodes an ubiquitin ligase that regulates proteasomal degradation of binding substrates (15, 16, 25). FBXW7 is a key regulator of several signaling pathways, including NOTCH, MYC and JUN (16), and FBXW7 mutation stimulates cell proliferation (16). The miR-27a binding site in the FBXW7 3’-UTR (nucleotides 502–508 and 1378–1384) (Fig. 2A, top) is highly conserved in different species. Using a luciferase reporter construct expressing the FBXW7 3’-UTR, we confirmed that FBXW7 is a direct target of miR-27a (Fig. 2A, bottom). We then used a lentiviral construct expressing anti-miR-27a and infected CRC cell lines and CCSCs. Western blot analysis confirmed that miR-27a knockdown significantly increases FBXW7 levels vs. control shRNA transduced cells (Fig. 2B). Altogether, these data are consistent with FBXW7 as a direct target of miR-27a.
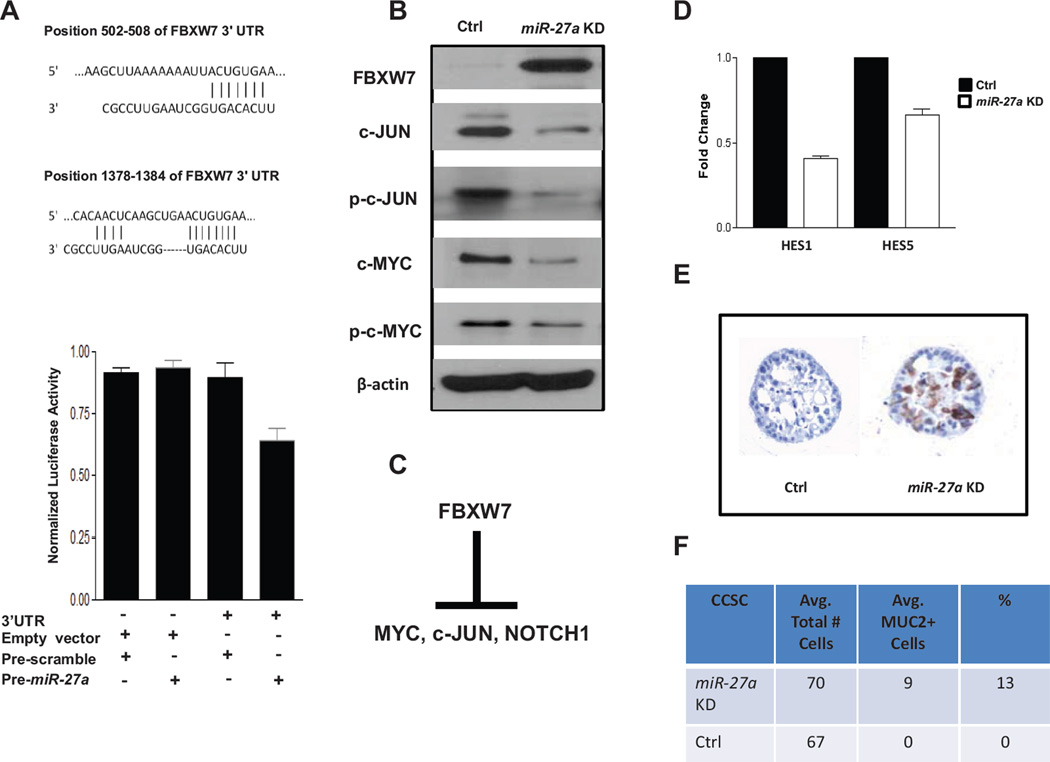
Figure 2.
FBXW7 is a direct target of miR-27a. JUN and MYC are downstream targets. A, schematic of miR-27a binding sites in FBXW7 mRNA 3’UTR sequences at nt (502–508 and 1378–1384) (top), miR-27a binding activity luciferase reporter assay in colon cancer stem cells CCSC (bottom). CCSC were transfected with a plasmid containing the FBXW7 miR-27a binding site fused to the 3' UTR of Firefly luciferase and co-transfected with plasmids driving expression of pre-miR-27a or a control insert sequence. Luciferase protein levels and activity are suppressed when a miR binds specifically to the 3' UTR target sequence. B, analysis of FBXW7, phospho and total c-JUN, phospho and total c-MYC protein levels. C, schematic of FBXW7 downstream targets under normal cellular conditions. D, qRT-PCR of NOTCH signaling downstream targets HES1 and HES5. E, bioassay of MUC2 immunocytochemistry+ cells. MUC2+ cells (brown HRP stain) indicate suppression of NOTCH signaling in CCSC. F, table with quantification of MUC2+ cells in miR-27a or control shRNA knockdown CCSC.
miR-27a Knockdown Increases FBXW7 Protein Levels and Downregulates MYC, JUN and NOTCH Signaling
c-MYC, c-JUN, Cyclin-E and several NOTCH pathway components are FBXW7 substrates (Fig. 2C) (28, 29). To test whether miR-27a regulates these downstream targets, we used lentiviral anti-miR-27a knockdown in CCSC (Supplemental Fig. S3B), which have active MYC, JUN and NOTCH signaling (20, 30). Compared with lentiviral expression of a control scrambled sequence shRNA, lentiviral miR-27a knockdown reduced protein levels of FBXW7, JUN and MYC. Similarly, phosphorylated c-JUN and c-MYC were also reduced, consistent with downregulated JUN and MYC signaling (Fig. 2B).
Next, we evaluated whether miR-27a knockdown in CCSC impairs NOTCH signaling
The downstream canonical NOTCH pathway target genes HES1 and HES5 are upregulated by active NOTCH signaling (31). Consistent with miR-27a upregulation of NOTCH signaling, CCSC with miR-27a knockdown had lower HES1 and HES5 levels than control cells (Fig. 2D). In CCSC, as well as normal intestinal stem cells, NOTCH signaling suppresses secretory lineage differentiation (20). To confirm that miR-27a knockdown functionally downregulates NOTCH, we performed immunostaining for the secretory lineage marker MUC2. Consistent with miR-27a knockdown downregulating NOTCH signaling, the number of MUC2+ CCSC increased vs. control shRNA infected cells (Fig. 2E and F). In contrast, CCSC levels of Cyclin-E levels, another FBXW7 target, were not affected by miR-27a knockdown (data not shown), suggesting additional signaling feedback pathways regulating Cyclin-E exists in CCSC.
miR-27a Knockdown Inhibits Colon Cancer Stem Cell Proliferation In Vitro and Tumor Formation In Vivo
Next, we tested the impact of miR-27a knockdown on FBXW7 levels, downstream MYC, JUN and NOTCH signaling and cell proliferation. For these experiments, we used two different methods to knockdown miR-27a, LNAs (locked nucleotide analog) and shRNA knockdown (Supplemental Fig. S3B and S3C). Both experimental approaches showed that lowering miR-27a levels consistently and significantly inhibited CCSC proliferation (Fig. 3A; Supplemental Fig. S3D). By comparison, miR-23a knockdown had a more modest effect on cell proliferation (Supplemental Fig. S4A). miR-27a knockdown also significantly impaired CCSC clonogenicity, while the impact of miR-23a knockdown had less effect (Fig. 3B and C; Supplemental Fig. S4B). Next, we tested the impact of miR-27a knockdown on xenograft tumor formation. For both CCSC and LoVo subcutaneous xenograft tumors, compared with lentiviral expression of a control, scrambled sequence shRNA, miR-27a knockdown significantly inhibited tumor growth (P=0.0004) (Fig. 3D–F). In contrast, miR-23a knockdown in CCSC did not significantly cause a reduction in xenograft tumor volume (Supplemental Fig. S4C).
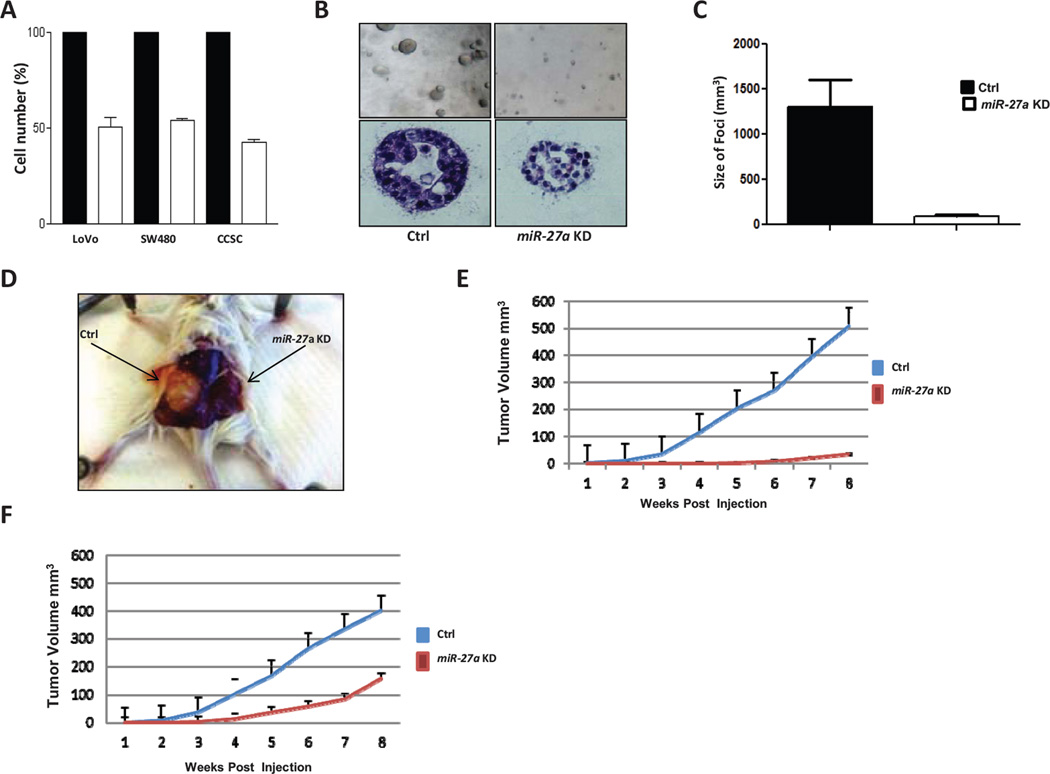
Figure 3.
miR-27a increases CRC cell proliferation, clonogenicity and xenograft tumor volume. A, MTT assay of colon cancer stem cell (CCSC), LoVo, and SW480 cells infected with lentivirus expressing either anti-miR-27a or scrambled control shRNA. In all three lines, anti-miR-27a shRNA significantly reduced the number of viable cells. B, representative photos of CCSC lentiviral transduced with either anti-miR-27a or control shRNA sequences and plated in 3D collagen culture. Top, light microscopy at 10X. Bottom, hematoxylin and eosin staining of CCSC colony at 40X. C, Reduced volume of collagen 3D culture colony size in anti-miR-27a vs. scrambled control sequence lentiviral transduced CCSC, p<0.001. D, photos of xenograft tumors from CCSC transduced with either control shRNA (left) or anti-miR-27a (right). E, growth curve of subcutaneous xenograft tumor volume in mice injected with anti-miR-27a expressing (red) or control sequence (blue) CCSC, p<0.001 by Student t test. F, growth curve of subcutaneous xenograft tumor volume in mice injected with anti-miR-27a expressing (red) or control sequence (blue) LoVo cells, p<0.001 by Student t test. 95% confidence interval error bars are shown.
miR-27a Overexpression In Vitro Promotes Cell Growth and Proliferation in DLD1 Cells
To understand in more detail its mechanistic role, we used lentiviral infection to express constitutively miR-27a in the DLD1 CRC cell line, which has low endogenous levels (Supplemental Figure S5). Complementary to shRNA knockdown studies, upregulation of miR-27a levels significantly increased DLD1 cell proliferation (p<0.01) and in vitro clonogenicity (Supplementary Fig. S6A–C). Overall, these data are consistent with a functional role of miR-27a to stimulate proliferation of both CCSC and CRC cells.
miR-23a Knockdown Reduces CRC and CCSC Cell Motility and Invasion by Upregulating MTSS1 and Downregulating SRC Signaling and Filopodia Formation
To understand the role of miR-23a in CRC progression, we performed in vitro migration and invasion assays in CRC and CCSC cells. Consistently, in both CRC and CCSC cells, lentiviral miR-23a knockdown caused a dramatic decrease in the number of cells migrating or invading to the lower Boyden assay chamber vs. control shRNA transduced cells (Fig. 4A–C). We used a computational approach to identify putative miR-23a targets (21–24), with putative miR-23a binding motifs in their 3’UTRs which could reveal the mechanism of miR-23a knockdown inhibition of cell motility. One high-ranking predicted target was Metastasis suppressor 1(MTSS1); (also known as missing in metastasis, MIM, or BEG4) a recently identified actin-binding protein (17, 19, 32). The MTSS1 3’-UTR contains a putative miR-23a binding site (nt 1949–1955) that is highly conserved across multiple species (Fig. 4D, top). Using a luciferase reporter construct expressing the MTSS1 3’-UTR, we confirmed that MTSS1 is a direct target of miR-23a (Fig. 4D, bottom). Next, using Western blotting we showed that miR-23a knockdown in CCSC significantly increases MTSS1 levels vs. control shRNA transduced cells (Fig. 4E).
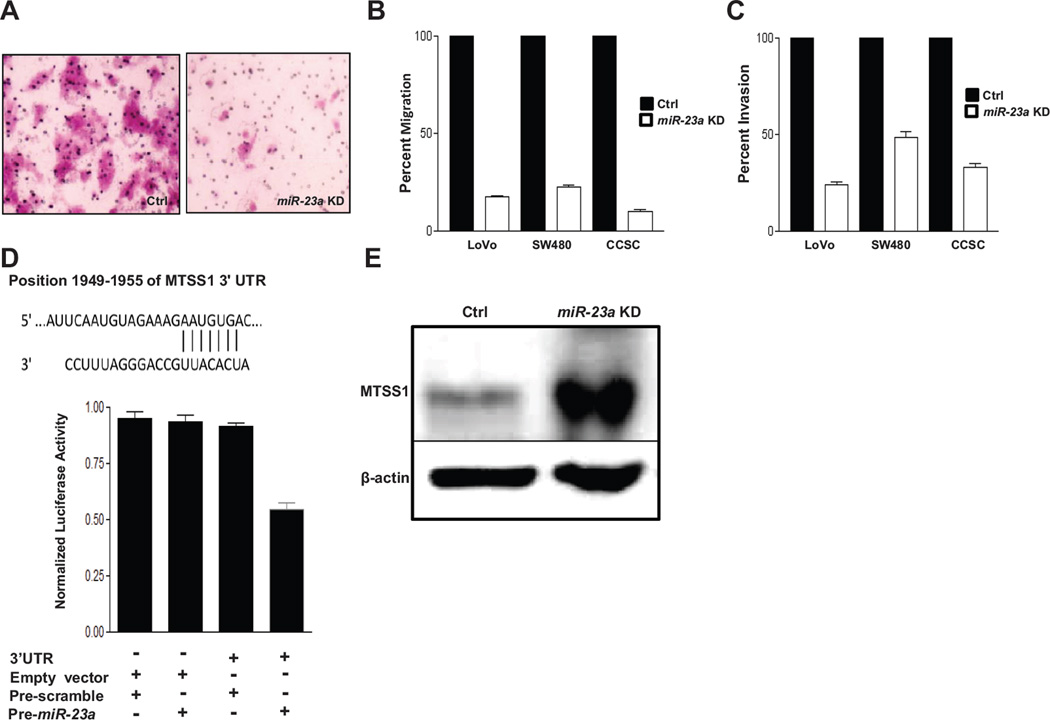
Figure 4.
miR-23a knockdown increases MTSS1 protein levels and inhibits CRC cell and CCSC migration and invasion. A, crystal violet staining of a transwell migration assay of CCSC transduced with lentivirus containing anti-miR-23a shRNA or control sequences. B,C, anti-miR-23a or control sequence lentiviral transduced LoVo, SW480, and CCSC migration and invasion, 95% CI error bars are shown. For all comparisons p <0.002 by Student t test. D, miR-23a binding site in MTSS1 mRNA 3’UTR (nt 1949–1955) (top), bottom, miR-23a binding activity luciferase reporter assay in CCSC. CCSC were transfected with a plasmid containing the MTSS1 miR-23a binding site fused to the 3' UTR of Firefly luciferase and co-transfected with plasmids driving expression of pre-miR-23a or a control insert sequence. Luciferase protein levels and activity are suppressed when a miR binds specifically to the 3' UTR target sequence. E, MTSS1 protein levels in CCSC infected with lentivirus expressing a control or anti-miR-23a shRNA.
MTSS1 is expressed in several embryonic tissues including the developing central nervous system [20]. There, MTSS1 interacts with SRC and cortactin to inhibit cell migration (33). Also consistent with a role in cell motility, MTSS1 is downregulated in several metastatic cancer cell lines (18, 34). Cell migration is accomplished by the formation of cellular protrusions including lamellipodia and filopodia (35). These protrusions result from actin filament (F-actin) rearrangement at the cell cortex by WASP/WAVE, RHO family proteins and other factors (35), which is an important mechanism for enabling cell motility. Small RHO family GTPases including CDC42, RAC, and RHO control the formation and extension of filopodia, lamellipodia, de novo actin polymerization and enable cell migration (36–38).
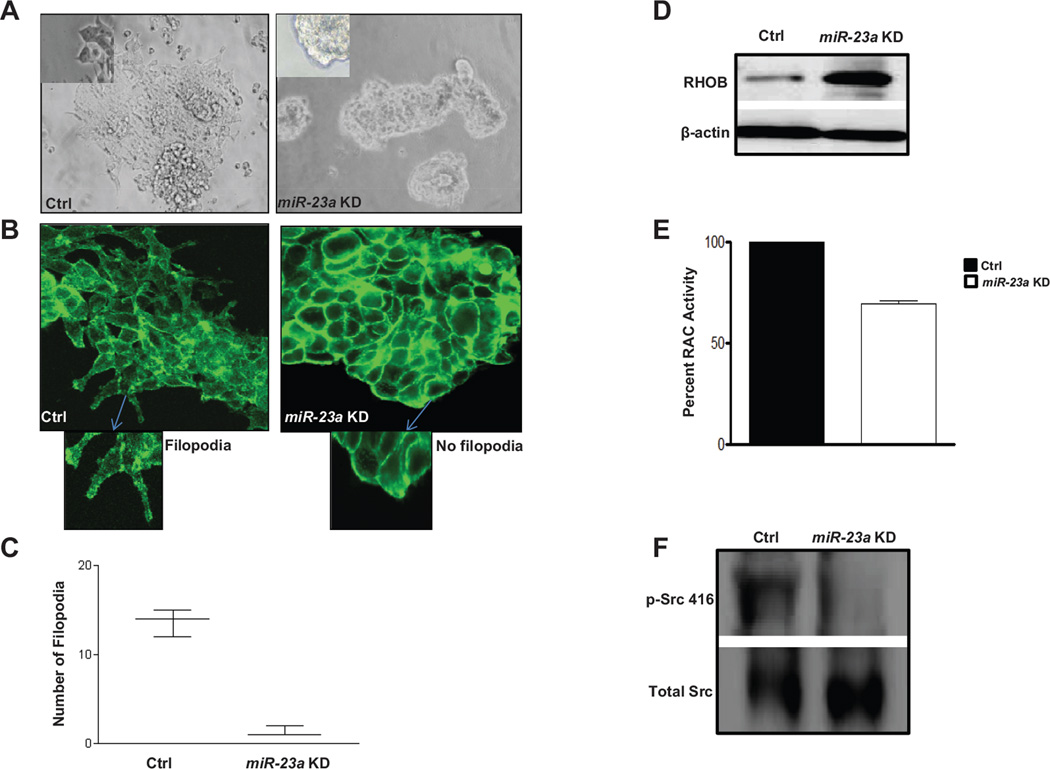
When plated onto laminin coated chamber slides, CCSC attach and extend filopodia along the surface. In contrast, CCSC with miR-23a knockdown do not attach well to laminin-coated plates (Fig. 5A). CCSC with lentiviral miR-23a knockdown also show morphological changes as compared to cells infected with a control lentivirus. These include extension of significantly fewer filopodia and growth with spherical morphology (Fig. 5A). To confirm that filopodia are being reduced in cells with miR-23a knockdown, we immunostained for phalloidin, which tracks the distribution of F-actin in filopodia. This revealed that miR-23a knockdown significantly reduced the number of phalloidin+ filopodia (Fig. 5B and C), consistent with a role for MTSS1 to inhibit CRC cell motility.
Figure 5.
Filopodia formation is dramatically decreased following knockdown of miR-23a in CCSC. A, light microscope image of CCSC transduced with lentivirus containing control 10X (left; inset (40X) indicate the presence of filopodia) or anti-miR-23a shRNAs 10X (right, inset (40X) showing absence of filopodia). B, F-actin staining (phalloidin) of CCSC with control (left, inset @ 40X showing filopodia formation) or anti-miR-23a lentiviral knockdown (right, inset @ 40X with no filopodia formation). C, mean number of filopodia per colony of CCSC infected with anti-miR-23a or control lentiviral shRNA. (Student t test p<0.001). D, RHOB levels are increased post restoring of MTSS1 protein by knockdown of miR-23a. E, Rac1 activity is decreased by 25% in CCSC expressing anti-miR-23a, tested using the G-LISA assay (Student t test p<0.05). F, Phosphorylated pSrc416 levels are decreased in the miR-23a KD cells.
RHOB is a mainly endosomal small GTPase that regulates actin organization, vesicle trafficking and RHO/RAC signaling. In multiple types of cancer cells, RHOB inhibits cellular invasion, and metastasis, and during malignant progression RHOB levels decrease (39). Because MTSS1 has been shown to directly affect RHO/RAC signaling (40), we assayed CCSC with lentiviral miR-23a knockdown for RHOB protein levels and RAC1 activity. CCSC with miR-23a knockdown and increased MTSS1 levels vs. control knockdown cells had increased levels of RHOB and reduced RAC1 activity (Fig. 5D and E), consistent with impaired cell motility.
MTSS1 has also been shown to regulate actin cytoskeleton dynamics and interact with cortactin (CTTN), a key substrate of the oncogenic SRC kinase and a major activator of actin branching and polymerization (41, 42). Consistent with a role for miR-23a regulation of MTSS1 and downstream inhibition of activated SRC, p-SRC (Thr416), is decreased in the miR-23a knockdown CCSC (Fig. 5F). In sum, these data are consistent with a mechanism whereby miR-23a downregulates MTSS1, which decreases RHOB and increases RAC1 and SRC activity. This in turn causes F-actin dysregulation and filopodia extension, which upregulates migration and invasion by CCSC and non-CCSC CRC cell lines.
miR-23a Overexpression In Vitro Promotes Cell Migration and Invasion in DLD1 Cells
DLD1 cells have low endogenous miR-23a levels (Supplemental Fig. S5). Similar to miR-27a overexpression studies, we used a lentiviral vector to overexpress miR-23a in DLD1 cells. DLD1 cells expressing high miR-23a levels showed a significant increase in migration and invasion ability as compared to the control cells (Supplemental Fig. S7A–C). Overall, these data are complementary to miR-23a shRNA knockdown studies and consistent with a role for miR-23a to promote migration and invasion.
miR-23a and miR-27a Inhibit CCSC Tumor Formation in a Mouse Model of Metastasis
Hematogenous injection of cells into the tail vein of immunodeficient mice is a commonly used in vivo assay of metastasis. We used this system to study the roles of miRs-23a and 27a in metastasis using CCSC expressing anti-miR-23a, anti-miR-27a or a control shRNA. Both miR-23a and miR-27a knockdown resulted in significantly fewer lung tumors vs. control CCSC (Fig. 6A–D). Also consistent with inhibition of metastatic tumor formation, knockdown of either miR-23a or 27a significantly increased overall survival of mouse hosts vs. cells expressing the control shRNA (Fig. 6E and F). In summary, these results are consistent with important in vivo roles for miR-23a and miR-27a in metastatic tumor formation. For miR-27a, the data are consistent with a mechanism of promoting cell proliferation, and for miR-23a a mechanism primarily caused by increased cell motility.
Figure 6.
Knockdown of miR-23a or miR-27a reduces tumor burden and increases survival. A, gross image of lung from mice with CCSC injected into tail vein, left is CCSC expressing control sequence, middle anti-miR-23a, right anti-miR-27a CCSC. B, Lung tumor multiplicity in tail vein metastasis assay using CCSC expressing control shRNA, anti-miR-23a or anti-miR-27a. C, image of H&E stained lung tumor from tail vein injected mice. Left is control shRNA, middle is anti-miR-23a, and right is anti-miR-27a. D, Lung tumor volume from either CCSC expressing control shRNA, anti-miR-23a or anti-miR-27a. E, Kaplan-Meir survival curve for tail vein injected mice with either CCSC expressing Ctrl shRNA (black line) or anti-miR-23a (red line). Control scrambled shRNA sequence. F, Kaplan-Meir survival curve for tail vein injected mice with either CCSC cells expressing Ctrl shRNA (black line) or anti-miR-27a (red line). Control scrambled shRNA sequence.
Analysis of CRC Gene Expression Microarray Datasets for Predicted miR-23a and miR-27a Target Genes
To further investigate our findings for miR-23a and miR-27a in CRC tumor progression, we examined relevant whole-genome CRC gene expression profiles annotated with clinical information from the Gene Expression Omnibus (GEO). While there were no gene expression (or microRNA) datasets specifically comparing preinvasive (adenomas and carcinoma in situ) to early stage CRCs available, we were able to analyze datasets generated by other investigators profiling the transitions from (a) normal colon to adenocarcinoma and (b) early to late stage CRC. These include human clinical samples from (i) GSE20916 (43), which compares normal colon to CRC adenocarcinoma and (ii) GSE14333 (44) and GSE17536 (45), which compare early (locally invasive) vs. late stage (metastatic) CRCs. The latter two datasets have been previously used by several investigators for CRC prognostication and (44, 45) the identification of an intestinal stem cell signature in poor prognosis CRC (46). We queried the changes in expression profiles of putative miR-23a and miR-27a target genes computationally predicted by different algorithms including PicTar, TargetScan, miRanda and DIANA-microT (21–24). These methods assess the complementarity of miR seed sequences to binding motifs in the 3’UTRs of their putative target genes. We pooled the individual predictions from each method to form a master list of putativetargets for miR-23a (1171 genes) and miR-27a (1179 genes). Globaltest analysis (47) of GSE20916 differential expression levels of predicted genes in the respective miR-23a and 27a master target lists showed consistent downregulation of both miR-23a and miR-27a predicted targets with statistically significant p-values in comparisons of adenocarcinoma vs. normal colon mucosa. Similarly, we confirmed upregulation of predicted miR-23a and miR-27a target expression levels with statistically significant p-values in both GSE14333 and GSE17536 clinical datasets in comparisons of late vs. early stage CRCs. These scores are summarized in (Supplemental Table S2 and Supplemental Fig. S8A–B) shows the distribution of the permuted test statistic of miR-23a and miR-27a in dataset GSE14333 from 10000 times permutation of samples. Finally, because of its potential role as a miR-23a target, we confirmed using immunohistochemistry that MTSS1 protein is expressed at high levels in primary CRCs from stage IV patients, as well as secondary liver metastases (Supplemental Figure S9A–D). Overall, these data corroborate our mouse and human tumor expression data and support potential mechanistic roles for miR-23a in primary CRCs from patients with early but not late stage disease. However, for miR-27a these data are less clear because predicted miR-27a target genes are upregulated in primary CRCs from patients with late vs. early stage disease.
DISCUSSION
Understanding the molecular mechanisms that regulate the transition from preinvasive to invasive CRC is critical to the development of novel approaches to arrest tumor progression and improve CRC patient outcomes. Here, we describe roles for miRs 23a and 27a in mechanisms that are associated with tumor progression. Importantly, both functional and patient CRC expression analyses are consistent with potentially important roles for miR-23a specifically to stimulate migration, invasion and promote the transition from indolent to invasive CRC.
miR-23a levels are upregulated during the evolution of mouse intestinal adenomas to adenocarcinomas, and, importantly, are specifically upregulated during the transition from pre-invasive (adenomas, carcinoma in situ) to locally invasive (stage I/II) primary CRC tumors. Subsequently, miR-23a levels decrease in primary CRCs from patients with cancer cells that have metastasized outside the colorectum (stage III/IV).
MTSS1 is a potentially important mechanistic target of miR-23a because in CRC patient tumors increased MTSS1 expression is strongly associated with metastatic disease (48). MTSS1 levels are lower in adenomas compared with CRC tumors. High MTSS1 levels correlate with stage III/IV disease, as well as lymph node metastasis, poor histomorphological differentiation and local tumor invasiveness. Strikingly, high MTSS1 expression levels in CRC tumors are associated with reduced patient 5-year overall survival, and in multivariate analysis, high MTSS1 expression is an independent poor prognostic indicator (48). Functionally, our studies show that MTSS1 is an important direct target for miR-23a, and that reduced MTSS1 levels accelerate mechanisms that promote CRC migration. miR-23a knockdown decreases filopodia formation, RAC1 signaling, SRC416 phosphorylation, cell motility and hematogenous metastasis, all of which are consistent with a causal role for miR-23a in invasive CRC. This finding complements previous studies proposing roles for MTSS1 as a metastasis suppressor in breast, prostate and bladder cancers (18, 19, 49). To validate our findings, we re-analyzed predicted miR-23a target gene expression levels in publically available datasets. Corroborating our data, expression levels of miR-23a predicted target genes including MTSS1 in these datasets are lower in CRCs from patients with early vs. late stage disease, and we confirmed using immunohistochemistry that MTSS1 is highly expressed in both primary CRCs and secondary metastases from patients with stage IV disease.
To further validate our findings, we searched for large scale gene expression and microRNA datasets specifically comparing pre-invasive (adenoma/carcinoma in situ) to invasive (stage I/lI) CRCs. However, to our knowledge these datasets are currently not available. Therefore, as genome wide datasets comparing pre-invasive to invasive CRC become available, it will be important in the future to analyze whether they further validate our findings of miR-23a target gene upregulation during this transition.
miR-27a levels are upregulated in mouse intestinal adenocarcinomas as well as in invasive and metastatic CRC. Functionally, in CRC stem cells and commonly used CRC cell lines, miR-27a can directly downregulate the tumor suppressor FBXW7 and promote cell proliferation. Consistent with these data, miR-27a knockdown and the subsequent increase in FBXW7 protein levels inhibits NOTCH, JUN and MYC signaling. Functionally, this causes CRC secretory lineage differentiation, as shown by expression of the colon goblet cell marker MUC2. Overall, these data are consistent with roles for miR-27a to promote general mechanisms associated with tumor progression. However, analysis of predicted miR-27a target gene expression levels in publically available large scale gene expression datasets show upregulation of predicted targets in primary CRCs from patients with late vs. early stage disease even though miR-27a levels do not decrease. While there are clearly many potential confounding factors that could cause this discrepancy (e.g. the lack of information in these datasets on which patients did or did not receive chemotherapy, cross-regulation by other microRNAs, stochastic noise, etc.), altogether they limit the specific conclusions that can be drawn at present about the role of miR-27a specifically in CRC patient tumor progression. Again, as genome wide datasets comparing pre-invasive to invasive CRC become available it will be important to investigate in more detail the potential role of miR-27a to promote the transition from pre-invasive to invasive CRC, as well as in other tumor types.
METHODS
Mice
All animal studies were performed under an approved Weill Cornell IACUC protocol. Wild-type (Wt), Apc1638N, Pms2+/−, Mlh3+/− and Apc1638N; Mlh3−/−; Pms2−/− mice have all been described previously and were maintained on the C57B/L6 genetic background (50). All lines of mice were necropsied when they became morbid or moribund. Sacrificed mice were surveyed for tumors and suspicious masses.
Mouse Tiling Array Studies
Genomic DNA was isolated from tumor and normal tissue from each mouse using the PUREGENE DNA Isolation kit (Gentra Systems, Minneapolis, MN). Nimblegen tiling array hybridization of mouse chromosomes 6–9 were performed at NimbleGen Systems Inc. (Madison, WI, USA). The 385K oligonucleotide tiling array produced by NimbleGen Systems Inc. (Madison, WI, USA) was used. Probe design, array fabrication, array CGH experiments including DNA labeling, hybridization, array scanning, data normalization and log2 copy-number ratio calculation were performed by NimbleGen Systems Inc. Array data were analyzed using the SignalMap Software version 1.8 (NimbleGen Systems Inc.).
MicroRNA Isolation and qRT-PCR
MicroRNA was extracted from cell lines and tissues using mirVANA spin columns (Ambion, Austin, TX, USA). All primary CRC tissues in this study were taken from colorectal adenomas and stage 0-IV CRCs collected by the New York Presbyterian Center for Advanced Digestive Care Colon Cancer Biobank, in accordance with the Weill Cornell IRB. Briefly, RT–PCR studies were performed using 10ng of total RNA and gene-specific PCR primers for microRNAs purchased from Life Sciences (Applied Biosystems). PCR cycling conditions used 32 cycles (95°C for 15 s, 60°C for 30 s) after an initial denaturation step (95°C for 3 min). Expression levels are the average of three or more independent experiments. All expression levels are normalized to the average of small nuclear U6 and RNU48 snRNA levels.
RNA Isolation and qRT-PCR
Total RNA was extracted using the Qiagen RNeasy kit and reverse transcribed using the ABI Reverse Transcription Kit. Gene expression was quantified on a BioRad realtime PCR analyzer (CFX96). Expression levels are the average of three or more independent experiments. All expression levels are normalized to GAPDH mRNA levels.
Culture of CCSC and Colon Cancer Cell Lines
CCSC lines used in this study are established cell lines from human colon cancer resections generated by our laboratory and have been described previously (19). CCSC were cultured in vitro in ultra-low attachment flasks with DMEM/F12 containing nonessential amino acids, antibiotic-antimycotic, N2 supplement (Invitrogen), B27 supplement (Invitrogen), heparin 4ug/ml (Sigma), EGF (40ng/ml), bFGF (20ng/ml) at 37°C at 5% CO2, essentially as previously described (19). CCSC lines were authenticated by KRAS, BRAF and SNP genotyping.
DLD1, HCT116, LoVo, RKO and SW480 were purchased from ATCC and cultured according to recommended media conditions at 37°C at 5%CO2, and no validation was performed.
LNA knockdown Cell Culture Studies
miR-23a and miR-27a LNA inhibitors were purchased from Exiqon (Vedbaak, Denmark) and are based on the miRCURY LNA™ microRNA Knockdown technology. Cells were transfected with miR-23a or -27a LNA inhibitors (final concentration of 50 nM) using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s recommended protocol. Control LNAs consisted of scrambled sequence 3′-fluorescein labeled miR LNAs (Exiqon, Vedbæk, Denmark). The sequences of the miR-23a, miR-27a inhibitors and the scrambled miR were: 5′GGAAATCCCTGGCAATGTGA-3’, 5′-CGGAACTTAGCCACTGTGA-3′ and 5′-GTGTAACACGTCTATACGCCCA-3′, respectively. Transfection efficiency was evaluated by fluorescence microscopy (Leica Microsystems).
Lentivirus Infection, Constitutive Expression and shRNA Knockdown Studies
CCSC and colon cancer cell lines (LoVo and SW480) were infected with lentivirus expressing shRNAs against either miR-23a, miR-27a (pMIRZIP-23a and pMIRZIP-27a), or control scrambled hairpin vector sequences under the control of constitutive H1 promoter (Systems Biosciences). DLD1 cells were infected with lentivirus constitutively expressing either miR-23a or miR-27a (pMIRH23aPA-1 and pMIRH27aPA-1) sequences. Lentiviral supernatants were used to infect cells with the addition of polybrene at 8ug/ml for 8h. The media was replaced with fresh media containing 2 ug/ml of puromycin, essentially as previously described (20).
Cell Viability Assays
Viability of cells was measured by using the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) reduction method. Cells at a density of 3.0 × l03/well were seeded into 96-well dishes in triplicate for each independent experiment. Cells were incubated with 2.5% MTT solution (5 mg/ml) for 3.5 h at 37 °C. DMSO was then used to dissolve the formazan product for spectrophotometric analysis at 540 nm.
Cell Migration, Invasion, and Clonogenicity Assays
To assay cell migration, the Boyden chamber assay was used. 1 × 105 of CCSC were seeded onto fibronectin-coated polycarbonate membrane inserts (6.5 mm in diameter with 8.0 µm pores) in a transwell apparatus (Costar) and cultured in CCSC media. 5% FBS was added to media in the lower chamber. After incubation for 12h at 37 °C in a CO2 incubator, the insert was washed with PBS and cells on the top surface of the insert were removed by wiping with a cotton swab. Cells were then fixed with methanol, stained with 0.4% crystal violet solution and inspected microscopically at 200× magnification. For the invasion assay the transwell membrane was coated with a 300 ng/µl Matrigel solution (BD, Franklin Lakes, NJ), and the cells were incubated for 24h at 37 °C. Cells that migrated to the bottom surface of the insert were fixed with methanol and stained with 0.4% crystal violet solution. Cells in five separate randomly chosen fields were counted. Clonogenicity assays were performed essentially as previously described (20). The value for each assay represents the mean ± SEM of triplicate measurements from at least 3 independent experiments.
Protein Isolation and Western blotting
CCSC spheres and cell pellets were lysed in ice cold NP40 lysis buffer (50mM Tris-HCI, pH7.5, 150mM NaCI, 1mM EDTA and 1% NP-40). Protein quantification was carried out using Bio-Rad protein quantification assay. Proteins were separated by SDS/PAGE and transferred to Immobilon-P PVDF (Millipore). Membranes were blocked with 5% nonfat dry milk and incubated overnight at 4°C with the indicated primary antibody. Detection was carried out by peroxidase based chemiluminescence (Amersham). Antibodies used are listed in Supplemental Methods.
Immunohistochemistry
Immunohistochemistry was performed on 5µM paraffin embedded sections from 3D Matrigel cultures as previously described (20). Paraffin embedded sections were deparaffinized in xylene and rehydrated in a graded alcohol series and water. The slides were heated in antigen retrieval solution and incubated with Anti-Myc-tag (1:500) or Anti-Muc2 (1:250) antibodies overnight at 4 °C. The ABC kit (Vector) was used to visualize antigen. Counter staining was done using hematoxylin. Immunofluorescence was performed on CCSC grown on laminin coated chamber slides that had been fixed with 3–4% paraformaldehyde for 10–20 minutes and permeabilized with 0.5% Triton X-100 for 2–10 minutes. Chamber slides were then stained with Alexa Fluor 488® phalloidin at a dilution of 1:250 and analyzed with fluorescence microscopy.
Subcutaneous Xenograft Assay
NOD/SCID (NOD) mice were obtained from the Jackson Labs (ME). All animals used were between 6 and 8 weeks of age and housed in micro-isolator cages in accordance with the institutional animal welfare guidelines of Weill Cornell Medical College. Mice were injected subcutaneously in the flank with either (1×106 CCSC or 0.5×106 LoVo) mixed in1:1 ratio with Matrigel (BD). Tumor size was measured weekly using calipers.
Tail Vein Metastasis Assay
1 × 106 of CCSC cells were injected into the tail vein of 6–8 weeks old NOG (NOD.Cg-Prkdcscid ll2rgtm1wjl/SzJ) mice obtained from the Jackson Labs (ME). Moribund mice were sacrificed immediately, necropsy performed and tumors harvested using a dissecting microscope. All animal protocols in this study were in accordance with the institutional animal welfare guideline of Weill Cornell Medical College.
Luciferase Assay
HEK293 cells were plated in 96 well plates at 5000 cells per well. On the second day, the cells were transfected with 1.0 µg of either (a) plasmid carrying the 3' UTR miR binding site for miR-23a or miR-27a cloned in pEZX-MT01 (GeneCopoeia), (b) empty vector or precursor miR expression clone sequences in pEZX-MR03 (GeneCopoeia), or (c) “scrambled” control sequences in pEZX-MR03, using Lipofectamine (Invitrogen) according to the manufacturer’s recommendations. Both Firefly luciferase and Renilla luciferase activities were measured 2 days after transfection and data was recorded on the GLOMAX system. Firefly luciferase activity was then normalized with Renilla luciferase activities in the same well. Luciferase activity was measured in triplicate with three different clones in at least four independent experiments and is presented as mean ±SEM.
miRCURY LNA™ microRNA Array Studies
Total RNA was extracted using the Qiagen RNeasy kit. MicroRNA profiling was performed at Exiqon (Vedbaek, Denmark). The hybridization was performed according to the miRCURY™ LNA array manual using a Tecan HS4800 hybridization station. After hybridization, the microarray slides were scanned and stored in an ozone-free environment (ozone level below 2.0 ppb) to prevent potential bleaching of the fluorescent dyes. The miRCURY™ LNA array microarray slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies, Inc., USA) and the image analysis was carried out using the ImaGene 8.0 software (BioDiscovery, Inc., USA).
Clinical CRC Gene Expression Microarray Dataset Analyses
To compare the expression of miR-23a and miR-27a predicted target genes between (a) normal colon vs. all CRCs and (b) primary tumors from stage I vs. stage IV CRC patients, three human whole-genome microarray datasets (GSE20916, GSE14333, and GSE17536) were downloaded from the public functional genomics data repository at the National Center for Biotechnology Information Gene Expression Omnibus. All of these three datasets used the Affymetrix HGU-133Plus2 chip. The GSE20916 dataset includes normal colon, adenocarcinoma and carcinoma array data (43). The GSE14333 dataset includes primary CRC tumor array data from 44 stage I and 61 stage IV CRC patients, respectively (44). The GSE17536 dataset includes primary CRC tumor array data from 24 stage I and 39 stage IV patients, respectively (45).
Genes targeted by miR-23a/miR-27a were predicted by several miR target prediction methods, including PicTar, TargetScan, DIANA-microT, and miRanda (21–24). The latter two methods were used with tight filter thresholds – a miTG score of 0.45 for DIANA-microT and a mirSVR score of −0.8 for miRanda. For each analysis of primary CRC tumor gene expression data, expression level changes in miR-23a and miR-27a predicted target genes were compared between (i) normal colon vs. adenocarcinoma and carcinoma combined or (ii) stage I vs. stage IV CRCs using the Globaltest algorithm (47). Briefly, Globaltest utilizes a generalized linear model with a random effect for gene set analysis. A score test statistic is used for testing the null hypothesis, and the p-value can be calculated from either from asymptotic distributions of the test statistics or by permutation distributions of the test statistics.
Statistical Data Analysis
All statistical data analysis was performed with GraphPad Prism 5 software (GraphPad Software, Inc., SanDiego, CA, USA), with the exception of the clinical CRC gene expression microarray dataset and Nimblegen tiling array analyses as described above.
Supplementary Material
SIGNIFICANCE.
Understanding the mechanisms regulating the transition from indolent adenomas to invasive and metastatic CRC is critical to improving patient outcomes. Our study highlights roles of miRs-23a and 27a in tumor progression and supports a potential mechanistic role for miR-23a in the transition from indolent to invasive CRC.
Acknowledgements
We would like to thank Drs. Eva Lee and Bogi Andersen for critical reading of this manuscript. This study was funded by a generous donation from Matthew Bell, NCI R01 CA098626, R01GM095990 and NCI R21 CA153049 to S.L. and an NCI Research Supplement to Promote Diversity in Health-Related Research to S.J. ZHG and JS gratefully acknowledge support from The HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine (ICB), and the computational resources of the Coffrin Center for Biomedical Information, (ICB) at Weill Cornell Medical College of Cornell University.
REFERENCES
- 1.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 2.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 3.Velculescu VE. Defining the blueprint of the cancer genome. Carcinogenesis. 2008;29:1087–1091. doi: 10.1093/carcin/bgn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markowitz SD, Dawson DM, Willis J, Willson JK. Focus on colon cancer. Cancer cell. 2002;1:233–236. doi: 10.1016/s1535-6108(02)00053-3. [DOI] [PubMed] [Google Scholar]
- 5.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2010;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 6.Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci U S A. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Din FV, Theodoratou E, Farrington SM, Tenesa A, Barnetson RA, Cetnarskyj R, et al. Effect of aspirin and NSAIDs on risk and survival from colorectal cancer. Gut. 2010;59:1670–1679. doi: 10.1136/gut.2009.203000. [DOI] [PubMed] [Google Scholar]
- 9.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schetter AJ, Nguyen GH, Bowman ED, Mathe EA, Yuen ST, Hawkes JE, et al. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:5878–5887. doi: 10.1158/1078-0432.CCR-09-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24-2 cluster and its implication in human diseases. Molecular cancer. 2010;9:232. doi: 10.1186/1476-4598-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chhabra R, Adlakha YK, Hariharan M, Scaria V, Saini N. Upregulation of miR-23a-27a-24-2 cluster induces caspase-dependent and -independent apoptosis in human embryonic kidney cells. PLoS One. 2009;4:e5848. doi: 10.1371/journal.pone.0005848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong KY, Owens KS, Rogers JH, Mullenix J, Velu CS, Grimes HL, et al. MIR-23A microRNA cluster inhibits B-cell development. Exp Hematol. 2010;38:629–640. doi: 10.1016/j.exphem.2010.04.004. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen PC, Kuraguchi M, Velasquez J, Wang Y, Yang K, Edwards R, et al. Novel roles for MLH3 deficiency and TLE6-like amplification in DNA mismatch repair-deficient gastrointestinal tumorigenesis and progression. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000092. e1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sancho R, Jandke A, Davis H, Diefenbacher ME, Tomlinson I, Behrens A. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology. 2010;139:929–941. doi: 10.1053/j.gastro.2010.05.078. [DOI] [PubMed] [Google Scholar]
- 16.Babaei-Jadidi R, Li N, Saadeddin A, Spencer-Dene B, Jandke A, Muhammad B, et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEKfor degradation. J Exp Med. 2011;208:295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saarikangas J, Mattila PK, Varjosalo M, Bovellan M, Hakanen J, Calzada-Wack J, et al. Missing-in-metastasis MIM/MTSS1 promotes actin assembly at intercellular junctions and is required for integrity of kidney epithelia. J Cell Sci. 2011;124:1245–1255. doi: 10.1242/jcs.082610. [DOI] [PubMed] [Google Scholar]
- 18.Parr C, Jiang WG. Metastasis suppressor 1 (MTSS1) demonstrates prognostic value and anti-metastatic properties in breast cancer. Eur J Cancer. 2009;45:1673–1683. doi: 10.1016/j.ejca.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 19.Xie F, Ye L, Ta M, Zhang L, Jiang WG. MTSS1: a multifunctional protein and its role in cancer invasion and metastasis. Front Biosci (Schol Ed) 2011;3:621–631. doi: 10.2741/s175. 2011:2011. [DOI] [PubMed] [Google Scholar]
- 20.Sikandar SS, Pate KT, Anderson S, Dizon D, Edwards RA, Waterman ML, et al. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer research. 2010;70:1469–1478. doi: 10.1158/0008-5472.CAN-09-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 23.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G, et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwatsuki M, Mimori K, Ishii H, Yokobori T, Takatsuno Y, Sato T, et al. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: clinical significance. International journal of cancer Journal international du cancer. 2009;126:1828–1837. doi: 10.1002/ijc.24879. [DOI] [PubMed] [Google Scholar]
- 26.Kemp Z, Rowan A, Chambers W, Wortham N, Halford S, Sieber O, et al. CDC4 mutations occur in a subset of colorectal cancers but are not predicted to cause loss of function and are not associated with chromosomal instability. Cancer research. 2005;65:11361–11366. doi: 10.1158/0008-5472.CAN-05-2565. [DOI] [PubMed] [Google Scholar]
- 27.Miyaki M, Yamaguchi T, lijima T, Takahashi K, Matsumoto H, Mori T. Somatic mutations of the CDC4 (FBXW7) gene in hereditary colorectal tumors. Oncology. 2009;76:430–444. doi: 10.1159/000217811. [DOI] [PubMed] [Google Scholar]
- 28.Reedijk M, Odorcic S, Zhang H, Chetty R, Tennert C, Dickson BC, et al. Activation of Notch signaling in human colon adenocarcinoma. International journal of oncology. 2008;33:1223–1229. doi: 10.3892/ijo_00000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodilla Vn, Villanueva A, Obrador-Hevia A, Robert-Moreno Ãl, FernÃjndez-Majada V, Grilli A, et al. Jaggedl is the pathological link between Wnt and Notch pathways in colorectal cancer. Proceedings of the National Academy of Sciences. 2009;106:6315–6320. doi: 10.1073/pnas.0813221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sikandar S, Dizon D, Shen X, Li Z, Besterman J, Lipkin SM. The class I HDAC inhibitor MGCD0103 induces cell cycle arrest and apoptosis in colon cancer initiating cells by upregulating Dickkopf-1 and non-canonical Wnt signaling. Oncotarget. 2010;1:596–605. doi: 10.18632/oncotarget.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development. 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- 32.Lee SH, Kerff F, Chereau D, Ferron F, Klug A, Dominguez R. Structural basis for the actin-binding function of missing-in-metastasis. Structure. 2007;15:145–155. doi: 10.1016/j.str.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glassmann A, Molly S, Surchev L, Nazwar TA, Hoist M, Hartmann W, et al. Developmental expression and differentiation-related neuron-specific splicing of metastasis suppressor 1 (Mtss1) in normal and transformed cerebellar cells. BMC developmental biology. 2007;7:111. doi: 10.1186/1471-213X-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nixdorf S, Grimm MO, Loberg R, Marreiros A, Russell PJ, Pienta KJ, et al. Expression and regulation of MIM (Missing In Metastasis), a novel putative metastasis suppressor gene, and MIM-B, in bladder cancer cell lines. Cancer letters. 2004;215:209–220. doi: 10.1016/j.canlet.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi K, Suzuki K. WAVE2, N-WASP, and Mena facilitate cell invasion via phosphatidylinositol 3-kinase-dependent local accumulation of actin filaments. Journal of cellular biochemistry. 2011;112:3421–3429. doi: 10.1002/jcb.23276. [DOI] [PubMed] [Google Scholar]
- 36.Kaibuchi K, Kuroda S, Amano M. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu Rev Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- 37.Li S, Chen BP, Azuma N, Hu YL, Wu SZ, Sumpio BE, et al. Distinct roles for the small GTPases Cdc42 and Rho in endothelial responses to shear stress. J Clin Invest. 1999;103:1141–1150. doi: 10.1172/JCI5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 39.Huang M, Prendergast GC. RhoB in cancer suppression. Histol Histopathol. 2006;21:213–218. doi: 10.14670/HH-21.213. [DOI] [PubMed] [Google Scholar]
- 40.Machesky LM, Johnston SA. MIM: a multifunctional scaffold protein. J Mol Med (Berl) 2007;85:569–576. doi: 10.1007/s00109-007-0207-0. [DOI] [PubMed] [Google Scholar]
- 41.Bershteyn M, Atwood SX, Woo WM, Li M, Oro AE. MIM and cortactin antagonism regulates ciliogenesis and hedgehog signaling. Dev Cell. 2010;19:270–283. doi: 10.1016/j.devcel.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin J, Liu J, Wang Y, Zhu J, Zhou K, Smith N, et al. Differential regulation of cortactin and N-WASP-mediated actin polymerization by missing in metastasis (MIM) protein. Oncogene. 2005;24:2059–2066. doi: 10.1038/sj.onc.1208412. [DOI] [PubMed] [Google Scholar]
- 43.Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5:e13091. doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 47.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 48.Wang D, Xu MR, Wang T, Li T, Zhu J. MTSS1 overexpression correlates with poor prognosis in colorectal cancer. J Gastrointest Surg. 2011;15:1205–1212. doi: 10.1007/s11605-011-1546-2. [DOI] [PubMed] [Google Scholar]
- 49.Loberg RD, Neeley CK, Adam-Day LL, Fridman Y, St John LN, Nixdorf S, et al. Differential expression analysis of MIM (MTSS1) splice variants and a functional role of MIM in prostate cancer cell biology. Int J Oncol. 2005;26:1699–1705. [PubMed] [Google Scholar]
- 50.Chen PC, Dudley S, Hagen W, Dizon D, Paxton L, Reichow D, et al. Contributions by MutL homologues Mlh3 and Pms2 to DNA mismatch repair and tumor suppression in the mouse. Cancer research. 2005;65:8662–8670. doi: 10.1158/0008-5472.CAN-05-0742. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.