Abstract
Dengue is a systemic viral infection transmitted between humans by Aedes mosquitoes1. For some patients dengue is a life-threatening illness2. There are currently no licensed vaccines or specific therapeutics, and substantial vector control efforts have not stopped its rapid emergence and global spread3. The contemporary worldwide distribution of the risk of dengue virus infection4 and its public health burden are poorly known2,5. Here we undertake an exhaustive assembly of known records of dengue occurrence worldwide, and use a formal modelling framework to map the global distribution of dengue risk. We then pair the resulting risk map with detailed longitudinal information from dengue cohort studies and population surfaces to infer the public health burden of dengue in 2010. We predict dengue to be ubiquitous throughout the tropics, with local spatial variations in risk influenced strongly by rainfall, temperature and the degree of urbanisation. Using cartographic approaches, we estimate there to be 390 million (95 percent credible interval 284-528) dengue infections per year, of which 96 million (67-136) manifest apparently (any level of clinical or sub-clinical severity). This infection total is more than three times the dengue burden estimate of the World Health Organization2. Stratification of our estimates by country allows comparison with national dengue reporting, after taking into account the probability of an apparent infection being formally reported. The most notable differences are discussed. These new risk maps and infection estimates provide novel insights into the global, regional and national public health burden imposed by dengue. We anticipate that they will provide a starting point for a wider discussion about the global impact of this disease and will help guide improvements in disease control strategies using vaccine, drug and vector control methods and in their economic evaluation. [285]
Dengue is an acute systemic viral disease that has established itself globally in both endemic and epidemic transmission cycles. Dengue virus infection in humans is often inapparent1,6 but can lead to a wide range of clinical manifestations, from mild fever to potentially fatal dengue shock syndrome2. The lifelong immunity developed after infection with one of the four virus types is type-specific1 and progression to more serious disease is frequently, but not exclusively, associated with secondary infection by heterologous types2,5. No effective antiviral agents yet exist to treat dengue infection and treatment therefore remains supportive2. Furthermore, no licensed vaccine against dengue infection is available, and the most advanced dengue vaccine candidate did not meet expectations in a recent large trial7,8. Current efforts to curb dengue transmission focus on the vector, using combinations of chemical and biological targeting of Aedes mosquitoes and management of breeding sites2. These control efforts have failed to stem the increasing incidence of dengue fever epidemics and expansion of the geographical range of endemic transmission9. While the historical expansion of this disease is well documented, the potentially large burden of ill-health attributable to dengue across much of the tropical and sub-tropical world remains poorly enumerated.
Knowledge of the geographical distribution and burden of dengue is essential for understanding its contribution to global morbidity and mortality burdens, in determining how to allocate optimally the limited resources available for dengue control and in evaluating the impact of such activities internationally. Additionally, estimates of both apparent and inapparent infection distributions form a key requirement for assessing clinical surveillance and for scoping reliably future vaccine demand and delivery strategies. Previous maps of dengue risk have used various approaches combining historical occurrence records and expert opinion to demarcate areas at endemic risk10-12. More sophisticated risk mapping techniques have also been implemented13,14, but the empirical evidence-base has since been improved, alongside advances in disease modelling approaches. Furthermore, no studies have used a continuous global risk map as the foundation for dengue burden estimation.
The first global estimates of total dengue virus infections were based on an assumed constant annual infection rate amongst a crude approximation of the population at risk (10% in 1 billion5 or 4% in 2 billion15), yielding figures of 80-100 million infections per year worldwide in 19885,15. As more information was collated on the ratio of dengue haemorrhagic fever to dengue fever cases and the ratio of deaths to dengue haemorrhagic fever cases, the global figure was revised to 50-100 million infections16,17, although larger estimates of 100-200 million have also been made10 (Figure 1). These estimates were intended solely as approximations but, in the absence of better evidence, the resulting figure of 50-100 million infections per year is widely cited and currently used by the World Health Organization (WHO). As the methods employed were informal, these estimates were presented without confidence intervals, and no attempt was made to assess geographical or temporal variation in incidence or the inapparent infection reservoir.
Figure 1. Global estimates of total dengue infections.
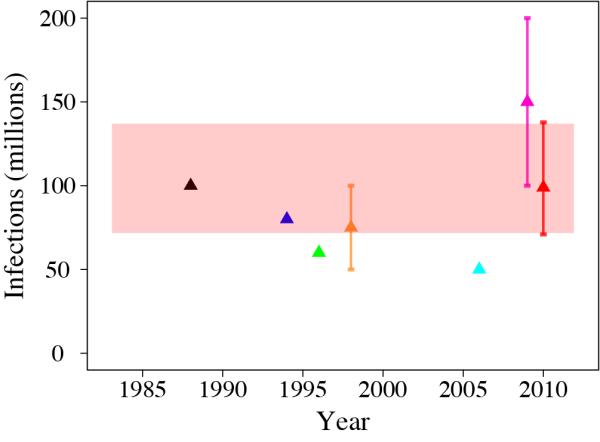
Comparison of previous estimates of total global dengue infections in individuals of all ages, 1985 to 2010:  Halstead et al. 19885,
Halstead et al. 19885,  Monath et al. 199415,
Monath et al. 199415,  Rodhain et al. 199617,
Rodhain et al. 199617,  Rigau-Perez et al. 199816,
Rigau-Perez et al. 199816,  TDR/WHO. scientific working group 200630,
TDR/WHO. scientific working group 200630,  Beatty et al. 200910,
Beatty et al. 200910,  apparent infections from this study. Estimates are aligned to the year of estimate and, if not stated, aligned to the publication date. Red shading marks the credible interval of our current estimate, for comparison. Error bars from ref. 10 and ref. 16 replicated the confidence intervals provided in these publications.
apparent infections from this study. Estimates are aligned to the year of estimate and, if not stated, aligned to the publication date. Red shading marks the credible interval of our current estimate, for comparison. Error bars from ref. 10 and ref. 16 replicated the confidence intervals provided in these publications.
Here we present the outcome of a new project to derive an evidence-based map of dengue risk and estimates of apparent and inapparent infections worldwide based on the global population in 2010. We compiled a database of 8,309 geo-located records of dengue occurrence from a systematic search, resulting from 2,838 published literature sources as well as newer online resources18 (see Supplementary Information A; the full bibliography4 and occurrence data are available from authors on request). Using these occurrence records we: chose a set of gridded environmental and socioeconomic covariates known, or hypothesised, to affect dengue transmission (see Supplementary Information B); incorporated recent work assessing the strength of evidence on national and sub-national-level dengue present/absent status4 (Figure 2A); and built a boosted regression tree (BRT) statistical model of dengue risk that addressed the limitations of previous risk maps (see Supplementary Information C) to define the probability of occurrence of dengue infection (dengue risk) within each 5km × 5km pixel globally (Figure 2B). The model was run 336 times to reflect parameter uncertainty and an ensemble mean map was created (see Supplementary Information C). We then combined this ensemble map with detailed longitudinal information on dengue infection incidence from cohort studies and built a non-parametric Bayesian hierarchical model to describe the relationship between dengue risk and incidence (see Supplementary Information D). Finally, we used the estimated relationship to predict the number of apparent and inapparent dengue infections in 2010 (see Supplementary Information E). Our definition of an apparent infection is consistent with that used by the cohort studies: an infection with sufficient severity to modify a person’s regular schedule, such as attending school. This definition encompasses any level of severity of the disease, including both clinical and sub-clinical manifestations.
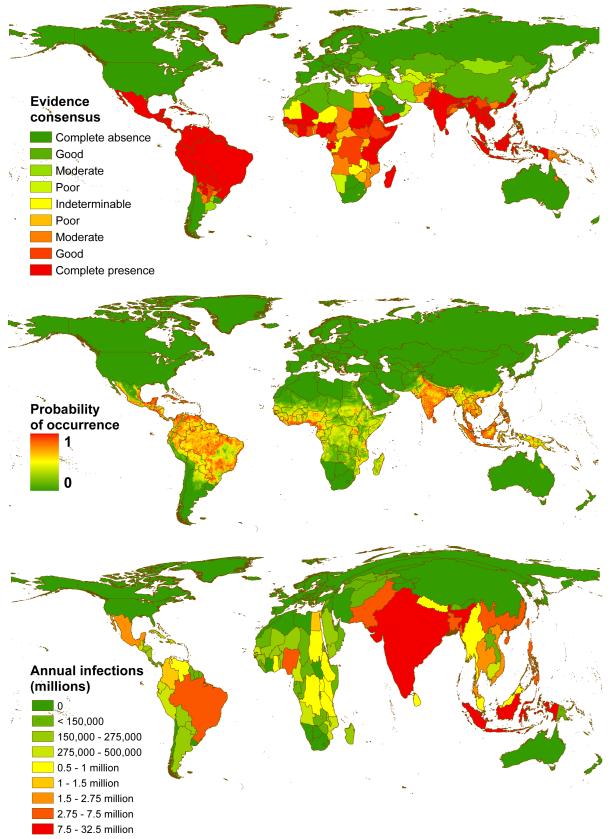
Figure 2. Global evidence consensus, risk and burden of dengue in 2010.
a, shows National and subnational evidence consensus on complete absence (green) through to complete presence (red) of dengue4. b, shows the probability of dengue occurrence at 5km × 5km spatial resolution of the mean predicted map (area under the receiver operator curve of 0.81 (±0.02 SD, n = 336)) from 336 boosted regression tree models. Areas with a high probability of dengue occurrence are shown in red and areas with a low probability in green. c, shows a cartogram of the annual number of infections for all ages as a proportion of national or sub-national (China) geographical area.
We predict that dengue transmission is ubiquitous throughout the tropics with the highest risk zones in the Americas and Asia (Figure 2B). Validation statistics indicated high predictive performance of the BRT ensemble mean map with area under the receiver operating characteristic (AUC) of 0.81 (±0.02 SD, n = 336) (see Supplementary Information C). Predicted risk in Africa, though more unevenly distributed than in other tropical endemic regions, is much more widespread than suggested previously. Africa has the poorest record of occurrence data and, as such, increased information from this continent would help to better define the spatial distribution of dengue within it and to improve derivative burden estimates. We found high levels of precipitation and temperature suitability for dengue transmission to be most strongly associated among the variables considered with elevated dengue risk, although low precipitation was not found to strongly limit transmission (see Supplementary Information C). Proximity to low-income urban and peri-urban centres was also linked to greater risk, particularly in highly connected areas, suggesting that human movement between population centres is an important facilitator of dengue spread. These associations have previously been cited9, but have not been demonstrated at the global scale and highlight the importance of including socioeconomic covariates when assessing dengue risk.
We estimate that there were 96 million apparent dengue infections globally in 2010 (Table 1). Asia bore 70% (67 [47-94] million infections) of this burden, and is characterised by large swathes of densely populated regions coinciding with very high suitability for disease transmission. India19,20 alone contributed 34% (33 [24-44] million infections) of the global total. The disproportionate infection burden borne by Asian countries is emphasized in the cartogram shown in Figure 2C. The Americas contributed 14% (13 [9-18] million infections) of apparent infections worldwide, of which over half occurred in Brazil and Mexico. Our results indicate that Africa’s dengue burden is nearly equivalent to that of the Americas (16 (11–22) million infections, 16% of the global total), representing a significantly larger burden than previously estimated. This disparity supports the notion of a largely hidden African dengue burden, being masked by symptomatically similar illnesses, under-reporting and highly variable treatment-seeking behaviour6,9,20. The countries of Oceania contributed less than 0.2% of global apparent infections.
Table 1.
Estimated burden of dengue in 2010, by continent
| Apparent | Inapparent | |
|---|---|---|
|
|
||
| Millions (credible interval) | Millions (credible interval) | |
| Africa | 15.7 (10.5 - 22.5) | 48.4 (34.3 - 65.2) |
| Asia | 66.8 (47.0 – 94.4) | 204.4 (151.8 – 273.0) |
| Americas | 13.3 (9.5 - 18.5) | 40.5 (30.5 - 53.3) |
| Oceania | 0.18 (0.11 - 0.28) | 0.55 (0.35 - 0.82) |
| Global | 96 (67.1 - 135.6) | 293.9 (217.0 – 392.3) |
We estimate that an additional 294 (217–392) million inapparent infections occurred worldwide in 2010. These mild ambulatory or asymptomatic infections are not detected by the public health surveillance system and have no immediate implications for clinical management. However, the presence of this huge potential reservoir of infection has profound implications for: (i) correctly enumerating economic impact (for example, how many vaccinations are needed to avert an apparent infection) and triangulating with independent assessments of disability adjusted life years (DALYs)21; (ii) elucidating the population dynamics of dengue viruses22; and (iii) hypothesising about population effects of future vaccine programmes23 (volume, targeting efficacy, impacts in combination with vector control), which will need to be administered to maximise cross-protection and minimize post-vaccination susceptibility.
The absolute uncertainties in the national burden estimates are inevitably a function of population size, with the greatest uncertainties in India, Indonesia, Brazil and China (see full rankings in Supplementary E table T4). In addition, comparing the ratio of the mean to the width of the confidence interval24 revealed the greatest contributors to relative uncertainty (see full rankings in Supplementary E table T4). These were countries with sparse occurrence points and low evidence consensus on dengue presence, such as Afghanistan or Rwanda (see Figure 2A), or those with ubiquitous high risk, such as Singapore or Djibouti, for which our burden prediction confidence interval is at its widest (see Figure SD2 in Supplementary D). Therefore, increasing evidence consensus and occurrence data availability in low consensus countries and assembling new cohort studies, particularly in areas of high transmission, will reduce uncertainty in future burden estimates. Our approach, uniquely, provides new evidence to help maximize the value and cost-effectiveness of surveillance efforts, by indicating where limited resources can be targeted to have their maximum possible impact in improving our knowledge of the global burden and distribution of dengue.
Our estimates of total infection burden (apparent and inapparent) are more than three times higher than the WHO predicted figure (Supplementary Information E). Our definition of an apparent infection is broad, encompassing any disruption to the daily routine of the infected individual, and consequently is an inclusive measurement of the total population affected adversely by the disease. Within this broad class, the severity of symptoms will affect treatment-seeking behaviours and the probability of a correct diagnosis in response to a given infection. Our definition is therefore more comprehensive than those of traditional surveillance systems, which, even in the most efficient system, report a much narrower range of dengue infections. By reviewing our database of longitudinal cohort studies, in which total infections in the community were documented exhaustively, we find that the biggest source of disparity between actual and reported infection numbers is the low proportion of individuals with apparent infections seeking care from formal health facilities (see Figure SE5 and Supplementary Information E for full analysis). Additional biases are introduced by misdiagnosis and the systematic failure of health management information systems to capture and report presenting dengue cases. By extracting the average magnitude of each of these sequential disparities from published cohort and clinical studies, we can recreate a hypothetical reporting chain with idealised reporting and arrive at estimates that are broadly comparable to those countries reported to the WHO. This is most clear in more reliable reporting regions such as the Americas. Systemic underreporting and low hospitalisation rates have important implications, for example, in the evaluation of vaccine efficacy based on reduced hospitalised caseloads. Inferences about these biases may be made from the comparison of estimated versus reported infection burdens in 2010, highlighting areas where particularly poor reporting might be strengthened (see Supplementary Information E).
We have strived to be exhaustive in the assembly of contemporary data on dengue occurrence and clinical incidence and have applied new modelling approaches to maximise the predictive power of these data. It remains the case, however, that the empirical evidence base for global dengue risk is more limited than that available, for example, for Plasmodium falciparum25 and P. vivax26 malaria. Records of disease occurrence carry less information than those of prevalence and, as databases of the latter become more widespread, future approaches should focus on assessing relationships between seroprevalence and clinical incidence as a means of assessing risk27. Additional cartographic refinements are also required to help differentiate endemic- from epidemic-prone areas, to determine the geographic diversity of dengue virus types and to predict the distributions of future risk under scenarios of socioeconomic and environmental change.
The global burden of dengue is formidable and represents a growing challenge to public health officials and policymakers. Success in tackling this growing global threat is, in part, contingent on strengthening the evidence base on which control planning decisions and their impact are evaluated. It is hoped that this evaluation of contemporary dengue risk distribution and burden will help to advance that goal. [1935].
Methods
Assembly of the occurrence database and its quality control
Occurrence data comprised of point or polygon locations of confirmed dengue infection presence derived from both peer-reviewed literature and HealthMap alerts18,31 (see Supplementary Information A). An occurrence was defined as one or more laboratory or clinically confirmed infection(s) of dengue occurring at a unique location (a 5km × 5km pixel) within one calendar year. All occurrence data underwent manual review and automatic quality control to ensure information fidelity and precise geo-positioning. In total, 9,648 and 1,622 occurrence locations were obtained from literature searches and HealthMap respectively. After the quality control procedures, our final dataset contained 8,309 occurrence locations (5,216 point locations and 3,093 small polygon centroids) spanning a period from 1960 to 2012. We assume any record of dengue occurrence, regardless of its age, represented an environment permissible for the disease, since dengue has expanded from a focal disease in Asia to a cosmopolitan disease of the tropics.
Explanatory covariates
We assembled gridded global data for a suite of eight explanatory covariates. The covariates were chosen based on factors known or hypothesised to contribute to suitability for dengue transmission (see Supplementary Information B). These covariates included: (i) annual maximum and minimum precipitation variables from a Fourier processed32 synoptic annual series interpolated from global meteorological stations33; (ii) a biological model combining the effects of temperature on the extrinsic incubation period of dengue virus and life-span of the Aedes aegypti vector to quantify the dengue-specific temperature suitability for transmission28,34,35; (iii) Fourier-processed annual average normalised difference vegetation index36 (iv); categorical demarcations of urban and peri-urban areas37; (v) an urban accessibility metric defining the travel time to nearest city of 50,000 people or more by land- or water-based travel38; and (vi) an indicator of relative poverty derived from the finest geographic scale data available for economic productivity and adjusted for purchasing power parity39. No covariate grids were shown to be adversely affected by multicollinearity (see Supplementary Information B) and were standardised to ensure identical spatial resolution, extent and boundaries. For point records, covariate values corresponded to the pixel value containing the location of the point. For polygon occurrence records, covariate values were averaged across the whole polygon.
Predicting the probability of occurrence (risk) of dengue transmission
We used a boosted regression tree (BRT) approach to establish a multivariate empirical relationship between the probability of occurrence of a dengue virus infection and the environmental conditions sampled at each site from the covariate suite. The BRT method has been shown to fit complicated response functions efficiently, while guarding against overfitting, and is therefore widely used for vector and disease distribution mapping40,41. The BRT approach combines regression trees42 with gradient boosting43, whereby an initial regression tree is fitted and iteratively improved upon in a forward stagewise manner (boosting) by minimising the variation in the response not explained by the model at each iteration (see Supplementary Information C).
Like other niche mapping approaches, the BRT models require not only presence data but also absence data defining areas of disease absence and potentially unsuitable environmental conditions at unsampled locations. Since data on absence of disease are not-definitive, pseudo-absence data estimate areas of disease absence instead. No consensus approach has been developed to optimise the generation of pseudo-absence data and we therefore created an evidence-based probabilistic framework for generating pseudo-absences, incorporating the main biasing factors in pseudo-absence generation, namely: (i) geographical extent; (ii) number; (iii) contamination bias; and (iv) sampling bias. To represent areas of absence, na pseudo-absence points29,44,45 were randomly generated based on dengue presence or absence certainty measures at a national or subnational level4. Pseudo-absence locations were restricted to a maximum distance μ from any recorded presence site46,47. Additionally, to compensate for “contamination” of true but unobserved presences within the generated pseudo-absences48, np pseudo-presence points were generated using the same procedure used to generate the pseudo-absences. Variation in the parameter set π = {μ, μa, μp} resulted in independent samples of the possible states of the real distribution, with all parameter combinations representing a null distribution of possible states. Therefore, rather than using an individual parameter combination from π, we created an ensemble49 of 336 BRT models spanning reasonable ranges in π and evaluated the central tendency as the mean across all 336 BRT models (see Supplementary Information C). The final ensemble BRT model was used to predict a global map of the probability of occurrence of dengue virus infection at a 5km × 5km resolution.
Estimation of dengue burden and populations at risk
Formal literature searches were conducted for serological dengue virus incidence surveys. Inclusion criteria were restricted to longitudinal surveys of seroconversion to dengue virus-specific antibodies carried out in parallel with active symptom surveillance in a defined cohort. The surveys were abstracted, standardised and geopositioned (see Supplementary Information D). In total, 54 dengue incidence surveys were collected. Of these, 39 contained information about the ratio of inapparent to apparent infections.
The empirical relationship between incidence and the probability of occurrence was represented using a Bayesian hierarchical model. We defined a negative binomial likelihood function50 with constant dispersion and a rate characterised by a highly flexible data-driven Gaussian process prior51. The Gaussian process prior was parameterised with a quadratic mean function and a squared exponential covariance function51. Uninformative hyperpriors were assigned hierarchically to the prior parameters and the full posterior distribution determined by Markov Chain Monte Carlo (MCMC) sampling52. The entire model was fitted separately for apparent and inapparent infection incidences, with missing inapparent to apparent ratio values imputed in the MCMC. Using human population gridded data for the year 201053, estimates of apparent and inapparent dengue infections were calculated nationally, regionally and globally. These estimates were then compared to national clinical cases reported to the WHO and differences between our cartographic estimates and the WHO surveillance estimates were reconciled in a comparative analysis addressing key factors in traditional surveillance underreporting (see Supplementary Information E). [973].
Supplementary Material
Acknowledgements
SIH is funded by a Senior Research Fellowship from the Wellcome Trust (#095066) which also supports SB and PWG. CPS is also funded by a Senior Research Fellowship from the Wellcome Trust (#084368). OJB is funded by a BBSRC Industrial CASE studentship. JPM, AWF, TJ, GRWW, CPS, TWS and SIH received funding from and with SB, PWG, OJB and JJF acknowledge the contribution of the International Research Consortium on Dengue Risk Assessment Management and Surveillance (IDAMS, #21803, http://www.idams.eu). This work was funded in part by EU grant 2011- 261504 EDENEXT and the paper is catalogued by the EDENEXT Steering Committee as EDENEXT. SIH and TWS also acknowledge funding support from the RAPIDD program of the Science & Technology Directorate, Department of Homeland Security, and the Fogarty International Center, National Institutes of Health.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions SIH and JJF conceived the research. SB and SIH drafted the manuscript. SB drafted the supplemental material with significant support on A (OJB, CLM), B (JPM, GRWW), C (PWG), D (OJB, TWS) and OJB wrote E. JSB and AGH provided Healthmap occurrence data and advice on its provenance. OJB reviewed all the occurrence data. SB did the modelling and analysis with advice from JMD, PWG and SIH. JPM created all maps. All authors discussed the results and contributed to the revision of the final manuscript.
Author Information Reprints and permission information is available at www.nature.com/reprints.
References
- 1.Simmons CP, Farrar JJ, van Vinh Chau N, Wills B. Dengue. N. Engl. J. Med. 2012;366:1423–1432. doi: 10.1056/NEJMra1110265. [DOI] [PubMed] [Google Scholar]
- 2.W.H.O . Dengue: guidelines for diagnosis, treatment, prevention and control. WHO/HTM/NTD/DEN/2009.1. World Health Organization; 2009. [PubMed] [Google Scholar]
- 3.Tatem AJ, Hay SI, Rogers DJ. Global traffic and disease vector dispersal. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6242–6247. doi: 10.1073/pnas.0508391103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady OJ, et al. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl. Trop. Dis. 2012;6:e1760. doi: 10.1371/journal.pntd.0001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halstead SB. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 6.Endy TP, et al. Determinants of inapparent and symptomatic dengue infection in a prospective study of primary school children in Kamphaeng Phet, Thailand. PLoS Negl. Trop. Dis. 2011;5:e975. doi: 10.1371/journal.pntd.0000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabchareon A, et al. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet. 2012 doi: 10.1016/S0140-6736(12)61428-7. [DOI] [PubMed] [Google Scholar]
- 8.Halstead SB. Dengue vaccine development: a 75% solution? Lancet. 2012 doi: 10.1016/S0140-6736(12)61510-4. [DOI] [PubMed] [Google Scholar]
- 9.Gubler DJ. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 1998;11:480–469. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beatty ME, Letson GW, Margolis HS. Estimating the global burden of dengue. Am. J. Trop. Med. Hyg. 2009;81(5 Suppl 1):231. [Google Scholar]
- 11.Van Kleef E, Bambrick H, Hales S. The geographic distribution of dengue fever and the potential influence of global climate change. TropIKA.net. 2009:1–22. [Google Scholar]
- 12.W.H.O . International travel and health: situation as on 1 January 2012. World Health Organization; 2012. [Google Scholar]
- 13.Hales S, de Wet N, Maindonald J, Woodward A. Potential effect of population and climate changes on global distribution of dengue fever: an empirical model. Lancet. 2002;360:830–834. doi: 10.1016/S0140-6736(02)09964-6. [DOI] [PubMed] [Google Scholar]
- 14.Rogers DJ, Wilson AJ, Hay SI, Graham AJ. The global distribution of yellow fever and dengue. Adv. Parasitol. 2006;62:181–220. doi: 10.1016/S0065-308X(05)62006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monath TP. Yellow fever and dengue-the interactions of virus, vector and host in the re-emergence of epidemic disease. Sem Virol. 1994;5:133–145. [Google Scholar]
- 16.Rigau-Perez JG, et al. Dengue and dengue haemorrhagic fever. Lancet. 1998;352:971–977. doi: 10.1016/s0140-6736(97)12483-7. [DOI] [PubMed] [Google Scholar]
- 17.Rodhain F. La situation de la dengue dans le monde. Bull Soc Pathol Exot. 1996;89:87–90. [PubMed] [Google Scholar]
- 18.Freifeld CC, Mandl KD, Reis BY, Brownstein JS. HealthMap: global infectious disease monitoring through automated classification and visualization of Internet media reports. J. Am. Med. Inform. Assoc. 2008;15:150–157. doi: 10.1197/jamia.M2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakravarti A, Arora R, Luxemburger C. Fifty years of dengue in India. Trans. R. Soc. Trop. Med. Hyg. 2012;106:273–282. doi: 10.1016/j.trstmh.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Kakkar M. Dengue fever is massively under-reported in India, hampering our response. Br. Med. J. 2012;345:e8574. doi: 10.1136/bmj.e8574. [DOI] [PubMed] [Google Scholar]
- 21.Murray CJL, et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. The Lancet. 2012;380:2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- 22.Cummings DA, et al. The impact of the demographic transition on dengue in Thailand: insights from a statistical analysis and mathematical modeling. PLoS Med. 2009;6:e1000139. doi: 10.1371/journal.pmed.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson MA, Hombach J, Cummings DA. Models of the impact of dengue vaccines: a review of current research and potential approaches. Vaccine. 2011;29:5860–5868. doi: 10.1016/j.vaccine.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hay SI, et al. Estimating the global clinical burden of Plasmodium falciparum malaria in 2007. PLoS Med. 2010;7:e1000290. doi: 10.1371/journal.pmed.1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gething PW, et al. A new world malaria map: Plasmodium falciparum endemicity in 2010. Malar. J. 2011;10:378. doi: 10.1186/1475-2875-10-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gething PW, et al. A long neglected world malaria map: Plasmodium vivax endemicity in 2010. PLoS Negl. Trop. Dis. 2012;6:e1814. doi: 10.1371/journal.pntd.0001814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders KL, Hay SI. Lessons from malaria control to help meet the rising challenge of dengue. Lancet Infect. Dis. 2012;12:977–984. doi: 10.1016/S1473-3099(12)70246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gething PW, et al. Modelling the global constraints of temperature on transmission of Plasmodium falciparum and P. vivax. Parasit. Vectors. 2011;4:92. doi: 10.1186/1756-3305-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chefaoui RM, Lobo JM. Assessing the effects of pseudo-absences on predictive distribution model performance. Ecol. Model. 2008;210:478–486. [Google Scholar]
- 30.TDR/W.H.O. Report of the Scientific Working Group on Dengue, 2006. TDR/SWG/08. TDR/World Health Organization; 2006. [Google Scholar]
- 31.Brownstein JS, Freifeld CC, Reis BY, Mandl KD. Surveillance sans frontieres: internet-based emerging infectious disease intelligence and the HealthMap project. PLoS Med. 2008;5:e151. doi: 10.1371/journal.pmed.0050151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scharlemann JPW, et al. Global data for ecology and epidemiology: a novel algorithm for temporal Fourier processing MODIS data. PLoS One. 2008;3:e1408. doi: 10.1371/journal.pone.0001408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005;25:1965–1978. [Google Scholar]
- 34.Focks DA, Haile DG, Daniels E, Mount GA. Dynamic life table model for Aedes aegypti (Diptera: Culcidae): analysis of the literature and model development. J. Med. Entomol. 1993;30:1003–1017. doi: 10.1093/jmedent/30.6.1003. [DOI] [PubMed] [Google Scholar]
- 35.Focks DA, Haile DG, Daniels E, Mount GA. Dynamic life table model for Aedes aegypti (Diptera: Culicidae): simulation and validation. J. Med. Entomol. 1993;30:1018–1028. doi: 10.1093/jmedent/30.6.1018. [DOI] [PubMed] [Google Scholar]
- 36.Hay SI, Tatem AJ, Graham AJ, Goetz SJ, Rogers DJ. Global environmental data for mapping infectious disease distribution. Adv. Parasitol. 2006;62:37–77. doi: 10.1016/S0065-308X(05)62002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hay SI, et al. A world malaria map: Plasmodium falciparum endemicity in 2007. PLoS Med. 2009;6:e1000048. doi: 10.1371/journal.pmed.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson A. Estimated travel time to the nearest city of 50,000 or more people in year 2000. Global Environment Monitoring Unit - Joint Research Centre of the European Commission; [accessed 01/01/2012]. 2008. ( http://bioval.jrc.ec.europa.eu/products/gam. [Google Scholar]
- 39.Nordhaus WD. Geography and macroeconomics: new data and new findings. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3510–3517. doi: 10.1073/pnas.0509842103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elith J, et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography. 2006;29:129–151. [Google Scholar]
- 41.Stevens KB, Pfeiffer DU. Spatial modelling of disease using data- and knowledge-driven approaches. Spat. Spatiotemporal Epidemiol. 2011;2:125–133. doi: 10.1016/j.sste.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Breiman L. Classification and regression trees. Chapman & Hall / CRC Press LLC; 1984. [Google Scholar]
- 43.Friedman JH. Greedy function approximation: a gradient boosting machine. Ann. Stat. 2001;29:1189–1232. [Google Scholar]
- 44.Stokland JN, Halvorsen R, Stoa B. Species distribution modelling. Effect of design and sample size of pseudo-absence observations. Ecol. Model. 2011;222:1800–1809. [Google Scholar]
- 45.Lobo JM, Tognelli MF. Exploring the effects of quantity and location of pseudo-absences and sampling biases on the performance of distribution models with limited point occurrence data. J. Nat. Conserv. 2011;19:1–7. [Google Scholar]
- 46.VanDerWal J, Shoo LP, Graham C, William SE. Selecting pseudo-absence data for presence-only distribution modeling: how far should you stray from what you know? Ecol. Model. 2009;220:589–594. [Google Scholar]
- 47.Barbet-Massin M, Jiguet F, Albert CH, Thuiller W. Selecting pseudo-absences for species distribution models: how, where and how many? Methods Ecol. Evol. 2012;3:327–338. [Google Scholar]
- 48.Ward G, Hastie T, Barry S, Elith J, Leathwick JR. Presence-only data and the EM algorithm. Biometrics. 2009;65:554–563. doi: 10.1111/j.1541-0420.2008.01116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Araújo MB, New M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 2007;22:42–47. doi: 10.1016/j.tree.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 50.Hilbe JM. Negative binomial regression. 2nd Edition Cambridge University Press; 2011. p. 251. [Google Scholar]
- 51.Banerjee S, Carlin BP, Gelfand AE. Hierarchical modeling and analysis for spatial data. Monographs on Statistics and Applied Probability 101. Chapman & Hall / CRC Press LLC; 2004. [Google Scholar]
- 52.Patil A, Huard D, Fonnesbeck CJ. PyMC: Bayesian stochastic modelling in Python. J. Stat. Softw. 2010;35:e1000301. [PMC free article] [PubMed] [Google Scholar]
- 53.Balk DL, et al. Determining global population distribution: methods, applications and data. Adv. Parasitol. 2006;62:119–156. doi: 10.1016/S0065-308X(05)62004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.