Abstract
Multiple genetic loci have been associated with body mass index (BMI) and obesity. The aim of this study was to investigate the effects of established adult BMI and childhood obesity loci in a Greek adolescent cohort. For this purpose, 34 variants were selected for investigation in 707 (55.9% females) adolescents of Greek origin aged 13.42 ± 0.88 years. Cumulative effects of variants were assessed by calculating a genetic risk score (GRS-34) for each subject. Variants at the FTO, TMEM18, FAIM2, RBJ, ZNF608 and QPCTL loci yielded nominal evidence for association with BMI and/or overweight risk (p < 0.05). Variants at TFAP2B and NEGR1 loci showed nominal association (p < 0.05) with BMI and/or overweight risk in males and females respectively. Even though we did not detect any genome-wide significant associations, 27 out of 34 variants yielded directionally consistent effects with those reported by large-scale meta-analyses (binomial sign p = 0.0008). The GRS-34 was associated with both BMI (beta = 0.17 kg/m2/allele; p < 0.001) and overweight risk (OR = 1.09/allele; 95% CI: 1.04–1.16; p = 0.001). In conclusion, we replicate associations of established BMI and childhood obesity variants in a Greek adolescent cohort and confirm directionally consistent effects for most of them.
Keywords: Obesity, BMI, common genetic variants, adolescents
Introduction
Prevalence of overweight and obesity in adolescents has increased over the last decades, and the observed rates in Greece are higher or comparable to those reported for other European countries (Lobstein & Frelut, 2003; Tzotzas et al., 2008). Although the increased obesity levels have been attributed to environmental changes, a strong genetic component has also been shown to contribute. Genome-wide association studies (GWAS) have been successful in identifying multiple genetic loci associated with BMI and/or obesity (Frayling et al., 2007; Scuteri et al., 2007; Loos et al., 2008; Thorleifsson et al., 2009; Willer et al., 2009; Speliotes et al., 2010; Bradfield et al., 2012).
Large GWAS meta-analyses, from the Genetic Investigation of Anthropometric Traits (GIANT) (Speliotes et al., 2010) and Early Growth Genetics (EGG) consortia (Bradfield et al., 2012), have identified multiple BMI-associated loci. The GIANT meta-analysis (Speliotes et al., 2010) of 249,796 individuals confirmed 14 established obesity susceptibility loci and identified 18 new loci associated with adult BMI. Twenty-three of the 32 variants also showed directionally consistent effects on children's BMI. The EGG meta-analysis replicated associations for seven established adult BMI variants and identified two novel loci robustly associated with increased childhood obesity risk (Bradfield et al., 2012).
In this study, we examined the effects of established adult BMI and childhood obesity associated loci reported by GIANT (Speliotes et al., 2010) and EGG (Bradfield et al., 2012) meta-analyses respectively on BMI and overweight risk in Greek adolescents. In total 34 single nucleotide polymorphisms (SNPs) were selected for investigation; 32 were based on the GIANT loci (FTO, MEM18, MC4R, GNPDA2, BDNF, NEGR1, SH2B1, ETV5, MTCH2, KCTD15, SEC16B, TFAP2B, FAIM2, NRXN3, RBJ, GPRC5BC, MAP2K5, QPCTL, TNNI3K, SLC39A8, FLJ35779, LRRN6C, TMEM160, FANCL, CADM2, PRKD1, LRP1B, PTBP2, MTIF3, ZNF608, RPL27A, NUDT3) (Speliotes et al., 2010); and two were based on the EGG loci (OLFM4, HOXB5) (Bradfield et al., 2012).
Materials and Methods
Our sample comprised 707 (55.9% females) adolescents of Greek origin aged 13.42 ± 0.88 years. Participants were drawn from the TEENAGE (TEENs of Attica: Genes and Environment) study. A random sample of 857 adolescent students (Table S1) attending public secondary schools located in the wider Athens area of Attica in Greece were recruited in the study from 2008 to 2010. Details of recruitment and data collection have been described elsewhere (Ntalla et al., in press). Prior to recruitment all study participants gave their verbal assent along with their parents’/guardians’ written consent forms. The study was approved by the Institutional Review Board of Harokopio University and the Greek Ministry of Education, Lifelong Learning and Religious Affairs.
DNA samples were genotyped using Illumina HumanOmniExpress BeadChips (Illumina, San Diego, CA, USA) at the Wellcome Trust Sanger Institute, Hinxton, UK. Genotype calling algorithm used was Illuminus (Teo et al., 2007). Sample exclusion criteria included: (i) sample call rate < 95%; (ii) samples with sex discrepancies; (iii) samples with genome-wide heterozygosity of <32% or >35%; (iv) duplicated/related samples identified by calculating the genome-wide pair-wise identity by descent (IBD) for each sample using PLINK (Purcell et al., 2007); from each pair with a π∧ > 0.2 the sample with the lower call rate was excluded; (v) samples with evidence of non-European descent as assessed by performing multidimensional scaling (MDS) in PLINK (Purcell et al., 2007) by combining the TEENAGE dataset with 1184 individuals from the HapMap phase III populations. SNP exclusion criteria included: (i) Hardy Weinberg Equilibrium (HWE) exact p < 0.0001, (ii) MAF < 1%, (iii) call rate < 95% (for SNPs with MAF ≥ 5%) or call rate < 99% for SNPs with MAF < 5%. Genotypes were imputed using the directly typed data and phased HapMap II genotype data from the 60 CEU HapMap founders using the program IMPUTE (Marchini & Howie, 2010).
Body weight (kg) was measured to the nearest 0.1 kg with the subjects barefoot and dressed in light clothing by the use of a weighing scale (Seca Alpha, Hamburg, Germany). Height was measured to the nearest 0.1 cm using a portable stadiometer with participants being barefoot with their shoulders in a relaxed position, their arms hanging freely and their head in a normal position, with the eyes looking straight ahead. BMI was calculated as weight (kg)/height squared (m2). Subjects were classified as normal weight, overweight and obese according to the age- and sex-specific criteria adopted by the International Obesity Task Force (IOTF) (Cole et al., 2000).
Before testing for associations, BMI was log-transformed to achieve normal distribution. The association of each variant with BMI was tested using linear regression. Overweight and obese subjects were classified into one category, and overweight risk was tested using logistic regression. Obesity risk was also tested with the use of logistic regression. All analyses were adjusted for age and sex and were carried out assuming an additive genetic model in SNPTEST (Marchini et al., 2007). In order to investigate whether some of the selected variants have sex-specific effects, stratification analyses by sex were also performed. In order to investigate the cumulative effects of variants, a genetic risk score, comprising all selected variants for investigation (GRS-34) was calculated for each subject by adding the effect alleles of the SNPs. For imputed variants, we used best-guess genotypes to calculate the score, using a genotype probability threshold of 0.8. Given that it has been proposed that allele weighting may have a limited effect (Janssens et al., 2007), we did not weight risk alleles for their individual effect sizes. Linear and logistic regression models were performed in STATA-11 (StataCorp, College Station, TX) to investigate the effects of the score on BMI and overweight risk respectively. Quanto v1.2.4 (http://hydra.usc.edu/gxe/) was used for power calculations. A two-sided p ≤ 0.05 was used as the threshold for nominal significance, and p ≤ 5 × 10−8 was used as the threshold for genome-wide significance.
Results
Three variants corresponding to previously identified GIANT loci (FTO, TMEM18, and FAIM2) yielded nominal evidence for association with both BMI (beta ± SE: 0.57 ± 0.19, p = 0.001; beta ± SE: 0.46 ± 0.24, p = 0.031; and beta ± SE: 0.49 ± 0.20, p = 0.015 respectively) and overweight risk (OR = 1.33, 95% CI: 1.06–1.67, p = 0.019; OR = 1.46, 95% CI: 1.08–1.97, p = 0.011; and OR = 1.29, 95% CI: 1.03–1.63, p = 0.025 respectively) in TEENAGE (Table 1). Variation at the QPCTL and ZNF608 loci yielded nominal evidence for association with BMI (beta ± SE: 0.54 ± 0.25, p = 0.026 and beta ± SE: 0.38 ± 0.25, p = 0.047 respectively), while the index variant at the RBJ locus was associated with overweight risk (OR = 1.32, 95% CI: 1.05–1.66, p = 0.017). The strongest association was observed at the FTO locus with BMI, which also accounted for the largest proportion of variation (0.96%) (Table 1). Obesity risk was nominally associated with variation at the FAIM2 locus (OR = 1.58, 95% CI: 1.05–2.40, p = 0.025), and with a variant at the BDNF locus (OR = 1.72, 95% CI: 1.00–2.95, p = 0.028) (Table S2). Despite the lack of statistically significant evidence for association for the majority of variants, overall 27 of the 34 GIANT and EGG loci yielded directionally consistent effects in TEENAGE (binomial sign test p = 0.0008) (Table 1).
Table 1.
TEENAGE association summary statistics for BMI and childhood obesity associated loci
| Consortial association summary statistics (GIANT and EGG) | TEENAGE association summary statistics | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BMI1 | Overweight risk3 | ||||||||||||||||
| Alleles | |||||||||||||||||
| SNP | Nearest gene | Chr | Position (bp) | Effect | Other | EAF | beta (or OR) | SE (or 95% CI) | p | EAF | beta2 | SE2 | Explained variation (%) | p | OR | 95% CI | P |
| GIANT (Speliotes et al., 2010)4 | |||||||||||||||||
| rs1558902 | FTO | 16 | 52361075 | A | T | 0.42 | 0.39 | 0.02 | 4.8E-120 | 0.48 | 0.57 | 0.19 | 0.96 | 0.001 | 1.33 | 1.06–1.67 | 0.019 |
| rs2867125 | TMEM18 | 2 | 612827 | C | T | 0.83 | 0.31 | 0.03 | 2.77E-49 | 0.80 | 0.46 | 0.24 | 0.95 | 0.031 | 1.46 | 1.08–1.97 | 0.011 |
| rs571312 | MC4R | 18 | 55990749 | A | C | 0.24 | 0.23 | 0.03 | 6.43E-42 | 0.26 | 0.04 | 0.22 | 0.93 | 0.941 | 1.00 | 0.77–1.30 | 0.938 |
| rs10938397 | GNPDA2 | 4 | 44877284 | G | A | 0.43 | 0.18 | 0.02 | 3.78E-31 | 0.42 | 0.11 | 0.20 | 0.65 | 0.490 | 0.99 | 0.79–1.25 | 0.855 |
| rs10767664 | BDNF | 11 | 27682562 | A | T | 0.78 | 0.19 | 0.03 | 4.69E-26 | 0.75 | 0.32 | 0.23 | 0.64 | 0.188 | 1.16 | 0.89–1.51 | 0.229 |
| rs2815752 | NEGR1 | 1 | 72585028 | A | G | 0.61 | 0.13 | 0.02 | 1.61E-22 | 0.72 | 0.04 | 0.22 | 0.30 | 0.913 | 0.94 | 0.73–1.21 | 0.698 |
| rs7359397 | SH2B1 | 16 | 28793160 | T | C | 0.4 | 0.15 | 0.02 | 1.88E-20 | 0.29 | -0.08 | 0.22 | 0.38 | 0.867 | 0.98 | 0.76–1.26 | 0.865 |
| rs9816226 | ETV5 | 3 | 187317193 | T | A | 0.82 | 0.14 | 0.03 | 1.69E-18 | 0.82 | 0.11 | 0.26 | 0.30 | 0.566 | 1.03 | 0.77–1.39 | 0.838 |
| rs3817334 | MTCH2 | 11 | 47607569 | T | C | 0.41 | 0.06 | 0.02 | 1.59E-12 | 0.41 | 0.02 | 0.20 | 0.33 | 0.720 | 0.81 | 0.64–1.02 | 0.083 |
| rs29941 | KCTD15 | 19 | 39001372 | G | A | 0.67 | 0.06 | 0.02 | 3.01E-09 | 0.67 | 0.05 | 0.21 | 0.35 | 0.840 | 1.19 | 0.94–1.52 | 0.158 |
| rs543874 | SEC16B | 1 | 176156103 | G | A | 0.19 | 0.22 | 0.03 | 3.56E-23 | 0.13 | 0.16 | 0.29 | 0.85 | 0.449 | 1.00 | 0.71–1.40 | 0.954 |
| rs987237 | TFAP2B | 6 | 50911009 | G | A | 0.18 | 0.13 | 0.03 | 2.9E-20 | 0.18 | 0.48 | 0.25 | 0.67 | 0.082 | 1.25 | 0.94–1.66 | 0.177 |
| rs7138803 | FAIM2 | 12 | 48533735 | A | G | 0.38 | 0.12 | 0.02 | 1.82E-17 | 0.37 | 0.49 | 0.20 | 0.91 | 0.015 | 1.29 | 1.03–1.63 | 0.025 |
| rs10150332 | NRXN3 | 14 | 79006717 | C | T | 0.21 | 0.13 | 0.03 | 2.75E-11 | 0.18 | 0.04 | 0.25 | 0.30 | 0.948 | 1.14 | 0.85–1.53 | 0.314 |
| rs713586 | RBJ | 2 | 25011512 | C | T | 0.47 | 0.14 | 0.02 | 6.17E-22 | 0.44 | 0.26 | 0.19 | 0.41 | 0.187 | 1.32 | 1.05–1.66 | 0.017 |
| rs12444979 | GPRC5BC | 16 | 19841101 | C | T | 0.87 | 0.17 | 0.03 | 2.91E-21 | 0.88 | 0.25 | 0.29 | 0.42 | 0.410 | 0.83 | 0.58–1.18 | 0.316 |
| rs2241423 | MAP2K5 | 15 | 65873892 | G | A | 0.78 | 0.13 | 0.02 | 1.19E-18 | 0.76 | 0.06 | 0.23 | 0.30 | 0.739 | 0.91 | 0.69–1.19 | 0.490 |
| rs2287019 | QPCTL | 19 | 50894012 | C | T | 0.8 | 0.15 | 0.03 | 1.88E-16 | 0.83 | 0.54 | 0.25 | 0.84 | 0.026 | 0.82 | 0.60–1.11 | 0.187 |
| rs1514175 | TNNI3K | 1 | 74764232 | A | G | 0.43 | 0.07 | 0.02 | 8.16E-14 | 0.40 | 0.02 | 0.19 | 0.41 | 0.740 | 0.97 | 0.77–1.22 | 0.831 |
| rs13107325 | SLC39A8 | 4 | 103407732 | T | C | 0.07 | 0.19 | 0.04 | 1.5E-13 | 0.10 | 0.51 | 0.32 | 0.40 | 0.084 | 1.33 | 0.93–1.90 | 0.122 |
| rs2112347 | FLJ35779 | 5 | 75050998 | T | G | 0.63 | 0.1 | 0.02 | 2.17E-13 | 0.60 | 0.04 | 0.19 | 0.36 | 0.704 | 1.01 | 0.80–1.27 | 0.898 |
| rs10968576 | LRRN6C | 9 | 28404339 | G | A | 0.31 | 0.11 | 0.02 | 2.65E-13 | 0.22 | -0.27 | 0.23 | 0.63 | 0.281 | 0.95 | 0.72–1.25 | 0.780 |
| rs3810291 | TMEM160 | 19 | 52260843 | A | G | 0.67 | 0.09 | 0.02 | 1.64E-12 | 0.68 | -0.09 | 0.21 | 0.38 | 0.753 | 0.91 | 0.72–1.16 | 0.420 |
| rs887912 | FANCL | 2 | 59156381 | T | C | 0.29 | 0.1 | 0.02 | 1.79E-12 | 0.28 | -0.15 | 0.21 | 0.36 | 0.550 | 0.97 | 0.75–1.24 | 0.728 |
| rs13078807 | CADM2 | 3 | 85966840 | G | A | 0.2 | 0.1 | 0.02 | 3.94E-11 | 0.22 | 0.25 | 0.23 | 0.84 | 0.280 | 1.11 | 0.84–1.45 | 0.551 |
| rs11847697 | PRKD1 | 14 | 29584863 | T | C | 0.04 | 0.17 | 0.05 | 5.76E-11 | 0.07 | 0.51 | 0.39 | 0.71 | 0.229 | 1.00 | 0.63–1.58 | 0.942 |
| rs2890652 | LRP1B | 2 | 142676401 | C | T | 0.18 | 0.09 | 0.03 | 1.35E-10 | 0.16 | 0.12 | 0.27 | 0.40 | 0.659 | 1.05 | 0.77–1.43 | 0.688 |
| rs1555543 | PTBP2 | 1 | 96717385 | C | A | 0.59 | 0.06 | 0.02 | 3.68E-10 | 0.50 | 0.20 | 0.19 | 0.50 | 0.154 | 1.15 | 0.92–1.44 | 0.220 |
| rs4771122 | MTIF3 | 13 | 26918180 | G | A | 0.24 | 0.09 | 0.03 | 9.48E-10 | 0.21 | -0.11 | 0.25 | 0.35 | 0.623 | 0.94 | 0.71–1.25 | 0.679 |
| rs4836133 | ZNF608 | 5 | 124360002 | A | C | 0.48 | 0.07 | 0.02 | 1.97E-09 | 0.53 | 0.38 | 0.20 | 0.49 | 0.047 | 1.15 | 0.91–1.44 | 0.213 |
| rs4929949 | RPL27A | 11 | 8561169 | C | T | 0.52 | 0.06 | 0.02 | 2.8E-09 | 0.37 | -0.27 | 0.21 | 0.82 | 0.245 | 0.97 | 0.76–1.22 | 0.783 |
| rs206936 | NUDT3 | 6 | 34410847 | G | A | 0.21 | 0.06 | 0.02 | 3.02E-08 | 0.24 | -0.02 | 0.23 | 0.32 | 0.928 | 0.86 | 0.66–1.13 | 0.298 |
| EGG (Bradfield et al., 2012)5 | |||||||||||||||||
| rs9568856 | OLFM4 | 13 | 52962982 | A | G | 0.16 | 1.22 | 1.14–1.29 | 1.82E-09 | 0.13 | 0.51 | 0.29 | 0.66 | 0.070 | 1.12 | 0.80–1.57 | 0.421 |
| rs9299 | HOXB5 | 17 | 44024429 | T | C | 0.65 | 1.14 | 1.09–1.20 | 3.54E-09 | 0.66 | 0.16 | 0.20 | 0.74 | 0.363 | 1.09 | 0.85–1.38 | 0.539 |
Chr = chromosome, bp = base pairs, EAF = effect allele frequency, SE = standard error, OR = odds ratio, CI = confidence interval.
Results were obtained using linear regression and logistic regression analysis assuming an additive effect while controlling for age and sex. Allelic test p, beta and SE, OR and 95% CIs are shown for each single SNP. Effect sizes (beta) and ORs are reported for the effect allele. Bold high-lighted loci yielded at least nominal evidence for association with BMI and/or overweight risk.
“BMI” refers to the linear regression analysis of each variant with BMI.
Effect sizes (beta) and SE are given for untransformed BMI (kg/m2).
“Overweight risk” refers to the binary trait analysis: normal weight subjects vs. overweight subjects (obese subjects were also classified as overweight).
Effect sizes in kg/m2 are referring to Stage 2 findings only; p are referring to Stage 1 and Stage 2 combined findings.
OR, CI and p are referring to the overall EGG consortium meta-analysis findings; EAF is referring to the discovery stage findings.
In the sex-stratified analysis, BMI and obesity were also nominally associated with a variant at the TFAP2B locus (beta ± SE: 1.22 ± 0.39, p = 0.002; OR = 1.96, 95% CI: 1.08–3.55, p = 0.026) (Table S3a) in males. In females, a variant at the NEGR1 locus also yielded nominal evidence of association with risk of obesity (OR = 3.15, 95% CI: 1.10–9.02, p = 0.018). Variants at MTCH2, LRRN6C and TMEM160 were also nominally associated with overweight or obesity risk. However, the direction of effects observed in our study for these variants was inconsistent with those reported from the GIANT consortium (Speliotes et al., 2010) (Table S3b).
The GRS-34, which was constructed to investigate the cumulative effects of the 34 variants under study, was significantly associated with BMI (beta = 0.17 kg/m2/allele; p < 0.001) and explained 3.2% of BMI variation. The difference in mean BMI between subjects with the higher GRS-34 (≥ 37 effect alleles) (2.0% of subjects) and those with the lower GRS-34 (≤ 22 effect alleles) (2.3% of subjects) was 4.3 kg/m2 (Fig. 1). Consistent with the observation for BMI, the association of the GRS-34 and risk of overweight showed that each additional risk allele was associated with an 1.09-fold increased odds of overweight (95% CI: 1.04–1.16; p = 0.001).
Figure 1.
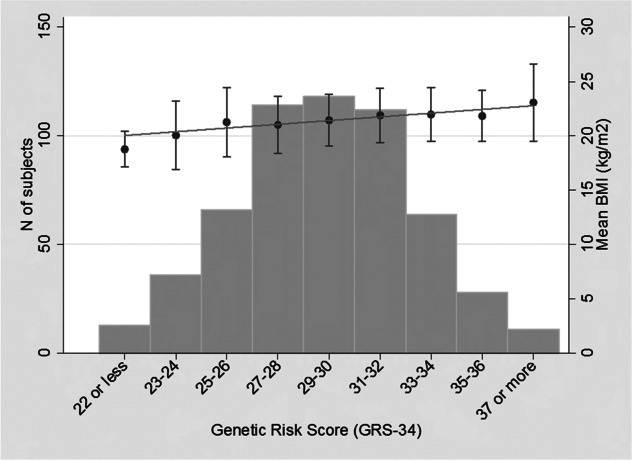
Distribution of the genetic risk score and cumulative effects of the risk alleles from 32 GIANT BMI and 2 EGG childhood obesity SNPs. Mean (±SE) values for BMI (kg/m2) are also presented.
Discussion
Our findings regarding the association of established adult BMI associated loci with BMI and/or risk of overweight in adolescents reflect previous reports either on single pediatric cohorts (den Hoed et al., 2010; Bradfield et al., 2012; Zhao et al., 2011) or large consortial meta-analyses (Bradfield et al., 2012). In the EYHS study, most of the BMI associated loci identified by GWAS in adults were also associated with anthropometric traits in children (den Hoed et al., 2010). In a case-control study of European American children, Zhao et al. (Zhao et al., 2011) reported evidence for association with common childhood obesity for nine of the 32 GIANT loci, while 28 of the 32 loci yielded consistent directional effects. The EGG consortium meta-analysis (Bradfield et al., 2012) also replicated robust associations for seven established adult BMI loci (FTO, TMEM18, POMC, MC4R, FAIM2, TNNI3K and SEC16B) with risk of childhood obesity; and identified two novel childhood obesity loci both of which had yielded directionally consistent effects in the GIANT meta-analysis of adult BMI (Speliotes et al., 2010). Consistency of directionality for the great majority of tested variants indicates that a large genetic component of BMI and obesity overlaps in children and adults (Bradfield et al., 2012).
Our study has a small sample size compared to both consortial meta-analyses, and power to confirm associations even at nominal significance level for all tested loci is relatively low. The effect sizes at the GIANT-identified loci require sample sizes that range from a few thousands to a few hundred thousand in order to achieve >80% power to detect association.
Despite the small effect sizes of established adult BMI associated variants, it has been shown that they have cumulative effects on BMI and obesity risk (Li et al., 2010). In our study, the combined effects of the 34 adult BMI and childhood obesity variants reported by the GIANT and EGG meta-analyses respectively were significantly associated with both BMI and risk of overweight, and the effect sizes are comparable to those reported in large meta-analyses of adults (Speliotes et al., 2010).
In this report, we investigated the individual and cumulative effects of established adult BMI and childhood obesity associated loci with BMI and overweight risk in a sample of Greek adolescents. We report evidence of nominal association for several loci and show that cumulatively tested variants are associated with both BMI and overweight risk. Our findings also support evidence for a large shared genetic component between adult and childhood BMI and obesity and validate the TEENAGE study as a cohort in which to study the genetics of anthropometric traits.
Acknowledgments
This research has been co-financed by the European Union (European Social Fund—ESF) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF)—Research Funding Program: Heracleitus II. Investing in knowledge society through the European Social Fund. This work was funded by the Wellcome Trust (098051).
We thank all study participants and their families as well as all volunteers for their contribution in this study. We thank the following staff from the Sample Management and Genotyping Facilities at the Wellcome Trust Sanger Institute for sample preparation, quality control and genotyping: Dave Jones, Doug Simpkin, Emma Gray, Hannah Blackburn, Sarah Edkins.
Author Contributions
GVD is the PI and EZ is the genetics PI for TEENAGE study. IN and GVD carried out the data collection. IN and PV contributed to database preparation. IN and NWR contributed to the phenotypic data QC. IN and KP performed the genetic data QC. NWR and LS were responsible for imputing the genotype data. IN and KP performed the statistical analyses. IN, EZ and GVD contributed to the interpretation of the data. All authors contributed to the writing and approval of the final version of the manuscript submitted for publication.
Conflicts of Interest
The authors declare no conflict of interest.
Supporting Information
Additional supporting information may be found in the online version of this article:
Table S1 Descriptive characteristics of all subjects from the TEENAGE study.
Table S2 TEENAGE results for BMI and childhood obesity associated loci with risk of obesity.
Table S3a TEENAGE association summary statistics for BMI and childhood obesity associated loci in males.
Table S3b TEENAGE association summary statistics for BMI and childhood obesity associated loci in females.
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organised for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Bradfield JP, Taal HR, Timpson NJ, Scherag A, Lecoeur C, Warrington NM, Hypponen E, Holst C, Valcarcel B, Thiering E, Salem RM, Schumacher FR, Cousminer DL, Sleiman PM, Zhao J, Berkowitz RI, Vimaleswaran KS, Jarick I, Pennell CE, Evans DM, St Pourcain B, Berry DJ, Mook-Kanamori DO, Hofman A, Rivadeneira F, Uitterlinden AG, Van Duijn CM, Van Der Valk RJ, De Jongste JC, Postma DS, Boomsma DI, Gauderman WJ, Hassanein MT, Lindgren CM, Magi R, Boreham CA, Neville CE, Moreno LA, Elliott P, Pouta A, Hartikainen AL, Li M, Raitakari O, Lehtimaki T, Eriksson JG, Palotie A, Dallongeville J, Das S, Deloukas P, Mcmahon G, Ring SM, Kemp JP, Buxton JL, Blakemore AI, Bustamante M, Guxens M, Hirschhorn JN, Gillman MW, Kreiner-Moller E, Bisgaard H, Gilliland FD, Heinrich J, Wheeler E, Barroso I, O'rahilly S, Meirhaeghe A, Sorensen TI, Power C, Palmer LJ, Hinney A, Widen E, Farooqi IS, Mccarthy MI, Froguel P, Meyre D, Hebebrand J, Jarvelin MR, Jaddoe VW, Smith GD, Hakonarson H, Grant SF. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44:526–531. doi: 10.1038/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TJ, Bellizzi MC, Flegal KM, Dietz WH. Establishing a standard definition for child overweight and obesity worldwide: International survey. BMJ. 2000;320:1240–1243. doi: 10.1136/bmj.320.7244.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Hoed M, Ekelund U, Brage S, Grontved A, Zhao JH, Sharp SJ, Ong KK, Wareham NJ, Loos RJ. Genetic susceptibility to obesity and related traits in childhood and adolescence: Influence of loci identified by genome-wide association studies. Diabetes. 2010;59:2980–2988. doi: 10.2337/db10-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, Mccarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens AC, Moonesinghe R, Yang Q, Steyerberg EW, Van Duijn CM, Khoury MJ. The impact of genotype frequencies on the clinical validity of genomic profiling for predicting common chronic diseases. Genet Med. 2007;9:528–535. doi: 10.1097/gim.0b013e31812eece0. [DOI] [PubMed] [Google Scholar]
- Li S, Zhao JH, Luan J, Luben RN, Rodwell SA, Khaw KT, Ong KK, Wareham NJ, Loos RJ. Cumulative effects and predictive value of common obesity-susceptibility variants identified by genome-wide association studies. Am J Clin Nutr. 2010;91:184–190. doi: 10.3945/ajcn.2009.28403. [DOI] [PubMed] [Google Scholar]
- Lobstein T, Frelut ML. Prevalence of overweight among children in Europe. Obes Rev. 2003;4:195–200. doi: 10.1046/j.1467-789x.2003.00116.x. [DOI] [PubMed] [Google Scholar]
- Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M, Freathy RM, Attwood AP, Beckmann JS, Berndt SI, Jacobs KB, Chanock SJ, Hayes RB, Bergmann S, Bennett AJ, Bingham SA, Bochud M, Brown M, Cauchi S, Connell JM, Cooper C, Smith GD, Day I, Dina C, De S, Dermitzakis ET, Doney AS, Elliott KS, Elliott P, Evans DM, Sadaf Farooqi I, Froguel P, Ghori J, Groves CJ, Gwilliam R, Hadley D, Hall AS, Hattersley AT, Hebebrand J, Heid IM, Lamina C, Gieger C, Illig T, Meitinger T, Wichmann HE, Herrera B, Hinney A, Hunt SE, Jarvelin MR, Johnson T, Jolley JD, Karpe F, Keniry A, Khaw KT, Luben RN, Mangino M, Marchini J, Mcardle WL, Mcginnis R, Meyre D, Munroe PB, Morris AD, Ness AR, Neville MJ, Nica AC, Ong KK, O'rahilly S, Owen KR, Palmer CN, Papadakis K, Potter S, Pouta A, Qi L, Randall JC, Rayner NW, Ring SM, Sandhu MS, Scherag A, Sims MA, Song K, Soranzo N, Speliotes EK, Syddall HE, Teichmann SA, Timpson NJ, Tobias JH, Uda M, Vogel CI, Wallace C, Waterworth DM, Weedon MN, Willer CJ, Wraight YuanX, Zeggini E, Hirschhorn JN, Strachan DP, Ouwehand WH, Caulfield MJ, Samani NJ, Frayling TM, Vollenweider P, Waeber G, Mooser V, Deloukas P, McCarthy MI, Wareham NJ, Barroso I, Jacobs KB, Chanock SJ, Hayes RB, Lamina C, Gieger C, Illig T, Meitinger T, Wichmann HE, Kraft P, Hankinson SE, Hunter DJ, Hu FB, Lyon HN, Voight BF, Ridderstrale M, Groop L, Scheet P, Sanna S, Abecasis GR, Albai G, Nagaraja R, Schlessinger D, Jackson AU, Tuomilehto J, Collins FS, Boehnke M, Mohlke KL. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, Mcvean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Ntalla I, Giannakopoulou M, Vlachou P, Giannitsopoulou K, Gkesou V, Makridi C, Marougka M, Mikou G, Ntaoutidou K, Prountzou E, Tsekoura A. Body composition, dietary profiles and eating behaviours according to dieting history in healthy Greek adolescents: The TEENAGE Study. Public Health Nutr, in press. [DOI] [PMC free article] [PubMed]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, Sham PC. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G, Dei M, Lai S, Maschio A, Busonero F, Mulas A, Ehret GB, Fink AA, Weder AB, Cooper RS, Galan P, Chakravarti A, Schlessinger D, Cao A, Lakatta E, Abecasis GR. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Allen HL, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, Van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proença C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grässler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jørgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, König IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaløy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimäki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, MAGIC, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O'Donnell CJ, O'Rahilly S, Ong KK, Oostra B, Paré G, Parker AN, Perola M, Pichler I, Pietiläinen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstråle M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tönjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kähönen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Grönberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Procardis Consortium. Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O'Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo YY, Inouye M, Small KS, Gwilliam R, Deloukas P, Kwiatkowski DP, Clark TG. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics. 2007;23:2741–2746. doi: 10.1093/bioinformatics/btm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- Tzotzas T, Kapantais E, Tziomalos K, Ioannidis I, Mortoglou A, Bakatselos S, Kaklamanou M, Lanaras L, Kaklamanos I. Epidemiological survey for the prevalence of overweight and abdominal obesity in Greek adolescents. Obesity (Silver Spring) 2008;16:1718–1722. doi: 10.1038/oby.2008.247. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, Mccarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, Mcardle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O'rahilly S, Purmann C, Rees MG, Ridderstrale M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YC, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Waterworth DM, Watkins N, Wellcome Trust Case Control Consortium. Witteman JC, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JN Genetic Investigation of ANthropometric Traits Consortium. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Bradfield JP, Zhang H, Sleiman PM, Kim CE, Glessner JT, Deliard S, Thomas KA, Frackelton EC, Li M, Chiavacci RM, Berkowitz RI, Hakonarson H, Grant SF. Role of BMI-associated loci identified in GWAS meta-analyses in the context of common childhood obesity in European Americans. Obesity (Silver Spring) 2011;19:2436–2439. doi: 10.1038/oby.2011.237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


