Abstract
Aneuploidy is a prominent phenotype of cancer. It refers to deviations from the normal number of chromosomes in a cell, as a result of whole-chromosome loss or gain. In most cases, aneuploidy is caused by mitotic errors due to defects in the mechanisms that have evolved to ensure faithful chromosome segregation, such as the spindle assembly checkpoint (SAC). The observation that SAC-deficient mice are tumor prone demonstrates that this checkpoint is important in suppressing tumor formation and suggests that aneuploidy can induce tumorigenesis. In this review, we will summarize our current knowledge about the cause of aneuploidy and discuss the cellular response to aneuploidy in the context of tumorigenesis.
1. Introduction
A hallmark of human cancers is genome instability a prominent form of which is chromosomal instability (CIN). CIN can be gains or losses of whole chromosomes, or translocation/deletion/duplication of chromosome segments. Alterations in chromosome number or aneuploidy are found in nearly all major human tumor types [1]. Examples include the loss of chromosome 10 in glioblastomas, often reflecting the inactivation of the tumor-suppressor gene PTEN [2], and the gain of chromosome 7 in papillary renal carcinomas, reflecting a duplication of a mutant MET oncogene [3]. However, most of the time, no specific molecular advantages can be associated with a particular gain or loss of a chromosome. In fact, cancer cells display gross abnormalities in their chromosome numbers. This gross alteration was recognized as early as 1914 by the German biologist Theodor Boveri who proposed that aneuploidy might be a cause of tumorigenesis [4].
The relationship between aneuploidy and tumor formation has been debated in the literature, given the fact that in normal subjects there are aneuploid cells present. For example, about 6% of mouse splenocytes were found to be aneuploid [5], and a significant percentage of mouse postmitotic neurons were reported to be aneuploid [6]. In humans, about 3% of stationary lymphocytes were aneuploid [7, 8] and a substantial portion of neurons showed aneuploidy, although the reported frequencies varied according to different studies [7–10]. This kind of genetic mosaicism was suggested to be a result of defects in chromosome segregation during mitosis [11]. It is very likely that aneuploid cells are present in all tissue types and because of their presence in low percentages they do not post any significant pathological danger including oncogenic transformation to the host. On the other hand, the discovery of mutations in BUBR1 and BUB1 in a subset of colon cancer cell lines [12] suggests a weakened spindle assembly checkpoint as the cause of CIN that contributes to the oncogenic process, which was further substantiated by the finding that BUBR1 was mutated in mosaic variegated aneuploidy, a rare human disorder characterized by an increased percentage of aneuploid cells (usually >25%) and predisposition to childhood cancers [13]. More recently, a series of mutant mouse strains that carry mutations in the spindle assembly checkpoint genes have been reported to incur spontaneous tumorigenesis (see below), lending a strong support to the argument that aneuploidy can induce oncogenesis.
In this review, we will summarize our current knowledge about the cause of aneuploidy and discuss the cellular response to aneuploidy in the context of tumorigenesis.
2. The Paths to Aneuploidy
The most likely place during the cell cycle where errors in the ploidy of a cell can occur is mitosis through which the duplicated genome is distributed into two daughter cells. A large number of biochemical pathways ensures faithful segregation of sister chromatids. Malfunction of these pathways is behind chromosome missegregation.
2.1. Disruption of the spindle assembly checkpoint
In S. cerevisiae, compromised SAC function results in elevated rates of chromosome loss, although the checkpoint is not essential for viability [14–17]. Subsequent studies in C. elegance, Drosophila, and mice all demonstrated the essentiality of SAC in the maintenance of chromosomal stability as well as the viability of the organisms [18–24]. High incidence of mitotic errors (e.g. lagging chromosomes) was observed in cells with mutations in SAC components, resulting in a dramatic increase in the percentage of aneuploid cells [19, 25].
It is worth noting that spindle assembly checkpoint is not “all-or-none”. A number of SAC components such as Mad1, Mad2, Bub1, and BubR1 are haploinsufficient in mice. The loss of one copy of these genes causes a partial loss of SAC function as manifested by the increased incidence of mitotic errors, but the remaining SAC function is sufficient to support the viability. Furthermore, not only can the loss of SAC components cause a disruption in the checkpoint, but also their overexpression, for example, the overexpression of Mad2 [26]. Mis-regulation of the spindle assembly checkpoint by mutation in or loss/gain expression of SAC components has been observed in a wide spectrum of human cancers. For example, BUBR1 (BUB1B) is mutated in MVA [13], overexpressed in oral squamous cell carcinoma [27], hepatocellular carcinoma [28], clear cell kidney carcinomas [29], bladder cancer [30], gastric cancer [31], and underexpressed in colon adenocarcinoma [32].
2.2. Centrosome abnormalities
Centrosome is the organizing center for microtubules in most eukaryotes and is critical for the formation of a bipolar-spindle microtubule structure during mitosis. Amplification of centrosomes is observed in nearly all human cancer types [33]. As such, it has been postulated that centrosome abnormalities are a cause of aneuploidy in cancer cells [34]. Indeed, super numerical centrosomes can result in multipolar divisions, leading to the distribution of chromosomes into more than two daughter cells which are bound to be aneuploid. It has also been noticed that a cell with additional centrosomes often tends to modify the multipolar spindle by coalescing all the centrosomes into two clusters to form a bipolar spindle [35]. However, this merge of centrosomes greatly increases the chance of merotelic attachments of microtubules [36, 37]. Merotelic attachment cannot be corrected by the spindle assembly checkpoint and is a major cause of aneuploidy in mammalian cells [38].
2.3. Alterations in microtubule-kinetochore dynamics
Aberrant dynamics of microtubule-kinetochore attachments is another common cause of aneuploidy. Due to the random nature of microtubule-kinetochore attachments, the initial attachment of kinetochores to spindle microtubules is an error-prone process. To achieve correct attachment, kinetochores are frequently attached to and detached from microtubules in early mitosis [39, 40], which helps eliminate erroneous attachments and generates a form of attachment which satisfies both occupancy and tension [41]. The attachment starts to be stabilized after chromosomes are being aligned at metaphase plate and becomes most stable after entering anaphase [40]. Disturbing the dynamics of microtubule-kinetochore attachments can cause errors in mitosis. One example is that the inhibition of Aurora B, a kinase which senses the tension between sister-chromatids and destabilizes mis-attachments, results in suppressed microtubule-kinetochore turnovers, increased merotelic attachments, and thus elevated incidence of lagging chromosomes [41]. The inhibition of Hec1, a target of Aurora B kinase, has been shown to enhance the stability of microtubule-kinetochore attachments, resulting in a significant increase in chromosome missegregation [42]. Given the aneuploid nature of cancer cells, it was not surprising that the attachments of microtubules to kinetochores were found to be more stable in certain cancer cells than in normal diploid cells [39].
2.4. Defects in chromosome cohesion
Sister chromatids are held together by cohesins. The core cohesin complex is composed of Smc1, Smc3, Scc1, and Scc3 [43] and is believed to form a ring-like structure enclosing the two sister chromatids [44]. Prior to anaphase, the majority of cohesin on chromosomal arms is removed by Plk1-and Aurora B–mediated phosphorylation of the cohesin subunit Scc3 [45–49], but the centromeric cohesins are protected from the phosphorylation and removal by a protein called shugoshin [50, 51]. Depletion of shugoshin results in premature separation of sister chromatids and aneuploidy in chromosome stable human colon cancer cells [52]. In addition to protecting the centromeric cohesins, shugoshin apparently also functions in maintaining the stability of centrosomes [53]. It has been found that shugoshin is down regulated in human colorectal cancers [52].
The final separation of sister chromatids at the onset of anaphase depends on separase-mediated cleavage of Scc1 [54, 55]. To prevent premature separation of sister chromatids, separase must be tightly regulated. In yeast, this occurs through direct inhibition by securin [16]. In vertebrates, inhibitory phosphorylation of separase provides an additional layer of regulation [56]. Securin is also named as PTTG (pituitary tumor transforming gene) due to the finding that this gene is overexpressed in pituitary and many other types of tumors [57]. The overexpression may interfere with the timely activation of separase, resulting in chromosome missegregation perhaps through non-disjunction of sisters.
3. Cellular Responses to Aneuploidy
Unlike DNA damage, once a cell becomes aneuploid, there is almost no way to correct the lost or the gained chromosomes. This cell and its progeny will remain as aneuploid and the aneuploidy may get worse if it loses or gains chromosomes again in subsequent mitoses. It is well established that cells respond to DNA damage by activating DNA damage checkpoint and initiating damage repair. Since aneuploidy cannot be repaired, do cells mount a response to it as well?
3.1. Aneuploid cells are genetically unfit
To determine if there are adverse effects associated with aneuploidy, Torres et al. [58] took the advantage of haploid budding yeasts and generated a series of haploid yeast strains with one or two additional chromosomes. No matter which chromosome is in excess, all of the aneuploid yeast strains displayed some common phenotypes including impaired growth and elevated energy metabolism. The slow growth and higher energy demand might be caused by the burden to maintain the extra DNA. However, the strains carrying human chromosome DNA in the form of YAC (yeast artificial chromosome) did not display growth disadvantage as the aneuploid yeasts. Instead, a more likely explanation is that the genes expressed from the extra chromosome causes imbalance in the stoichiometry of protein complexes. The extra proteins have to be turned-over, leading to the increased energy metabolism and slow growth. Thus, compared to the euploid yeast cells, the aneuploid cells are genetically unfit. This genetic unfitness can also be applied to mammalian cells as evidenced by the impaired proliferation of mouse embryonic fibroblasts derived from embryos that were trisomy for chromosome 1, 13, 16, or 19 [59].
3.2. Activation of p53
The loss of Mad2 causes embryonic lethality around blastocyst stage in mice [21], which fueled the argument that the spindle assembly checkpoint might be essential for cell viability. However, Burds et al. [60] managed to obtain mouse cells that lacked both Mad2 and p53, indicating that as in budding yeasts SAC is not essential for cell viability. This result also suggests that p53 might be activated by aneuploidy or the lack of spindle assembly checkpoint function. A large number of cellular stresses including DNA damage, hypoxia, heat or cold shock can activate p53. One possibility of p53 activation in the cells with defects in the spindle assembly checkpoint is that the p53 protein somehow monitors the functionality of the checkpoint, instead of being activated by aneuploidy. By transiently disrupting the spindle checkpoint via siRNA-mediated depletion of MAD2 in HCT116 cells, Li et al. [61] demonstrated that p53 responded to aneuploidy, not the defects in spindle assembly checkpoint. A similar conclusion was reached independently by Thompson et al. [62].
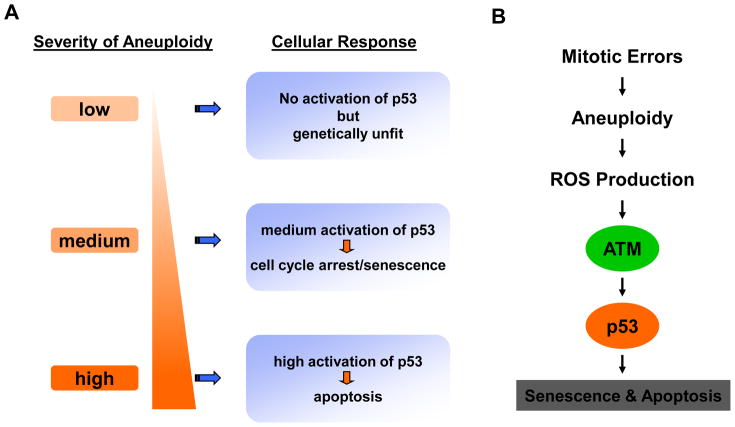
The degree of p53 activation seems to fall in a range and there is a correlation between the level of p53 activation and the severity of the aneuploidy [61], suggesting that different levels of aneuploidy activate p53 with different strengths and may result in different outcomes (Fig. 1A). Highly aneuploid cells are expected to die of p53-mediated apoptosis. Indeed, the MEFs derived from embryos carrying the homozygous mutation in Cdc20 that disrupted Mad2 inhibition (Cdc20AAA) could not be maintained in culture because of apoptotic cell death [5]. The culture could be rescued, however, if p53 was deleted from the cells [61].
Figure 1. Cellular responses to aneuploidy.
A. The severity of aneuploidy correlates with the level of p53 activation. B. The aneuploidy checkpoint.
3.3. The aneuploid checkpoint
How does aneuploidy induce p53 activation then? Li et al. [61] searched for the kinase that activates p53 in aneuploid cells and it turned out to be ATM. However, there was no canonical DNA damage checkpoint activation in these cells as there was no detectable CHK1 or CHK2 activation [61]. Given the alterations in energy metabolism seen in aneuploid budding yeasts and mouse embryonic fibroblasts [58, 59], it was reasoned whether aneuploidy caused an increase in energy demand which could then lead to an increased production of reactive oxygen species (ROS). The surge of ROS would in turn activate ATM and p53. Indeed, the levels of ROS in aneuploid cells were elevated when compared to that of control cells and treating these aneuploid cells with ROS scavenger N-acetyl cysteine (NAC) not only suppressed ATM and p53 activation but also greatly improved their growth [61]. The recent demonstration that ROS can directly activate ATM via forcing dimerization (disulfide-cross linking) and hence the activation of ATM in the absence of DNA double strand breaks [63] provides a direct link between ROS and ATM activation.
Based on these results, we propose that there is an aneuploidy checkpoint (Fig. 1B) which limits the proliferation of aneuploid cells. The strength of the activation of this checkpoint (as measured by p53 activation) depends on the level of ROS which therefore serves as a gauge on the severity of the aneuploidy of a cell. It is unclear at present what is the threshold for p53 activation nor is it known whether certain combinations of chromosomes activate p53 more than other combinations.
4. Aneuploidy and Tumorigenesis
The association of aneuploidy and tumorigenesis has long been recognized [4]. However, it has been a chicken-and-egg issue when considering the role of aneuploidy in tumorigenesis. Some argued that aneuploidy was no more than a byproduct in the process of tumor formation, while others suggested that aneuploidy may cause carcinogenesis all by itself. Nonetheless, it is becoming evident that aneuploidy contributes to oncogenic transformation.
4.1. Why are most human cancer cells aneuploid?
Given that aneuploid cells are genetically unfit comparing to euploid cells, it is a bit puzzling why most human cancer cells are aneuploid. It is well established that genome instability caused by replication errors or defects in DNA damage repair plays a driving role in the process of oncogenesis. It is understandable that these errors or defects in repair can lead to mutations that either inactivate tumor suppressors or activate oncogenes. What benefits does aneuploidy provides to cancerous or even pre-cancerous cells then, amid the apparent disadvantage (unfitness)? Although there are a few cases where a gain or loss of a particular chromosome can be linked with the gain of oncogenes or the loss of tumor suppressors, in most cases, there isn’t a clear link. In fact, cancerous cells within a tumor often display heterogeneity in their karyotypes [64]. On the other hand, it is also not entirely clear if the aneuploidy-associated unfitness is true for all possible sorts of chromosome combinations, especially in mammalian cells. In addition, we have to recognize that unfitness is a relative term. Comparing to their normal neighbors, pre-cancerous and cancerous cells are actually more fit in terms of proliferation, since a majority of the cells in our body are restricted from dividing again. Further, it is certainly possible that some combinations of chromosomes may not confer any unfitness or may even provide growth advantages under certain circumstances.
In a cancer-unrelated study of evolvability, Rancati et al. [65] demonstrated that aneuploidy could afforded yeasts with higher evolvability even to the point of “inventing” new mechanisms of cytokinesis. It is highly likely that aneuploidy also affords cancer cells with the ability to evolve and to adapt. This ability may very well exceed what the other forms of genome instability can provide, simply because of the sheer scale of genes being affected under aneuploidy. Thus, the aneuploid pre-cancerous and cancerous cells may better survive and gain growth advantages, especially if there are other mutations acquired which may alleviate the unfitness. Indeed, the loss-of-function mutation in the deubiquitinating enzyme Ubp6 improves the fitness of multiple aneuploid yeast strains [66].
4.2. Can aneuploidy induce tumorigenesis?
Given the prominence of aneuploidy in human cancers [1], it has been debated whether aneuploidy can induce tumorigenesis. Opposite to the argument that aneuploidy is merely a byproduct of the oncogenic processes, one view proposes that aneuploidy can induce tumorigenesis all by itself [67–69]. An aneuploid cell, randomly generated due to occasional mitotic errors or induced by carcinogens, has an unbalanced production of mitotic regulators due to the aneuploid chromosome composition. Therefore, each and every succeeding divisions will be error-prone, leading to a chain reaction that generates ever more unstable and severe aneuploid cells (aneuploidization). Eventually, a cell with cancer-specific combinations and rearrangements of chromosomes will emerge. Thus, cancer occurs as a result of abnormal dosages of thousands of un-mutated aneuploid genes. However, such a view contradicts the finding that there are certain percentages of aneuploid cells present in our body and that not all mice with a disrupted spindle assembly checkpoint develop spontaneous tumors despite the presence of quite high percentages of aneuploid cells in the adult animals. On the other hand, the fact that spontaneous tumors did develop in these SAC mutant strains of mice does indicate that aneuploidy contributes to tumorigenesis, if not inducing tumorigenesis all by itself.
In the last decade, a number of mouse strains have been generated which carry deletions of or mutations in the genes functioning in the spindle assembly checkpoint. In most cases, the homozygous mutant animals could not survive the embryogenesis. Although the heterozygous animals are viable and apparently healthy, they inevitably generate aneuploid cells (at varying rates, depending on the genes being mutated) [5, 18, 19, 23, 26, 70–74]. The percentage of aneuploidy in the adult animals can reach up to 35% (assayed with splenocytes). Not surprisingly, these mice do develop spontaneous tumors. However, again, the tumor phenotype (latency and penetrance) varies in a wide spectrum amongst these strains of mice. At one end of the spectrum, there are examples of no tumor induction, as seen in the mice with reduced expression of Bub3 [70] or BubR1 [19]. In the middle, haploinsufficiency of Mad2 [23], Mad1 [74] or CENP-E [72] caused a mild increase in the rate of spontaneous tumors. At the other end of the spectrum, Bub1−/H (null over hypomorphic) [71], Cdc20+/AAA [5] and Mad2 over-expressing mice [26] all displayed high rates of spontaneous tumorigenesis. Such variations in the tumor phenotype are likely caused by differences in the percentage of aneuploidy, the genetic defect, and the genetic background of the strains.
The fact that the tumors developed in the SAC mutant mice are usually late onset and the penetrance seldom reaches 100% suggests that aneuploidy itself may not be a tumor initiator but rather a promoting factor. Consistent with that notion, mutations in SAC components showed cooperation with chemical carcinogens [75].
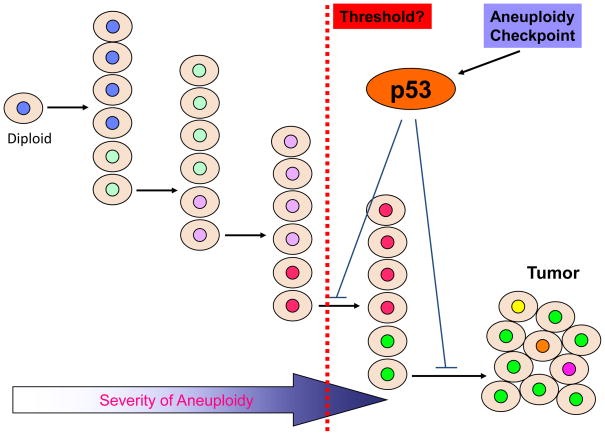
It must be made clear that aneuploidy is a term referring to any deviation from the diploid number of chromosomes. Therefore there is a severity issue: how much deviation a particular aneuploidy is. In the life span of an organism, there will be random mitotic errors that occur at some low frequencies despite all the mechanisms that are there to ensure faithful chromosome segregation. The so-produced aneuploid cells are very unlikely to give rise to ever more aneuploid descendants so to impose a significant oncogenic risk, because the original random mitotic error may never happen again. Even when the spindle checkpoint function is compromised and there is a clear increase in the rate of mitotic errors, there are no substantial increases in tumorigenesis as seen in the mice with the reduced expression of Bub3 [70] or BubR1 [19]. Thus, we believe for the tumor to form, continuous erroneous mitoses are necessary (Fig. 2), which can only happen when the mitotic error rate is substantial, at least higher than that reached with the reduced expression of Bub3 or BubR1. However, at some point (threshold), p53 is activated by the aneuploidy checkpoint to prevent further deterioration in ploidy and the formation of a full-fledged tumor (Fig. 2).
Figure 2. Induction of tumorigenesis by aneuploidy.
The emergence of a transformed aneuploid cell requires multiple rounds of erroneous mitoses that produced ever more aneuploidy. In addition, the p53 pathway must be inactivated. It is unclear what is the threshold in terms of the severity of aneuploidy for p53 activation, nor is it clear if a particular combination of chromosomes activates p53 more than another combination.
4.3. The aneuploidy checkpoint limits tumorigenesis in SAC-deficient mice
The low penetrance of the tumor phenotype in SAC-deficient mice argues that either additional mutations are needed or the cancer-causing aneuploidy is an extremely rare event such that it can never happen in all of the mutant animals. At present, there is no evidence for either arguments, but inactivation of the aneuploidy checkpoint seems necessary for tumor development. Li et al. examined 5 tumor samples isolated from Cdc20+/AAA mice and all five seemed to have lost the p53 response [61]. Furthermore, when Atm or p53 deletion was combined with a SAC mutation, the double mutant mice develop tumors at much earlier ages and the penetrance is 100% [61], indicating that the aneuploidy checkpoint is important in preventing aneuploidy-induced oncogenic transformation.
5. Summary
It is becoming clear that aneuploidy is not a mere byproduct of tumorigenesis. It is positively selected for during oncogenic transformation as it provides the precancerous cells the ability to evolve into ever more malignant and to adapt the “harsh” environment. However, the mechanisms behind the aneuploidy in cancer cells are difficult to dissect out because a myriad of factors could be at fault, although progresses have been made. Defects in spindle assembly checkpoint per se may not even be the major cause of aneuploidy in human cancers, as mutations in SAC components are not common.
We may never know the exact reason behind the chromosomal instability in each and every cancer. However, such chromosomal instability may provide opportunities for therapeutic intervention. One strategy is to induce even more aneuploidy in cancers so that the genome becomes so unstable that the cell viability is compromised [76]. Similarly, in aneuploid tumor cells with amplified centrosomes, an inhibition of the nonessential motor protein HEST could prevent the coalescence of the extra centrosomes, resulting in multi-polar division and lethality [77]. Exploiting the chromosomal instability of cancer cells will certainly be an active area of research in the future.
Acknowledgments
This work is supported by grants from National Cancer Institute to PZ (CA122623 and CA116097). The authors apologize for any omissions of the literature due to the space limit.
References
- 1.Mertens F, Johansson B, Mitelman F. Isochromosomes in neoplasia. Genes, chromosomes & cancer. 1994;10:221–30. doi: 10.1002/gcc.2870100402. [DOI] [PubMed] [Google Scholar]
- 2.Wang SI, Puc J, Li J, Bruce JN, Cairns P, Sidransky D, et al. Somatic mutations of PTEN in glioblastoma multiforme. Cancer research. 1997;57:4183–6. [PubMed] [Google Scholar]
- 3.Zhuang Z, Park WS, Pack S, Schmidt L, Vortmeyer AO, Pak E, et al. Trisomy 7-harbouring non-random duplication of the mutant MET allele in hereditary papillary renal carcinomas. Nature genetics. 1998;20:66–9. doi: 10.1038/1727. [DOI] [PubMed] [Google Scholar]
- 4.Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. In: Harris Henry., translator. Journal of cell science. Suppl 1. 121 . 2008. pp. 1–84. [DOI] [PubMed] [Google Scholar]
- 5.Li M, Fang X, Wei Z, York JP, Zhang P. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. The Journal of cell biology. 2009;185:983–94. doi: 10.1083/jcb.200904020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osada T, Kusakabe H, Akutsu H, Yagi T, Yanagimachi R. Adult murine neurons: their chromatin and chromosome changes and failure to support embryonic development as revealed by nuclear transfer. Cytogenetic and genome research. 2002;97:7–12. doi: 10.1159/000064037. [DOI] [PubMed] [Google Scholar]
- 7.Pack SD, Weil RJ, Vortmeyer AO, Zeng W, Li J, Okamoto H, et al. Individual adult human neurons display aneuploidy: detection by fluorescence in situ hybridization and single neuron PCR. Cell cycle (Georgetown, Tex. 2005;4:1758–60. doi: 10.4161/cc.4.12.2153. [DOI] [PubMed] [Google Scholar]
- 8.Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, et al. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–80. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yurov YB, Iourov IY, Monakhov VV, Soloviev IV, Vostrikov VM, Vorsanova SG. The variation of aneuploidy frequency in the developing and adult human brain revealed by an interphase FISH study. J Histochem Cytochem. 2005;53:385–90. doi: 10.1369/jhc.4A6430.2005. [DOI] [PubMed] [Google Scholar]
- 10.Kingsbury MA, Friedman B, McConnell MJ, Rehen SK, Yang AH, Kaushal D, et al. Aneuploid neurons are functionally active and integrated into brain circuitry. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6143–7. doi: 10.1073/pnas.0408171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang AH, Kaushal D, Rehen SK, Kriedt K, Kingsbury MA, McConnell MJ, et al. Chromosome segregation defects contribute to aneuploidy in normal neural progenitor cells. J Neurosci. 2003;23:10454–62. doi: 10.1523/JNEUROSCI.23-32-10454.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, et al. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–3. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 13.Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nature genetics. 2004;36:1159–61. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 14.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–31. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 15.Wassmann K, Benezra R. Mitotic checkpoints: from yeast to cancer. Current opinion in genetics & development. 2001;11:83–90. doi: 10.1016/s0959-437x(00)00161-1. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) The Journal of cell biology. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto A, Guacci V, Koshland D. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. The Journal of cell biology. 1996;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. The Journal of cell biology. 2003;160:341–53. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nature genetics. 2004;36:744–9. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 20.Basu J, Bousbaa H, Logarinho E, Li Z, Williams BC, Lopes C, et al. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. The Journal of cell biology. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–45. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 22.Kitagawa R, Rose AM. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nature cell biology. 1999;1:514–21. doi: 10.1038/70309. [DOI] [PubMed] [Google Scholar]
- 23.Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–9. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Liu T, Fang Y, Xie S, Huang X, Mahmood R, et al. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 2004;103:1278–85. doi: 10.1182/blood-2003-06-2158. [DOI] [PubMed] [Google Scholar]
- 25.Schliekelman M, Cowley DO, O’Quinn R, Oliver TG, Lu L, Salmon ED, et al. Impaired Bub1 function in vivo compromises tension-dependent checkpoint function leading to aneuploidy and tumorigenesis. Cancer research. 2009;69:45–54. doi: 10.1158/0008-5472.CAN-07-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizzardi C, Torelli L, Barresi E, Schneider M, Canzonieri V, Biasotto M, et al. BUBR1 expression in oral squamous cell carcinoma and its relationship to tumor stage and survival. Head Neck. 2010 doi: 10.1002/hed.21532. [DOI] [PubMed] [Google Scholar]
- 28.Liu AW, Cai J, Zhao XL, Xu AM, Fu HQ, Nian H, et al. The clinicopathological significance of BUBR1 overexpression in hepatocellular carcinoma. J Clin Pathol. 2009;62:1003–8. doi: 10.1136/jcp.2009.066944. [DOI] [PubMed] [Google Scholar]
- 29.Pinto M, Vieira J, Ribeiro FR, Soares MJ, Henrique R, Oliveira J, et al. Overexpression of the mitotic checkpoint genes BUB1 and BUBR1 is associated with genomic complexity in clear cell kidney carcinomas. Cell Oncol. 2008;30:389–95. doi: 10.3233/CLO-2008-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto Y, Matsuyama H, Chochi Y, Okuda M, Kawauchi S, Inoue R, et al. Overexpression of BUBR1 is associated with chromosomal instability in bladder cancer. Cancer Genet Cytogenet. 2007;174:42–7. doi: 10.1016/j.cancergencyto.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 31.Grabsch H, Takeno S, Parsons WJ, Pomjanski N, Boecking A, Gabbert HE, et al. Overexpression of the mitotic checkpoint genes BUB1, BUBR1, and BUB3 in gastric cancer--association with tumour cell proliferation. J Pathol. 2003;200:16–22. doi: 10.1002/path.1324. [DOI] [PubMed] [Google Scholar]
- 32.Shin HJ, Baek KH, Jeon AH, Park MT, Lee SJ, Kang CM, et al. Dual roles of human BubR1, a mitotic checkpoint kinase, in the monitoring of chromosomal instability. Cancer cell. 2003;4:483–97. doi: 10.1016/s1535-6108(03)00302-7. [DOI] [PubMed] [Google Scholar]
- 33.Fukasawa K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005;230:6–19. doi: 10.1016/j.canlet.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 34.Duensing S, Munger K. Centrosome abnormalities, genomic instability and carcinogenic progression. Biochimica et biophysica acta. 2001;1471:M81–8. doi: 10.1016/s0304-419x(00)00025-1. [DOI] [PubMed] [Google Scholar]
- 35.Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- 36.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. The Journal of cell biology. 2001;153:517–27. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol. 2009;19:1937–42. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. The Journal of cell biology. 1995;131:721–34. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16:1711–8. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 42.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–82. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 43.Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Molecular cell. 2002;9:773–88. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 44.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–77. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 45.Gimenez-Abian JF, Sumara I, Hirota T, Hauf S, Gerlich D, de la Torre C, et al. Regulation of sister chromatid cohesion between chromosome arms. Curr Biol. 2004;14:1187–93. doi: 10.1016/j.cub.2004.06.052. [DOI] [PubMed] [Google Scholar]
- 46.Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS biology. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Losada A, Hirano M, Hirano T. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes & development. 2002;16:3004–16. doi: 10.1101/gad.249202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, et al. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Molecular cell. 2002;9:515–25. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- 49.Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 50.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–7. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 51.Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–78. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 52.Iwaizumi M, Shinmura K, Mori H, Yamada H, Suzuki M, Kitayama Y, et al. Human Sgo1 downregulation leads to chromosomal instability in colorectal cancer. Gut. 2009;58:249–60. doi: 10.1136/gut.2008.149468. [DOI] [PubMed] [Google Scholar]
- 53.Wang X, Yang Y, Duan Q, Jiang N, Huang Y, Darzynkiewicz Z, et al. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Developmental cell. 2008;14:331–41. doi: 10.1016/j.devcel.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 55.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–86. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 56.Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW. Dual inhibition of sister chromatid separation at metaphase. Cell. 2001;107:715–26. doi: 10.1016/s0092-8674(01)00603-1. [DOI] [PubMed] [Google Scholar]
- 57.Salehi F, Kovacs K, Scheithauer BW, Lloyd RV, Cusimano M. Pituitary tumor-transforming gene in endocrine and other neoplasms: a review and update. Endocr Relat Cancer. 2008;15:721–43. doi: 10.1677/ERC-08-0012. [DOI] [PubMed] [Google Scholar]
- 58.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science (New York, NY. 2007;317:916–24. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 59.Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science (New York, NY. 2008;322:703–9. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burds AA, Lutum AS, Sorger PK. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11296–301. doi: 10.1073/pnas.0505053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li M, Fang X, Baker DJ, Guo L, Gao X, Wei Z, et al. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14188–93. doi: 10.1073/pnas.1005960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. The Journal of cell biology. 2010;188:369–81. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science (New York, NY. 2010;330:517–21. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 64.Ye CJ, Liu G, Bremer SW, Heng HH. The dynamics of cancer chromosomes and genomes. Cytogenetic and genome research. 2007;118:237–46. doi: 10.1159/000108306. [DOI] [PubMed] [Google Scholar]
- 65.Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–93. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Duesberg P, Fabarius A, Hehlmann R. Aneuploidy, the primary cause of the multilateral genomic instability of neoplastic and preneoplastic cells. IUBMB Life. 2004;56:65–81. doi: 10.1080/15216540410001667902. [DOI] [PubMed] [Google Scholar]
- 68.Duesberg P, Li R. Multistep carcinogenesis: a chain reaction of aneuploidizations. Cell cycle (Georgetown, Tex. 2003;2:202–10. [PubMed] [Google Scholar]
- 69.Duesberg P, Rasnick D. Aneuploidy, the somatic mutation that makes cancer a species of its own. Cell Motil Cytoskeleton. 2000;47:81–107. doi: 10.1002/1097-0169(200010)47:2<81::AID-CM1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 70.Baker DJ, Jeganathan KB, Malureanu L, Perez-Terzic C, Terzic A, van Deursen JM. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. The Journal of cell biology. 2006;172:529–40. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. The Journal of cell biology. 2007;179:255–67. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 73.Dai W, Wang Q, Liu T, Swamy M, Fang Y, Xie S, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer research. 2004;64:440–5. doi: 10.1158/0008-5472.can-03-3119. [DOI] [PubMed] [Google Scholar]
- 74.Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, Peloponese JM, Jr, et al. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer research. 2007;67:160–6. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- 75.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–87. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Janssen A, Kops GJ, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19108–13. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes & development. 2008;22:2189–203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]