Abstract
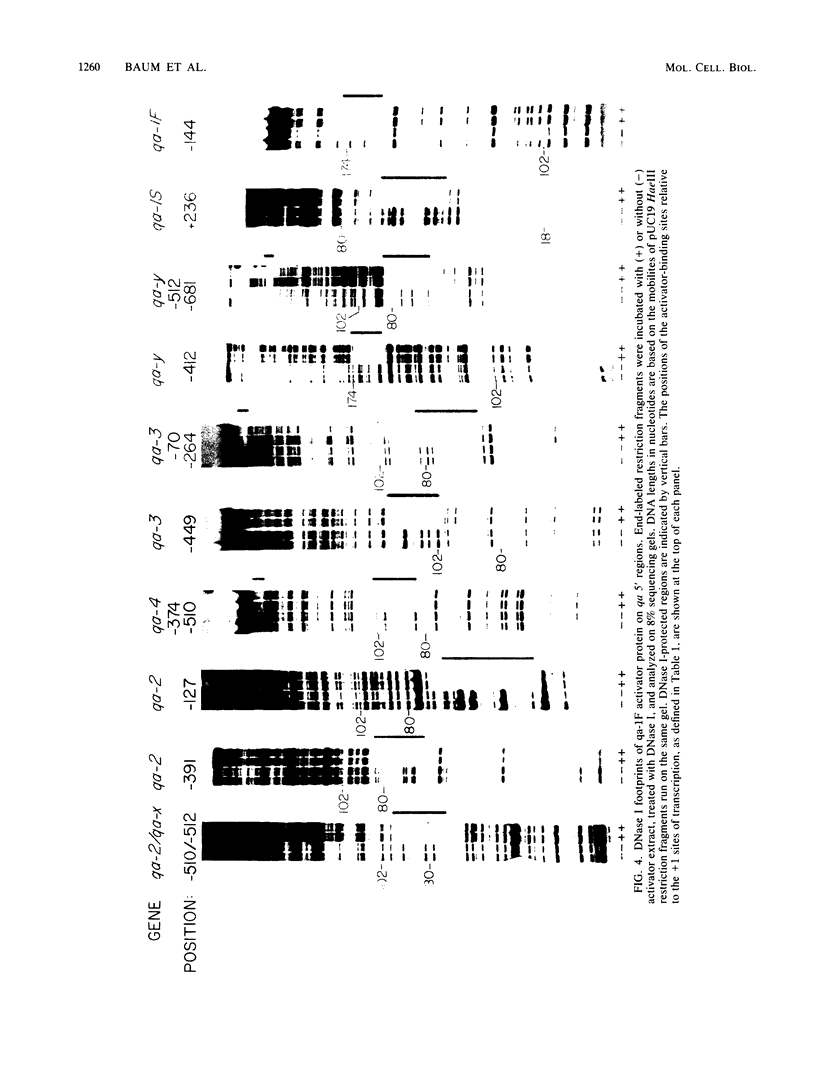
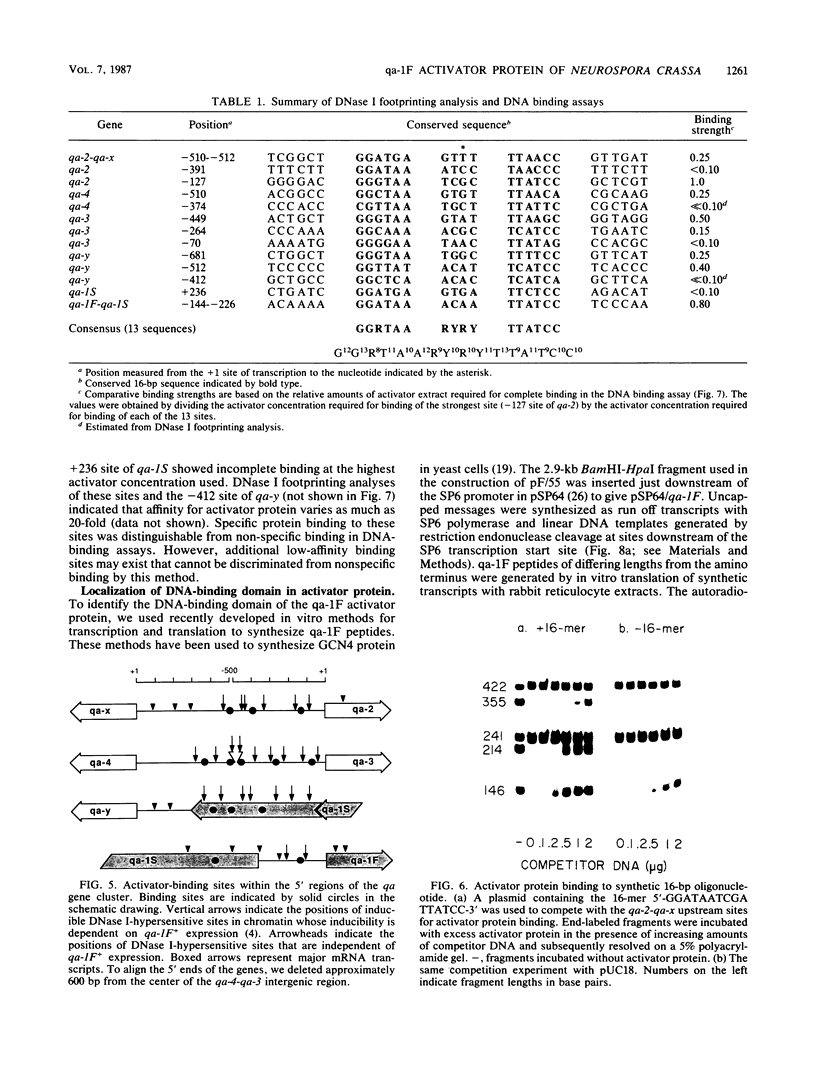
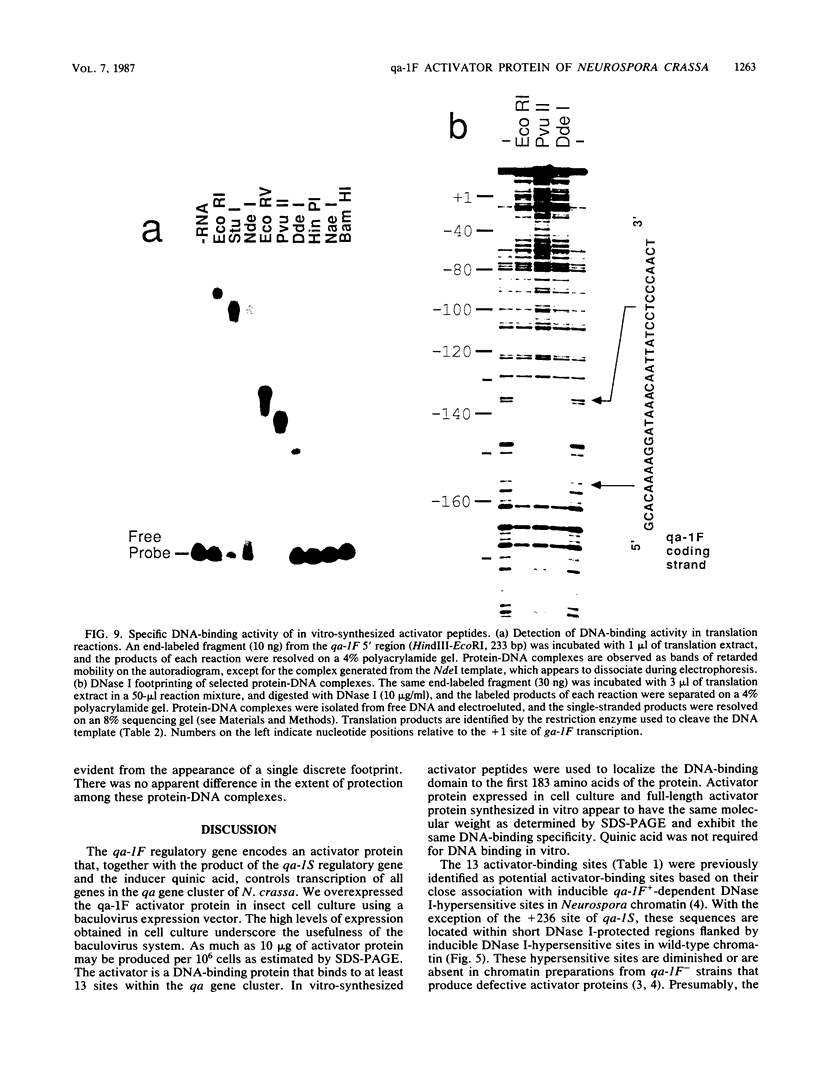
The qa-1F regulatory gene of Neurospora crassa encodes an activator protein required for quinic acid induction of transcription in the qa gene cluster. This activator protein was expressed in insect cell culture with a baculovirus expression vector. The activator binds to 13 sites in the gene cluster that are characterized by a conserved 16-base-pair sequence of partial dyad symmetry. One site is located between the divergently transcribed qa-1F and qa-1S regulatory genes, corroborating prior evidence that qa-1F is autoregulated and controls expression of the qa-1S repressor. Multiple upstream sites located at variable positions 5' to the qa structural genes appear to allow for greater transcriptional control by qa-1F. Full-length and truncated activator peptides were synthesized in vitro, and the DNA-binding domain was localized to the first 183 amino acids. A 28-amino acid sequence within this region shows striking homology to N-terminal sequences from other lower-eucaryotic activator proteins. A qa-1F(Ts) mutation is located within this putative DNA-binding domain.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alton N. K., Buxton F., Patel V., Giles N. H., Vapnek D. 5'-Untranslated sequences of two structural genes in the qa gene cluster of Neurospora crassa. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1955–1959. doi: 10.1073/pnas.79.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerer G. Expression of genes in yeast using the ADCI promoter. Methods Enzymol. 1983;101:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- Baum J. A., Giles N. H. DNase I hypersensitive sites within the inducible qa gene cluster of Neurospora crassa. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6533–6537. doi: 10.1073/pnas.83.17.6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J. A., Giles N. H. Genetic control of chromatin structure 5' to the qa-x and qa-2 genes of Neurospora. J Mol Biol. 1985 Mar 5;182(1):79–89. doi: 10.1016/0022-2836(85)90029-4. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Lue N. F., Kornberg R. D. A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J. 1986 Mar;5(3):603–608. doi: 10.1002/j.1460-2075.1986.tb04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case M. E., Giles N. H. Genetic evidence on the organization and action of the qa-1 gene product: a protein regulating the induction of three enzymes in quinate catabolism in Neurospora crassa. Proc Natl Acad Sci U S A. 1975 Feb;72(2):553–557. doi: 10.1073/pnas.72.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen P. D., Miller L. K. Temporal regulation of baculovirus RNA: overlapping early and late transcripts. J Virol. 1985 May;54(2):392–400. doi: 10.1128/jvi.54.2.392-400.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geever R. F., Case M. E., Tyler B. M., Buxton F., Giles N. H. Point mutations and DNA rearrangements 5' to the inducible qa-2 gene of Neurospora allow activator protein-independent transcription. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7298–7302. doi: 10.1073/pnas.80.23.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geever R. F., Murayama T., Case M. E., Giles N. H. Rearrangement mutations on the 5' side of the qa-2 gene of Neurospora implicate two regions of qa-1F activator-protein interaction. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3944–3948. doi: 10.1073/pnas.83.11.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles N. H., Case M. E., Baum J., Geever R., Huiet L., Patel V., Tyler B. Gene organization and regulation in the qa (quinic acid) gene cluster of Neurospora crassa. Microbiol Rev. 1985 Sep;49(3):338–358. doi: 10.1128/mr.49.3.338-358.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the "TATA box". Proc Natl Acad Sci U S A. 1984 Dec;81(24):7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Huiet L. Molecular analysis of the Neurospora qa-1 regulatory region indicates that two interacting genes control qa gene expression. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1174–1178. doi: 10.1073/pnas.81.4.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer B., Guyonvarch A., Hubert J. C. Yeast regulatory gene PPR1. I. Nucleotide sequence, restriction map and codon usage. J Mol Biol. 1984 Dec 5;180(2):239–250. doi: 10.1016/s0022-2836(84)80002-9. [DOI] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984 Feb;4(2):260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F., Dubois E., Descamps F. Nucleotide sequence of the ARGRII regulatory gene and amino acid sequence homologies between ARGRII PPRI and GAL4 regulatory proteins. Eur J Biochem. 1986 May 15;157(1):77–81. doi: 10.1111/j.1432-1033.1986.tb09640.x. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. K., Trimarchi R. E., Browne D., Pennock G. D. A temperature-sensitive mutant of the baculovirus Autographa californica nuclear polyhedrosis virus defective in an early function required for further gene expression. Virology. 1983 Apr 15;126(1):376–380. doi: 10.1016/0042-6822(83)90487-7. [DOI] [PubMed] [Google Scholar]
- Miyamoto C., Smith G. E., Farrell-Towt J., Chizzonite R., Summers M. D., Ju G. Production of human c-myc protein in insect cells infected with a baculovirus expression vector. Mol Cell Biol. 1985 Oct;5(10):2860–2865. doi: 10.1128/mcb.5.10.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V. B., Schweizer M., Dykstra C. C., Kushner S. R., Giles N. H. Genetic organization and transcriptional regulation in the qa gene cluster of Neurospora crassa. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5783–5787. doi: 10.1073/pnas.78.9.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock G. D., Shoemaker C., Miller L. K. Strong and regulated expression of Escherichia coli beta-galactosidase in insect cells with a baculovirus vector. Mol Cell Biol. 1984 Mar;4(3):399–406. doi: 10.1128/mcb.4.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Johnston S. A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986 Oct 10;14(19):7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer M., Case M. E., Dykstra C. C., Giles N. H., Kushner S. R. Identification and characterization of recombinant plasmids carrying the complete qa gene cluster from Neurospora crassa including the qa-1+ regulatory gene. Proc Natl Acad Sci U S A. 1981 Aug;78(8):5086–5090. doi: 10.1073/pnas.78.8.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. E., Fraser M. J., Summers M. D. Molecular Engineering of the Autographa californica Nuclear Polyhedrosis Virus Genome: Deletion Mutations Within the Polyhedrin Gene. J Virol. 1983 May;46(2):584–593. doi: 10.1128/jvi.46.2.584-593.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. E., Summers M. D., Fraser M. J. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol Cell Biol. 1983 Dec;3(12):2156–2165. doi: 10.1128/mcb.3.12.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. E., Vlak J. M., Summers M. D. Physical Analysis of Autographa californica Nuclear Polyhedrosis Virus Transcripts for Polyhedrin and 10,000-Molecular-Weight Protein. J Virol. 1983 Jan;45(1):215–225. doi: 10.1128/jvi.45.1.215-225.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler B. M., Geever R. F., Case M. E., Giles N. H. Cis-acting and trans-acting regulatory mutations define two types of promoters controlled by the qa-1F gene of Neurospora. Cell. 1984 Feb;36(2):493–502. doi: 10.1016/0092-8674(84)90242-3. [DOI] [PubMed] [Google Scholar]
- Vaughn J. L., Goodwin R. H., Tompkins G. J., McCawley P. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae). In Vitro. 1977 Apr;13(4):213–217. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]










