Summary
In biology as in real estate, location is a cardinal organizational principle that dictates the accessibility and flow of informational traffic. An essential question in nuclear organization is the nature of the address code—how objects are placed and later searched for and retrieved. Long noncoding RNAs (IncRNAs) have emerged as key components of the address code, allowing protein complexes, genes, and chromosomes to be trafficked to appropriate locations and subject to proper activation and deactivation. LncRNA-based mechanisms control cell fates during development, and their dysregulation underlies some human disorders caused by chromosomal deletions and translocations.
Introduction
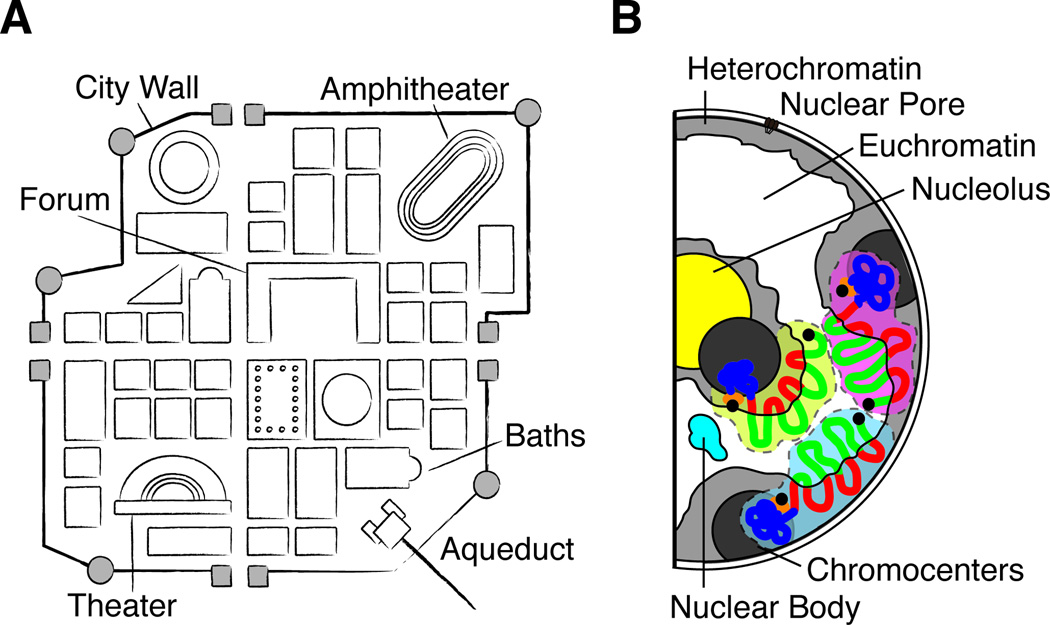
From a single cell to an entire organism, spatial positioning is a key problem in biology. It is well appreciated that robust systems sort and distribute macromolecules, a property essential for the function of cells and tissues (Shevtsov and Dundr, 2011; Wolpert, 2011). A historical example illustrates the general utility of spatial organization. As the Roman Empire expanded and the Romans were faced with the need to construct cities in new lands, they developed a city prototype that included a group of answers to the many practical problems related to the creation and maintenance of a city (Figure 1A). This was a universal plan of simple execution. City walls protected the citizens from attack and delimited the city. At the center stood the forum, where the business and political activities of the city were concentrated. Fountains were placed throughout the city to supply water, and other spaces were dedicated to organize daily activities, such as amphitheaters, temples, and baths. Thus, a group of structures analogous in function was always present, in an organization that follows the original prototype (Grimal and Woloch, 1983).
Figure 1. Comparison between a Roman city and the cell nucleus reveals the importance of spatial organization.
(A) Depiction of the basic features of a Roman city. City walls delimit the city, with gates at the two main roads that intersect at the center of the city. The Forum was the business and political center of the city and many buildings provided specific functions essential for city life. (B) Schematic representation of the typical nuclear organization during interphase. Each chromosome occupies a discrete territory. Euchromatin localizes to the interior regions of the nucleus and the densely compacted Heterochromatin localizes near the nuclear envelope. Many specialized functions are executed in distinct regions in the nucleus, known as nuclear bodies. One example is the nucleolus, where ribosomes are assembled. Adapted from (Solovei et al., 2009).
Just like the Roman city, the nucleus of the eukaryotic cell is a highly organized space (Figure 1B). Evolution gave rise to a ‘nuclear’ prototype that provides answers to the many challenges the cell has to respond to maintain homeostasis and growth, though subject to developmental specialization (Solovei et al., 2009). Chromosomes are not randomly organized in the nucleus, and during interphase each chromosome occupies a discrete territory (reviewed in (Cremer and Cremer, 2010)). Furthermore, while the densely compacted heterochromatin is localized at the nuclear envelope, euchromatin localizes to the interior regions of the nucleus. Gene expression is also localized, and occurs mostly at nuclear center. In addition, active genes that are co-regulated are often found forming clusters. During development, individual loci such as immunoglobulin or Hox genes are known to change position within the nucleus according to their transcriptional status (reviewed in (Misteli, 2007)). Large portions of the genome are partitioned into topological domains of chromatin interaction ranging from hundreds of kilobases to megabase (the resolution of current methods), within which the genes tend to be more co-regulated (Dixon et al., 2012; Nora et al., 2012). The complex task of gene expression—ensuring the proper timing, space, and rate of expression—involves noncoding regions of the genome, chromatin modifications and the arrangement of chromosomes and nuclear domains. Here we review the evidence that IncRNAs are a rich source of molecular addresses in the eukaryotic nucleus.
Biogenesis and Characteristics
Efforts over the last decade revealed that a large fraction of the noncoding genome is transcribed. Extensive annotation of IncRNA has been performed in multiple model organisms (reviewed in (Rinn and Chang, 2012)) and there is now evidence that, while 2% of genome encodes for proteins (2004), primary transcripts cover 75% of the human genome, with processed transcripts covering 62.1% of the genome (Djebali et al., 2012). In this review we focus on a particular class of noncoding transcripts known as long noncoding RNAs (IncRNAs), and the roles they play in nuclear organization.
LncRNAs are currently defined as transcripts of greater than 200 nucleotides without evident protein coding function (Rinn and Chang, 2012). It is important to note that IncRNA is a broad definition that encompasses different classes of RNA transcripts, including enhancer RNAs, snoRNA hosts, intergenic transcripts, and transcripts overlapping other transcripts in either sense or antisense orientation. LncRNAs predominantly localize to the nucleus and have on average a lower level of expression than protein coding genes, although details vary for different classes (Djebali et al., 2012; Ravasi et al., 2006). Multiple studies have shown that IncRNA expression is more cell type specific that protein coding genes (Cabili et al., 2011; Djebali et al., 2012; Ravasi et al., 2006). At the DNA and chromatin level, IncRNA loci are similar to mRNA loci, but IncRNAs show a bias for having just one intron and a trend for less efficient co-transcriptional splicing (Derrien et al., 2012; Tilgner et al., 2012). Although IncRNAs are under lower selective pressure than protein coding genes, sequence analysis shows that InRNAs are under higher selective pressures than ancestral repeat sequences which are considered to be under neutral selection. Interestingly, the promoters of IncRNAs are the region of the IncRNA gene under higher selective pressure, displaying levels of selection comparable to the promoters of protein coding genes (Derrien et al., 2012; Guttman et al., 2009; Marques and Ponting, 2009; Orom et al., 2010; Ponjavic et al., 2007). This analysis has also revealed a high number of correlated positions between IncRNA in sequence alignments, an observation that fits the hypothesis that IncRNAs are under selective pressure to maintain a functional RNA structure (Derrien et al., 2012). Comparison between mammalian and zebrafish IncRNAs revealed that short stretches of conserved sequence are functionally important and that location and structure of IncRNAs can be conserved, even in the absence of strong sequence conservation. The ability to induce a loss of function phenotype by blocking the short conserved motif in addition to the ability to rescue loss of function of two IncRNAs with the addition of human and mouse IncRNAs (Ulitsky et al., 2011), demonstrates that these ‘in silico’ observations are of biological significance.
Sequence analysis of IncRNAs, focusing on presence and size of open reading frames, as well as codon conservation frequency has been used to exclude protein coding potential. Ribosome profiling, a method that enumerates transcripts associated with ribosomes, had detected many IncRNAs, but it was unclear if these IncRNAs are just being scanned similar to 5’ untranslated regions or actually productively engaged in translation (Ingolia et al., 2011). Comparison of RNA-seq data to tandem mass spectrometry data for two cell lines suggests that ~92% of the annotated IncRNAs do not yield detectable peptides in these cell lines (Banfai et al., 2012; Derrien et al., 2012). While the differences between these two studies may stem from measuring two different endpoints, they suggest that IncRNAs have low translational potential even when ribosomes attempt to decode them. Current annotations suggest that the actual number of IncRNAs exceeds that of protein coding genes. (Derrien et al., 2012).
The repertoire of roles performed by IncRNAs is growing; as there is now evidence that IncRNA participate in multiple networks regulating gene expression and function. Several characteristics of IncRNAs make them the ideal system to provide the nucleus with a system of molecular addresses. LncRNAs, unlike proteins, can function both in cis, at the site of transcription, or in trans. An RNA-based address code may be deployed more rapidly and economically than a system that relies only on proteins. LncRNAs do not need to be translated, and do not require transport between the cytoplasm and the nucleus. LncRNAs can also interact with multiple proteins, enabling scaffolding functions and combinatorial control (Wang and Chang, 2011). As such, the act of transcription can rapidly create an anchor that will lead to the formation, or remodeling, of nuclear domains, through the recruitment or sequestration of proteins already present in the nuclear compartment. Using IncRNAs allows cells to create addresses that are regional-, locus- or even allele-specific (Lee, 2009). At the regional level IncRNAs can influence the formation of nuclear domains, the transcriptional status of an entire chromosome and can participate in the interaction of two different chromosomal regions. At a more fine-grained level, IncRNA can control the chromatin state and activity of a chromosomal locus or specific gene. We explore each of these concepts below with recently published examples.
Locus Control of Gene Regulation
Cells can use noncoding RNAs to modulate gene expression by changing the accessibility of gene promoters. These mechanisms can be used to fine tune gene expression in response to environmental conditions or to silence a gene as part of a developmental program.
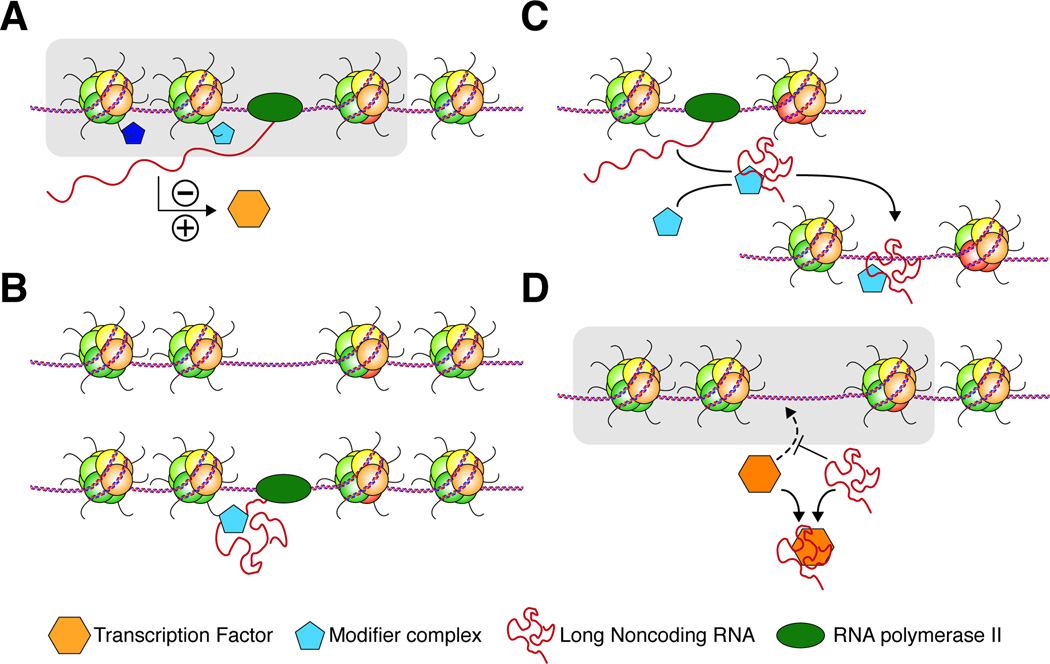
First, the act of ncRNA transcription itself can be purposed for regulatory function. For example, transcription through a regulatory sequence, such as a promoter, can block its function, a mechanism termed transcriptional interference (Figure 2A) first identified in yeast (Martens et al., 2004). In such instances, the IncRNA promoter is finely tuned to receive appropriate inputs to exert regulatory function; the IncRNA product is typically a faithful biomarker of transcriptional interference in action but is not required for its success. In conditions that limit vegetative growth, diploid S. cerevisiae cells enter sporulation, a differentiation program that results in the formation of haploid daughter cells. Entry into meiosis has catastrophic consequences in haploid cells, and is therefore inhibited via a transcriptional interference mechanism. A transcription factor in haploid cells activates the expression of IRT1(SUT643), a noncoding RNA that overlaps the promoter of IME1, the master regulator of sporulation. Transcription of IRT1 establishes a repressive chromatin state at the IME1 promoter through the recruitment of histone methyltransferase Set2 and the histone deacetylase Set3 (van Werven et al., 2012). The use of noncoding transcription to control chromatin modification is a wide spread strategy. The Set3 histone deacetylase has also been implicated in the modulation gene induction kinetics during changes of carbon source. Transcription of ncRNAs that overlap the regulated genes leads to the establishment of H3K4me2, which recruits Set3 and leads to the deacetylation of the gene promoter. Deacetylation of the promoter results in delayed or reduced induction of the regulated genes. This mechanism is also involved in the inhibition of cryptic promoters (Kim et al., 2012). Expression of GAL10-ncRNA, driven by Reb1, leads to deacetylation across the GAL1-10 promoter, facilitating glucose repression of GAL1-10 (Houseley et al., 2008).
Figure 2. Functional modules of IncRNAs in the nucleus.
(A) The act of transcription at noncoding regions can modulate gene expression through the recruitment of chromatin modifiers to the site of transcription. These complexes can create a local chromatin environment that facilitates or blocks the binding of other regulators. (B) LncRNAs can function in cis, recruiting protein complexes to their site of transcription, thus creating a locus specific address. Cells can use this mechanism to repress gene or activate gene expression. (C) LncRNAs can function in trans and recruit protein complexes to chromatin loci away from their site of transcription. (D) LncRNAs can bind and sequester transcription factors away from their target chromosomal regions.
In mammalian imprinting, the noncoding RNA Air (also known as Aim), is expressed from the paternal chromosome and involved in silencing the paternal alleles of multiple genes. The promoter of one of these genes, Igf2r, overlaps with the Air transcriptional unit, and is silenced by transcriptional interference (Latos et al., 2012).
Transcriptional interference can also be used to activate gene expression, by inhibiting the action of repressor elements, functioning as an anti-silencing mechanism. In Drosophila embryogenesis, transcription through Polycomb response elements (PRE) alters the function of these elements, blocking the establishment of repressive chromatin (Schmitt et al., 2005).
Second, IncRNAs can silence or activate gene expression in cis, acting on neighboring genes of the IncRNA locus. Some of the first studied examples of IncRNA function involve dosage compensation and genomic imprinting, where IncRNAs provide allele-specific gene regulation to differentially control two copies of the same gene within one cell (reviewed in this issue[sym1]) (Figure 2B). Several such IncRNAs are now recognized to interact with and recruit histone modification complexes, including Xist (recruits PRC2 for H3K27me3 and RYBP-PRC1 for H2A ubiquitylation), and Kcnq1ot1 (recruits G9a for H3K9me3 and PRC2) (Pandey et al., 2008; Tavares et al., 2012; Zhao et al., 2010). The Air IncRNA (the transcription of which inhibits Igfr2) recruits and targets G9a and H3K9me3 to silence more distantly located gene on the paternal chromosome (Nagano et al., 2008); hence one IncRNA gene can employ multiple mechanisms to regulate nearby and distantly located genes. In genome-wide studies, numerous IncRNAs have now been found to interact with chromatin modification complexes (Guil et al., 2012; Guttman et al., 2011; Khalil et al., 2009; Zhao et al., 2010). In the plant A. thaliana, two cold-inducible IncRNAs COOLAIR and COLDAIR are embedded antisense or intronic to the flowering control locus gene FLC, and they help to recruit PRC2 to stably silence FLC in a cold-dependent manner, a key mechanism to ensure the proper flowering time after winter termed vernalization (reviewed in (Ietswaart et al., 2012)). In an analogous fashion, DNA damage induces a IncRNA from the promoter of cyclin D1 gene (CCND1); this IncRNA binds to TLS protein to allosterically inhibit histone acetyltransferase in cis, which suppresses CCND1 transcription (Wang et al., 2008).
DNA methylation can occur as a long term silencing mechanism downstream of repressive histone modifications, and IncRNAs may also guide DNA methylation in addition to histone modification. The ribosomal DNA (rDNA) loci are tandemly repeated in the genome, with some copies being transcriptionally active while others are silenced by DNA methylation and histone modifications. Each ribosomal DNA transcribes rRNA separated by intergenic spacers (IGSs) as a polycistronic unit, and IGS can be process to 150–250 nt fragments termed promoter (p)RNAs (reviewed in (Bierhoff et al., 2010)). pRNA serves as a platform to recruit the de novo cytosine methylase DNMT3 and the NoRC complex containing poly-ADP ribose polymerase-1 (PARP-1) to promote silencing of rDNA (Guetg et al., 2012; Mayer et al., 2006). Notably, a stretch of 20nt in pRNA binds the rDNA promoter, forming a RNA:DNA:DNA triplex (Schmitz et al., 2010). This triplex structure is proposed to recruit DNMT3, and also serves as the specific recognition mechanism between IncRNA and genomic DNA—a model likely applies to other IncRNA-DNA interactions (Martianov et al., 2007).
A distinct family of IncRNAs serves to activate gene expression. Many active enhancer elements transcribe IncRNAs, termed eRNAs (De Santa et al., 2010; Kim et al., 2010), and several IncRNAs are required to activate gene expression, which are termed enhancer-like RNAs (Orom et al., 2010). Evf is a cis-acting IncRNA that is required for activation of Dlx5/6 genes and generation of GABAergic interneurons in vivo (Martianov et al., 2007). A key mechanism of IncRNA specificity in cis is the higher order chromosomal configuration (Wang et al., 2011). The noncoding RNA HOTTIP is expressed from the 5’ end tip of the HoxA locus and drives histone H3 lysine 4 trimethylation and gene transcription of HoxA distal genes through the recruitment of the WDR5/MLL complex (Wang et al., 2011). Endogenous HOTTIP is brought to its target genes by chromosomal looping, and ectopic HOTTIP only activates transcription when it is artificially tethered to the reporter gene (Wang et al., 2011). The MLL complex is also recruited to the Hox locus by the noncoding RNA Mistral, located between Hoxa6 and Hoxa7. Mistral directly interacts with MLL1, leading to changes at the chromatin level that activate Hoxa6 and Hoxa7 (Bertani et al., 2011). Hence, IncRNA interaction with MLL/Trx complexes and likely additional proteins will define their function in enforcing active chromatin states and gene activation.
Third, IncRNAs can control chromatin states at distantly located genes (i.e. in trans) both for gene silencing and activation (Figure 2C). These IncRNAs bind to some of the same effector chromatin modification complexes, but target them to genomic loci genome-wide. For instance, human HOTAIR IncRNA binds to PRC2 and LSD1 complexes and couples H3K27 methylation and H3K4 demethylation activity to hundreds of sites genome-wide (Chu et al., 2011; Tsai et al., 2010). HOTAIR is located in the HOXC locus and is regulated in an anatomic position-specific fashion. Linc-p21 is induced by p53 during DNA damage, and recruits hnRNPK via physical interaction to mediate p53-mediated gene repression (Huarte et al., 2010). Linc-p21 also has a recently recognized role in translational control (Yoon et al., 2012). In contrast, PANDA, another IncRNA induced by p53, acts as a decoy by binding to the transcription factor NF-YA and preventing NF-YA from activating genes encoding cell death proteins (Hung et al., 2011) (Figure 2D). LncRNA-mediated activation can also occur in trans. Jpx, a X-linked IncRNA that activates Xist expression, is important for X chromosome inactivation in female cells, and Jpx deletion can be rescued by Jpx supplied in trans (Tian et al., 2010).
Nuclear Domains
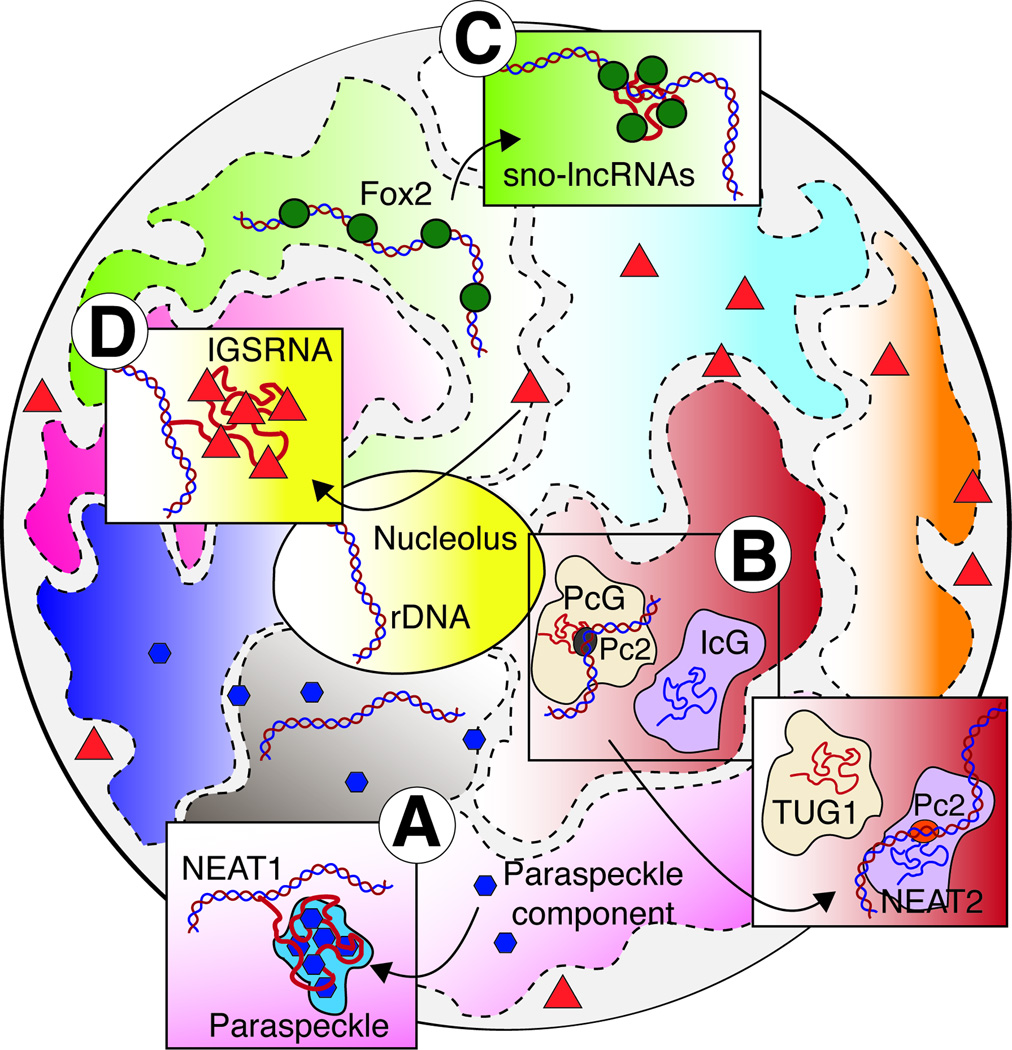
The concept of IncRNA recruitment of factors to genes may be more properly considered a two-way street, with genes being moved into specific cytotopic locations by IncRNAs. One type of molecular address can be found in the formation of nuclear domains. These are regions of the nucleus where specific functions are performed. Unlike cellular organelles, these domains are not membrane delimited. They are instead characterized by the components that form them. These domains are believed to form through molecular interactions between its components. Once a stable interaction is found, the components remain associated. These domains are often formed around the sites of transcription of RNA components, which function as molecular anchors (reviewed in (Dundr and Misteli, 2010)). The noncoding RNA NEAT1, an essential component of the Paraspeckle, is a well-characterized example of how noncoding RNAs can function as structural components of nuclear bodies. Upon transcription of NEAT1, diffusible components of this domain nucleate at the site of NEAT1 accumulation, leading to the formation of the Paraspeckle (Figure 3A) (Chen and Carmichael, 2009; Clemson et al., 2009; Mao et al., 2011; Sasaki et al., 2009; Shevtsov and Dundr, 2011; Sunwoo et al., 2009).
Figure 3. Schematic representation of the cell nucleus showing the nucleolus and chromosomal territories.
(A) Protein components of the Paraspeckle diffused throughout the nucleoplasm aggregate upon the transcription of NEAT1 forming the Paraspeckle nuclear domain. (B) Pc2 differentially binds MALAT1/NEAT2 or TUG1 depending on methylation status. Methylated Pc2 interacts with TUG1, bringing associated growth control genes to a repressive environment; the polycome body (PcG). Unmethylated Pc2 interacts with MALAT1/NEAT2 at the Interchromatin granule (ICG), where gene expression is permitted. (C) Expression of IncRNAs with snoRNA ends from the Prader-Willi syndrome locus function as sinks for the FOX2 protein, leading to a redistribution of this splicing factor in this nuclear region. (D) In response to cellular stress, transcription of specific IGSRNAs leads to the retention of targeted proteins at the nucleolus. Different types of stress lead to the retention of different proteins, through the expression of specific noncoding RNAs.
Nuclear domains can be dynamically regulated in a RNA-dependent fashion. In response to serum stimulation, the demethylase KDM4C is recruited to the promoters of genes controlled by the cell cycle-specific transcription factor E2F, where it demethylates Polycomb protein Pc2. While methylated Pc2 interacts with the noncoding RNA TUG1, a component of Polycomb bodies, unmethylated Pc2 interacts with the noncoding RNA MALAT1/NEAT2, a component of Interchromatin granules. Therefore, changes in the methylation status of Pc2 lead to the relocation of growth-control genes from an environment that inhibits gene expression, the Polycomb body, to a domain permissive of gene expression, the Interchromatin granule (Figure 3B). Interestingly, the reading ability of Pc2 is modulated by the noncoding RNA it is interacting with. When bound to TUG1, Pc2 reads H4R3me2s and H3K27me2 while it reads H2AK5ac and H2AK13ac when interacting with MALAT1/NEAT2 (Yang et al., 2011). These interplays control the growth factor-dependent expression of cell cycle genes in vitro, but it came as a surprise that mouse knockouts of either NEAT1 or MALAT1/NEAT2 had no little overt phenotype (Eissmann et al., 2012; Nakagawa et al., 2012; Nakagawa et al., 2011; Zhang et al., 2012). Clearly, the question of redundancy or compensation in vivo needs to be addressed in the future.
Unusual processing mechanisms may explain the localization activity of certain IncRNAs. An imprinted region in chromosome 15 (15q11-q13) that had been implicated in the Prader-Willi syndrome (PWS) hosts multiple intron derived IncRNAs with small nucleolar RNAs at their ends—so called sno-lncRNAs. It is probable that the presence of structured snoRNAs at the ends of IncRNAs stabilizes these molecules, which have no 5’ cap or polyA tail. These RNAs are retained in the nucleus and localize to, or remain near, their sites of transcription. Knockdown of sno-lncRNAs has little effect on the expression of nearby genes, suggesting that it does not affect gene expression in cis. Instead, these sno-IncRNAs seem to create a ‘domain’ where the splicing factor Fox2 is enriched. These sno-lncRNAs contain multiple binding sites for Fox2, and altering the level of sno-lncRNAs led to a redistribution of Fox2 in the nucleus and changes in mRNA splicing patterns. Hence, the sno-lncRNAs appear to function as Fox2 sinks, participating in the regulation of splicing in specific subnuclear domains (Yin et al., 2012) (Figure 3C). Similarly, formation of a blunt-ended triplex RNA structure at the 3’ end of MALAT1/NEAT2 IncRNA, which lacks a polyA tail, stabilizes the IncRNA and presumably limits its export to the cytoplasm (Brown et al., 2012; Wilusz et al., 2012). Viral nuclear IncRNAs have also adapted this strategy and hides its 3’ polyA tail in a triplex RNA structure to prevent decay (Mitton-Fry et al., 2010; Tycowski et al., 2012).
Gene Control through Sequestration
In contrast to the model of nuclear domains that concentrate and thereby facilitate molecular interactions, spatial control can also separate reactants until the moment is right. For example, certain environmental stresses trigger the retention of select proteins in the nucleolus, away from their normal site of action. The retention at the nucleolus requires a signal sequence and the expression of specific noncoding RNAs expressed from large intergenic spacer (IGS) of the rDNA repeats. IGS ncRNAs turn out to gate the responses to cellular stress. Unique IGS ncRNAs are transcriptionally induced by specific stressors, functioning as baits for proteins with specific signal sequences, and interfering with a specific IGSRNA does not affect the function of other IGSRNAs (Audas et al., 2012) (Figure 3D).
In S. pombe both mRNAs and IncRNAs function together to form heterochromatin and sequester genes in the control of meiosis. During vegetative growth the expression of meiotic genes is repressed through selective elimination of meiotic mRNAs. Meiotic genes contain within their transcripts a region, known as determinant of selective removal (DSR), that determines their degradation. This sequence is recognized by Mmi1, which promotes both mRNA degradation (Harigaya et al., 2006) as well as formation of facultative heterochromatic islands (Zofall et al., 2012). Hence, aberrant nascent mRNAs can function in an IncRNA-like fashion to tether the formation for heterochromatin. Furthermore, during vegetative growth, Mei2p, an RNA binding protein crucial for entry in meiosis, is kept in an inactive form. When cells commit to the meiosis expression program, Mei2p accumulates in its active form and sequesters Mmi1 to a structure known as Mei2 dot, where Mmi1 function is inhibited. The Mei2 dot forms at the sme2 locus, at the site of transcription of two noncoding RNAs, meiRNA-S and meiRNA-L, which are necessary for the formation of the Mei2 dot structure, and therefore entry in meiosis (Yamamoto, 2010).
Higher order chromosomal interactions
An intriguing possibility is that IncRNAs can regulate the three dimensional structure of the chromosomes by facilitating the interaction of specific chromosomal loci. The act of transcription itself can influence gene expression and genome organization by promoting chromatin modifications, recruiting gene active regions to common transcription factories or by exposing the DNA strands to enzymatic activity. Hence, the presence of multiple IncRNA genes in a region may help chromosomal loci adopt distinct conformation with transcriptional activation. For example in the Hox loci, collinear expression of Hox mRNA genes and Hox IncRNAs along the chromosome is associated with the progressive recruitment of those chromosomal segments into a tightly interacting domain distinct from the transcriptionally silent portion of the loci (Noordermeer et al., 2011; Wang et al., 2011). A similar phenomenon was first appreciated in the (3 globin locus and intergenic transcripts from its locus control regions (Ashe et al., 1997). Transcription-coupled looping is likely related to the fact that the Mediator complex that links transcription factors to basal transcription machinery promotes long range enhancer-promoter interactions (Kagey et al., 2010). A similar transcription-directed mechanism has also been proposed to guide DNA recombination of lymphocyte receptor genes over megabases (Verma-Gaur et al., 2012). The IncRNA transcripts are useful readouts of the chromosomal configuration, but are not necessarily required for the chromosomal interactions.
LncRNAs can also regulate chromosome structure through direct mechanisms. High throughput chromosomal conformation assays revealed that the active and inactive X chromosomes adopt quite distinct conformations. The inactive X (Xi) is coated by the Xist IncRNA, which is required for choosing the inactive X chromosome. Importantly, conditional knockout of Xist has demonstrated that the folding of inactive X requires the Xist RNA. After Xist deletion, the Xi chromosome adopts a conformation more similar to that of the active X chromosome (Xa) without reactivation of Xi gene expression. Hence, Xist appears to regulate X chromosome structure through mechanisms other that the relocation of active genes to transcriptional factories (Splinter et al., 2011). One intriguing clue is that conditional Xist deletion also led to loss of PRC2 and H3K27me3 marks. The conformations of the two X chromosomes appear to be regulated by distinct mechanisms, because PRC2 is dispensable for the topological domains of Xa (Nora et al., 2012). Whether one of several Xa-expressed IncRNA controls Xa conformation remains to be seen.
LcnRNAs can also regulate the interaction between chromosomes, a concept exemplified by S. pombe meiosis. In order for chromosomes to properly segregate in meiosis and prevent aneuploidy, homologous chromosomes must interact and generate stable associations. The sme2 locus plays a key role in the mutual identification of homologous chromosomes during meiosis, in addition to its role in the mitosis/meiosis switch discussed above. The meiRNA-L transcript accumulates at the sme2 locus and is necessary for the robust chromosomal pairing (Ding et al., 2012). These studies suggest that noncoding RNAs can be components of a cis-acting pairing factor that allows homologous chromosomes to identify each other.
Cytoplasmic functions
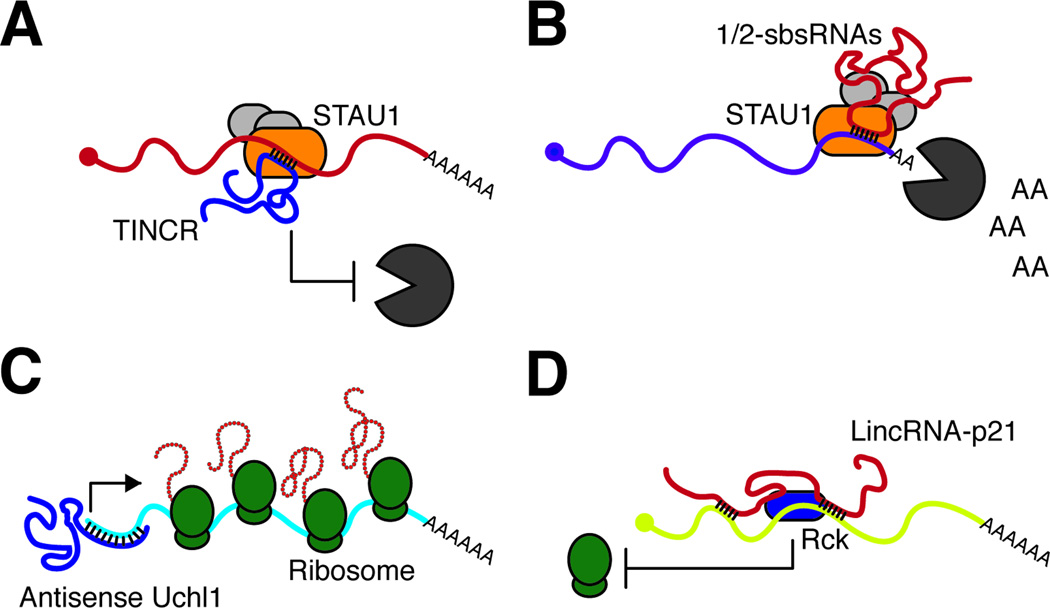
The ultimate function of mRNAs is to be translated, and like other steps of gene expression, multiple layers of post-transcriptional regulation exist in the cytoplasm (Figure 4). LncRNAs can also ‘identify’ mRNAs in the cytoplasm and modulate their ‘life cycle’. Recent works demonstrated that IncRNAs impact both the mRNA half-life and translation of mRNAs. The IncRNA TINCR (terminal differentiation-induced ncRNA) is induced during epidermal differentiation and is required for normal induction of key mediators of epidermal differentiation. TINCR localizes to the cytoplasm where it interacts with Staufen 1 protein (STAU1) to promote the stability of mRNAs containing the TINCR box motif (Kretz et al., 2012) (Figure 4A). Hence, the TINCR mechanism is the diametric opposite of post-transcriptional silencing by small regulatory RNAs like siRNA or miRNAs. STAU1 can also be programmed by other IncRNAs to facilitate mRNA degradation. The half-STAU1-binding site RNAs (1/2-sbsRNAs) contain Alu elements that bind to Alu elements in the 3’UTR of actively transcribed targets genes, generating a STAU1-binding site. These mRNAs are therefore identified as STAU1-mediated messenger RNA decay (SMD) targets (Gong and Maquat, 2011) (Figure 4B). In addition, a recently identified class of IncRNA impacts gene expression by promoting translation of targets mRNAs. Expression of Antisense Uchl1 RNA leads to an increase in Uchl1 protein level without any change at the mRNA level. Antisense Uchl1 IncRNA is composed by a region that overlaps with the first 73 nucleotides of Uchl1 and two embedded repetitive sequences, one of which (SINEB2) is required for the ability of the IncRNA to induce protein translation. Under stress conditions where cap-dependent translation is inhibited, antisense Uchl1 IncRNA, previously enriched in the nucleus, moves into the cytoplasm and hybridizes with Uchl1 mRNA to enable cap-independent translation of Uchl1. In other words, the IncRNA acts like a mobile internal ribosomal entry element to promote selective translation. Other SINEB2 containing antisense IncRNAs may function in a similar way (Carrieri et al., 2012) (Figure 4C). Conversely, lincRNA-p21 can inhibit the translation of target mRNAs. In the absence of HuR, lincRNA-p21 is stable and interacts with the mRNAs CTNNB1 and JUNB and translational repressor Rck, repressing the translation of the targeted mRNAs (Yoon et al., 2012) (Figure 4D). These emerging examples illustrate that IncRNAs can provide a rich palette of regulatory capacities in the cytoplasm.
Figure 4. LncRNAs regulate gene expression in the cytoplasm.
(A) The IncRNA TINCR interacts with STAU1 and target mRNAs containing the TINCR box motif, promoting their stability. (B) LncRNAs of the 1/2-sbsRNAs class hybridize with 3’ UTR containing Alu elements, and promotes the degradation of these target mRNAs. (C) Under stress conditions the IncRNA Antisense to Uchl1 moves from the nucleus to the cytoplasm and binds the 5’ end of the Uchl1 mRNA to promote its translation under stress conditions. (D) LincRNA-p21 interacts with and targets RcK to mRNAs resulting in translation inhibition.
Human Diseases
Considering the wide range of roles that IncRNAs play in cellular networks, it is not surprising that noncoding RNAs have been implicated in disease. Genome wide association studies have revealed that only 7% of disease or trait-associated single nucleotide polymorphisms (SNPs) reside in protein coding exons, while 43% of trait/disease associated SNP are found outside of protein coding genes (Hindorff et al., 2009). In addition to the example of sno-lncRNAs in Prader-Willi syndrome discussed above, several recent discoveries of IncRNAs in Mendelian disorders illustrate the emerging recognition of IncRNAs in human diseases.
Facioscapulohumeral muscular dystrophy (FSHD) is the third most common myopathy and is predominantly caused by a contraction in copy number of the D4Z4 repeats mapping to 4q35. The D4Z4 repeat is the target of several chromatin modifications, including H3K9me3 and H3K27me3, which are reduced in FSHD patients. Cabianca et al. found that long array of D4Z4 repeats recruit Polycomb complexes to promote the formation of a repressive chromatin state that inhibits the expression of genes at 4q35. Loss of D4Z4 repeats results in derepression of DBE-T, a novel IncRNA that functions in cis and localizes to the FSHD locus. DBE-T recruits ASH1L (a component of MLL/TrX complex) leading to improper establishment of active chromatin and expression of genes from 4q35 (Cabianca et al., 2012). Hence, DBE-T is a IncRNA that functions as a locus control element by promoting active chromatin domain, and FSHD results from IncRNA “promoter mutations” that perturb DBE-T regulation.
HELLP syndrome (Hemolysis, Elevated Liver enzymes, Low Platelets) is a recessively inherited life threatening pregnancy complication. Linkage analysis narrowed the HELLP locus to a gene desert between C12orf48 and IGF1 on 12q23.2, where a single 205 kb capped and polyadenylated IncRNA is transcribed (van Dijk et al., 2012). Knockdown of this IncRNA revealed a role in the transition from G2 to mitosis and trophoblast cell invasion, although the precise mechanism is still unclear. Notably, morpholino oligonucleotides complementary to the mutation site in HELLP IncRNA boosted IncRNA level and reversed the gene expression and cell invasion defects.
Similarly, deletions in a coding-gene desert at 16q24.1 lead to Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACD/MPV) (Szafranski et al., 2012). This region contains a distant enhancer of FOXF1, a key regulator of lung development. This enhancer element interacts with FOXF1 in human pulmonary microvascular endothelial cells but not in lymphoblasts, suggesting that FOXF1 expression in the lung endothelium is regulated at the chromatin structure levels. In addition to transcription factor binding sites, the focal deletion includes two IncRNA expressed specifically in the lung. An intriguing possibility is that the expression of these IncRNAs, which happens specifically in the lung, contributes to the establishment of a chromatin loop that brings the enhancer in close proximity to FOXF1.
Chromosomal translocations lead to inheritable structural and genetic changes, and as such are relevant causes of genetic disease. One way chromosomal translocations can lead to disease is through disruption of the higher order chromatin organization and the cis-regulatory landscape. Recently, two different translocations have been identified in brachydactyly Type E (BDE) that implicate IncRNA dysregulation (Maass et al., 2012). These translocations affect a regulatory region that interacts in cis with PTHLH and in trans with SOX9. Interestingly, this region is home to a IncRNA whose expression is important for the proper expression of PTHLH and SOX9. Depletion of this IncRNA (DA125942) resulted in downregulation of PTHLH and SOX9. The IncRNA interacts with both loci, and the occupancy is reduced in chromatin originated from BDE patients. This study demonstrates how IncRNAs and chromatin higher order organization collaborate in the regulation of gene expression.
Recognition of the roles of IncRNAs in human disease has unveiled new diagnostic and therapeutic opportunities. LncRNAs are expressed in a more tissue specific fashion than mRNA genes, a pattern that has been found to hold true in pathologic states such as cancer (Brunner et al., 2012). LncRNA measurements could hence trace cancer metastases or circulating cancer cells to their origins. In addition, a strong connection between IncRNAs and cancer has been clearly established, as many IncRNAs are dysregulated in human cancers. The IncRNA HOTAIR in overexpressed in breast, colon, pancreas and liver cancers and over expression of HOTAIR has been shown to drive breast cancer metastasis in vivo (Gupta et al., 2010; Gutschner and Diederichs, 2012). LncRNAs appear to be more structured and stable than mRNA transcripts, which facilitate their detection as free nucleic acids in body fluid such as urine and blood—knowledge already put to good use in clinically approved tests for prostate cancer (Fradet et al., 2004; Shappell, 2008; Tinzl et al., 2004). Aberrant IncRNAs can be knocked down in vivo using oligonucleotide “drugs” (Modarresi et al., 2012; Wheeler et al., 2012), which should spur advance in IncRNA genetics and therapeutics.
CONCLUSION
LncRNAs are well poised to be molecular address codes, particularly in the nucleus. On one hand, transcription of IncRNAs is often exquisitely regulated, reflecting the particular developmental stage and external environment that the cell has experienced. On the other, the capacity of IncRNAs to function as guides, scaffolds, and decoys endow them with enormous regulatory potential in gene expression and for spatial control within the cell. These outstanding properties of long RNAs have already been leveraged to make designer RNA scaffolds for synthetic cell circuits (Delebecque et al., 2011). Many questions remain to be addressed in this rapidly expanding field. First, the in vivo function of most IncRNAs has not been determined. An extensive catalog of IncRNAs has recently been described available for several model organisms (Nam and Bartel, 2012; Pauli et al., 2012; Ulitsky et al., 2011), opening the door of a wide array of powerful techniques to be used in the in vivo study of IncRNAs that will complement the study of human IncRNAs. In addition, detailed knowledge of structure-function relationship in IncRNAs is still lacking, which prohibits the de novo prediction of IncRNA domains and functions that we take granted for protein coding transcripts. New technologies to deconvolute RNA structure and function (Martin et al., 2012; Wan et al., 2012), probe RNA-chromatin interactions (Chu et al., 2011; Simon et al., 2011), and track RNA movement in real time (Paige et al., 2011) will be crucial for understanding IncRNAs and realizing their therapeutic potential.
Acknowledgement
We thank members of Chang lab for discussion and apologize to colleagues whose works are not discussed due to space limitation. Supported by NIH and California Institute for Regenerative Medicine (H.Y.C.). P.J.B. is the Kenneth G. and Elaine A. Langone Fellow of the Damon Runyon Cancer Research Foundation. H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell. 2012;45:147–157. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bierhoff H, Schmitz K, Maass F, Ye J, Grummt I. Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harb Symp Quant Biol. 2010;75:357–364. doi: 10.1101/sqb.2010.75.060. [DOI] [PubMed] [Google Scholar]
- Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3'-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A. 2012;109:19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R, Montgomery K, Varma S, Gilks T, Guo X, et al. Transcriptional profiling of IncRNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13:R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, Gabellini D. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149:819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. Long non-coding antisense RNA controls Uchll translation through an embedded SINEB2 repeat. Nature. 2012 doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harb PerspectBiol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding DQ, Okamasa K, Yamane M, Tsutsumi C, Haraguchi T, Yamamoto M, Hiraoka Y. Meiosis-specific noncoding RNA mediates robust pairing of homologous chromosomes in meiosis. Science. 2012;336:732–736. doi: 10.1126/science.1219518. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zornig M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9 doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradet Y, Saad F, Aprikian A, Dessureault J, Elhilali M, Trudel C, Masse B, Piche L, Chypre C. uPM3, a new molecular urine test for the detection of prostate cancer. Urology. 2004;64:311–315. doi: 10.1016/j.urology.2004.03.052. discussion 315–316. [DOI] [PubMed] [Google Scholar]
- Gong C, Maquat LE. IncRNAs transactivate STAUl-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimal P, Woloch M. Roman cities = Les villes romaines. Madison, Wis.: University of Wiscons; 1983. in Press. [Google Scholar]
- Guetg C, Scheifele F, Rosenthal F, Hottiger MO, Santoro R. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol Cell. 2012;45:790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- Guil S, Soler M, Portela A, Carrere J, Fonalleras E, Gomez A, Villanueva A, Esteller M. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012;19:664–670. doi: 10.1038/nsmb.2315. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Diederichs S. The Hallmarks of Cancer: A long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk o, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ietswaart R, Wu Z, Dean C. Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 2012;28:445–453. doi: 10.1016/j.tig.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesion connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Xu Z, Clauder-Munster S, Steinmetz LM, Buratowski S. Set3 HDAC Mediates Effects of Overlapping Noncoding Transcription on Gene Induction Kinetics. Cell. 2012;150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2012 doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, et al. Airn transcriptional overlap, but not its IncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass PG, Rump A, Schulz H, Stricker S, Schulze L, Platzer K, Aydin A, Tinschert S, Goldring MB, Luft FC, et al. A misplaced IncRNA causes brachydactyly in humans. J Clin Invest. 2012 doi: 10.1172/JCI65508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- Martin L, Meier M, Lyons SM, Sit RV, Marzluff WF, Quake SR, Chang HY. Systematic reconstruction of RNA functional motifs with high-throughput microfluidics. Nat Methods. 2012 doi: 10.1038/nmeth.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244–1247. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malatl is not an essential component of nuclear speckles in mice. Rna. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193:31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam JW, Bartel DP. Long noncoding RNAs in C. elegans. Genome Res. 2012 doi: 10.1101/gr.140475.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. The dynamic architecture of Hox gene clusters. Science. 2011;334:222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnqlotl antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Prestel M, Paro R. Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shappell SB. Clinical utility of prostate carcinoma molecular diagnostic tests. Rev Urol. 2008;10:44–69. [PMC free article] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, Kaaij LJ, van Ijcken W, Gribnau J, Heard E, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranski P, Dharmadhikari AV, Brosens E, Gurha P, Kolodziejska KE, Ou Z, Dittwald P, Majewski T, Mohan KN, Chen B, et al. Small non-coding differentially-methylated copy-number variants, including IncRNA genes, cause a lethal lung developmental disorder. Genome Res. 2012 doi: 10.1101/gr.141887.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for IncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinzl M, Marberger M, Horvath S, Chypre C. DD3PCA3 RNA analysis in urine-a new perspective for detecting prostate cancer. Eur Urol. 2004;46:182–186. doi: 10.1016/j.eururo.2004.06.004. discussion 187. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Borah S, Shi M, Steitz JA. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep. 2012;2:26–32. doi: 10.1016/j.celrep.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk M, Thulluru HK, Mulders J, Michel OJ, Poutsma A, Windhorst S, Kleiverda G, Sie D, Lachmeijer AM, Oudejans CB. HELLP babies link a novel lincRNA to the trophoblast cell cycle. J Clin Invest. 2012 doi: 10.1172/JCI65171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A. Transcription of Two Long Noncoding RNAs Mediates Mating-Type Control of Gametogenesis in Budding Yeast. Cell. 2012;150:1170–1181. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma-Gaur J, Torkamani A, Schaffer L, Head SR, Schork NJ, Feeney AJ. Noncoding transcription within the Igh distal VH region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1208398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Qu K, Ouyang Z, Kertesz M, Li J, Tibshirani R, Makino DL, Nutter RC, Segal E, Chang HY. Genome-wide Measurement of RNA Folding Energies. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3' ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012 doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L. Positional information and patterning revisited. J Theor Biol. 2011;269:359–365. doi: 10.1016/j.jtbi.2010.10.034. [DOI] [PubMed] [Google Scholar]
- Yamamoto M. The selective elimination of messenger RNA underlies the itosis-meiosis switch in fission yeast. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:788–797. doi: 10.2183/pjab.86.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Long Noncoding RNAs with snoRNA Ends. Mol Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al. The IncRNA Malatl is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SI. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]